Abstract
SREBP cleavage activating protein (SCAP), a membrane-bound glycoprotein, regulates the proteolytic activation of sterol regulatory element binding proteins (SREBPs), which are membrane-bound transcription factors that control lipid synthesis in animal cells. SCAP-stimulated proteolysis releases active fragments of SREBPs from membranes of the endoplasmic reticulum and allows them to enter the nucleus where they activate transcription. Sterols such as 25-hydroxycholesterol inactivate SCAP, suppressing SREBP proteolysis and turning off cholesterol synthesis. We here report the isolation of Chinese hamster ovary cells with a point mutation in SCAP (Y298C) that renders the protein resistant to inhibition by 25-hydroxycholesterol. Like the previously described D443N mutation, the Y298C mutation occurs within the putative sterol-sensing domain, which is part of the polytopic membrane attachment region of SCAP. Cells that express SCAP(Y298C) continued to process SREBPs in the presence of 25-hydroxycholesterol and hence they resisted killing by this sterol. In wild-type Chinese hamster ovary cells the N-linked carbohydrate chains of SCAP were mostly in the endoglycosidase H-sensitive form when cells were grown in medium containing 25-hydroxycholesterol. In contrast, when cells were grown in sterol-depleted medium, these chains were converted to an endoglycosidase H-resistant form. 25-Hydroxycholesterol had virtually no effect in cells expressing SCAP(D443N) or SCAP(Y298C). The relation between this regulated carbohydrate processing to the SCAP-regulated proteolysis of SREBP remains to be explored.
Keywords: sterol regulatory element binding protein/cholesterol/sterol-sensing domain/Golgi apparatus/endoplasmic reticulum
SREBP cleavage-activating protein (SCAP) is a central regulator of lipid synthesis and uptake in animal cells (reviewed in ref. 1). Located in the endoplasmic reticulum (ER), SCAP stimulates the proteolytic cleavage of sterol regulatory element binding proteins (SREBPs), a family of three closely related membrane-bound transcription factors that activate the synthesis of cholesterol and unsaturated fatty acids and their uptake from plasma via the low density lipoprotein receptor (2). The activity of SCAP is abolished by sterols, a regulatory mechanism that maintains a constant lipid composition of cell membranes (1).
The SREBPs are tripartite proteins of ≈1,100 aa that are bound to membranes of the ER in a hairpin fashion (1). The NH2-terminal domain of ≈480 aa and the COOH-terminal domain of ≈560 aa project into the cytosol. They are joined by a membrane attachment domain consisting of two membrane-spanning helices and a short hydrophilic loop of 31 aa that projects into the ER lumen. The NH2-terminal domain of SREBP is a transcription factor of the basic helix–loop–helix leucine zipper family. In sterol-depleted cells, it is released from the ER membrane by two-step proteolysis, which begins when an enzyme called site-1 protease cleaves SREBP at site 1, which is a leucine-serine bond in the luminal loop (3). This first cleavage separates SREBP into halves and allows a second protease to cleave at site 2, which is a leucine-cysteine bond that is located within the first membrane-spanning helix (4). This second cleavage releases the NH2-terminal segment, which enters the nucleus where it activates transcription of target genes.
The activity of site-1 protease is controlled by SCAP, a polytopic protein of the ER membrane that exists in a tight complex with an SREBP (2, 5). SCAP has two domains. The NH2-terminal domain of ≈730 aa is believed to contain eight membrane-spanning helices that anchor it in the ER membrane. The COOH-terminal domain of ≈550 aa projects into the cytosol. This domain contains five WD repeats, which are sequences of about 40 aa in length that are found in many proteins where they mediate protein–protein interactions (6). The WD repeat domain of SCAP forms a complex with the COOH-terminal domain of an SREBP (7, 8). Disruption of this complex abolishes cleavage of SREBP at site 1. We have postulated that the SCAP/SREBP complex serves as the site of attachment of a luminal protease, site-1 protease, which initiates the processing of SREBP (8).
Genetic evidence has led to the hypothesis that sterols regulate the cleavage of SREBPs by interacting, directly or indirectly, with the polytopic membrane attachment domain of SCAP (2, 9). This domain contains a segment of ≈170 aa that has been called the sterol-sensing domain, which includes five of the eight membrane-spanning helices of SCAP (helices 2–6). Similar sequences have been found in three other proteins that are postulated to interact with sterols. These include the sterol-regulated enzyme 3-hydroxy-3-methylglutaryl (HMG) CoA reductase (2, 10); the Niemann Pick-type C1 protein, which is necessary for export of cholesterol from lysosomes (11); and the developmental protein Patched (12), which serves as a receptor for Hedgehog, the only protein known to have a covalently attached cholesterol moiety (13). Recently another member of this family, related to Patched, has been described (14), bringing the total number of sterol sensor-containing proteins to five.
Support for the concept of a sterol-sensing domain in SCAP came from the identification of a Chinese hamster ovary (CHO) cell line with a point mutation that changes an aspartic acid to an asparagine within this domain (D443N mutation) (2). These cells, designated 25-RA, were obtained through chemical mutagenesis and selected for growth in medium devoid of cholesterol, but containing 25-hydroxycholesterol (15). This medium is lethal for wild-type cells because 25-hydroxycholesterol inhibits the site-1 cleavage of SREBPs, apparently by suppressing the activity of SCAP (2). By blocking cleavage and nuclear entry of SREBPs, 25-hydroxycholesterol turns off cholesterol synthesis, but it cannot replace cholesterol in cell membranes, thereby causing cells to die from cholesterol deprivation. The 25-RA cells survive in 25-hydroxycholesterol because the D443N substitution renders SCAP partially resistant to the suppressive action of 25-hydroxycholesterol (2). The cells continue to cleave SREBPs, and therefore they continue to produce sufficient cholesterol to survive. Remarkably, we found the same nucleotide substitution in two other cell lines that were isolated independently, but in a similar fashion (9).
In the current paper we report the isolation of an additional 25-hydroxycholesterol resistant cell line, designated sterol regulatory defective-9 (SRD-9). This cell line lacks the D443N mutation, but it has another point mutation in the sterol-sensing domain of SCAP, namely the substitution of a cysteine for a tyrosine at position 298 (Y298C). Tyr-298 is conserved in all five known sterol-sensing domains. We show by transfection that this substitution is sufficient to confer a 25-hydroxycholesterol-resistant phenotype. Furthermore, we show that 25-hydroxycholesterol causes a shift in the processing of the N-linked carbohydrates of wild-type SCAP. In sterol-depleted CHO cells, these carbohydrates are partially resistant to digestion with endoglycosidase (endo) H. Growth in 25-hydroxycholesterol shifts the pattern in the direction of endo H sensitivity. This change is blunted in cells with the D443N or Y298C mutant forms of SCAP. These findings raise the possibility that sterols block the movement of SCAP at some point between the ER and the cis/medial Golgi complex and that this regulation is defective in the 25-hydroxycholesterol-resistant cells.
MATERIALS AND METHODS
Reagents.
We obtained hexyl-β-d-glucopyranoside from Calbiochem-Novabiochem, and endo H and peptide N-glycosidase F (PNGase F) from New England Biolabs. Fetal and newborn calf lipoprotein-deficient serum (d > 1.215 g/ml) and human low density lipoprotein (d = 1.019–1.063 g/ml) were prepared as described (16). Other reagents were obtained from previously reported sources (5, 17).
Cell Culture and Transfection.
All cells were grown in monolayer at 37°C in an atmosphere of 8–9% CO2. CHO-7 cells, a clone of CHO-K1 cells adapted to growth in lipoprotein-deficient serum (17), were grown in medium A (a 1:1 mixture of Ham’s F-12 medium and DMEM containing 100 units/ml of penicillin and 100 μg/ml of streptomycin sulfate) supplemented with 5% (vol/vol) newborn calf lipoprotein-deficient serum. SRD-1 cells (18) and 25-RA cells (2, 15), mutant subclones of CHO cells, were maintained in medium A supplemented with 5% newborn calf lipoprotein-deficient serum and 1 μg/ml of 25-hydroxycholesterol. Cells were set up for experiments on day 0 at a density of 5 × 105 cells/100-mm dish, refed on day 2, and harvested on day 3.
Human embryonic kidney 293 cells were set up on day 0 at a density of 5.5 × 105 cells/100-mm dish in medium B (DMEM containing 100 units/ml of penicillin and 100 μg/ml of streptomycin sulfate) supplemented with 10% fetal calf serum. On day 2, cells were transfected with the indicated plasmids by using an MBS kit (Stratagene) as described (19). Three hours after transfection, the cells were switched to fresh medium as indicated, incubated overnight, and harvested on day 3.
Isolation of Sterol-Resistant SRD-9 Cells.
SRD-9 cells were isolated from pools of CHO-7 cells that had been mutagenized with nitrosoethylurea and selected in medium containing lipoprotein-deficient serum and 0.3 μg/ml of 25-hydroxycholesterol as described (17). Surviving colonies were isolated with cloning cylinders, cloned by dilution plating, and further selected with 1 μg/ml of 25-hydroxycholesterol. One surviving clone, designated SRD-9, was expanded and maintained in medium A supplemented with 5% newborn calf lipoprotein-deficient serum and 1 μg/ml of 25-hydroxycholesterol.
Cloning of Mutant SCAP from SRD-9 Cells.
cDNA synthesis, isolation of genomic DNA, PCR, and single-strand conformational polymorphism (SSCP) analysis of SCAP were carried out as described (9). Total RNA from CHO-7, 25-RA, and SRD-9 cells was used as a template for reverse transcription with a SCAP-specific primer (9). From the resulting cDNA, PCR fragments containing codons 270–348 were amplified with a pair of primers (5′-AACCTGGTCCACGTGCACTTC-3′ and 5′-TCTCACCGCCATTGAGTGTG-3′) in the presence of [32P]dCTP and subjected to SSCP analysis (20) on 7% PAGE. Comparison of the band pattern revealed a band in the sample from SRD-9 cells that was not present in the other two cell lines. This result was confirmed by SSCP analysis of genomic DNA from the same cell lines by using a different set of primers (5′-CTCATGCTCCTGCACCCTAGC-3′ and 5′-GAAGTGGGTGTCATGGAAAGC-3′). To determine the nature of the mutation in SCAP from SRD-9 cells, unlabeled PCR fragments amplified from both cDNA and genomic DNA were cloned into a plasmid, pNoTA/T7 (5 Prime →3 Prime), and several independent clones were sequenced on both strands with an Applied Biosystems model 377 DNA Sequencer.
Recombinant Plasmids.
Expression vectors pTK, pTK-HSV-BP1a, pTK-HSV-BP2, pTK-HSV-SCAP-T7, and pTK-HSV-SCAP-T7(D443N) have been described (2, 21). A vector expressing epitope-tagged mutant SCAP(Y298C) was constructed as follows: total RNA from SRD-9 cells was used as a template for reverse transcription with a SCAP-specific primer as described (9). From the resulting cDNA, a PCR fragment containing codons 133–539 was amplified with a pair of primers (5′-TACTGAGAGACAGCTCAGGG-3′ and 5′-AGCCCTGCTGGGTCTGTGTA-3′) and subcloned into pNoTA/T7. Several independent clones were analyzed for the presence of the Y298C mutation by single-strand conformational polymorphism analysis. The insert of one positive plasmid, pAN233–3, was sequenced in its entirety and cut with BstEII and FseI. The resulting insert was used to replace the corresponding fragment of pTK-HSV-SCAP-T7 to generate pTK-HSV-SCAP-T7(Y298C).
pTK3-SCAP, a vector expressing wild-type hamster SCAP driven by the herpes simplex virus (HSV) thymidine kinase promoter, was created as follows: the SpeI–BstEII fragment of pTK-HSV-SCAP (5) (encompassing the two HSV epitope tags plus the N-terminal 150 codons of SCAP) was replaced with a PCR-generated fragment containing an SpeI restriction site, followed by a Kozak consensus sequence (ACCATG) (22), followed by codons 2–150 of SCAP to generate pTK2-SCAP. Subsequently, pTK2-SCAP was digested with SpeI, blunt-ended with Klenow fragment, and religated to generate pTK3-SCAP. To generate pTK3-SCAP(Y298C) and pTK3-SCAP(D443N), the BstEII–FseI fragment of pTK3-SCAP was replaced with the corresponding fragments of pTK-HSV-SCAP-T7(Y298C) and pTK-HSV-SCAP-T7(D443N), respectively.
Trypsin Digestion of Membranes.
Cell monolayers were scraped into the culture medium, centrifuged at 1,000 g for 5 min at 4°C, resuspended in 1 ml of PBS, and centrifuged again as above. Cell pellets from individual dishes were resuspended in 0.4 ml of buffer E (10 mM Hepes-KOH, pH 7.4/10 mM KCl/1.5 mM MgCl2/5 mM sodium EDTA/5 mM sodium EGTA/250 mM sucrose), passed through a 22-gauge needle 20 times, and centrifuged at 1,000 g for 5 min at 4°C. The supernatants were centrifuged at 1.5 × 104 g for 10 min at 4°C. The resulting membrane pellets were resuspended in 0.1 ml of buffer F (buffer E containing 100 mM NaCl) and rocked at 4°C for 5 min. Aliquots (5 μl) were added to 5 μl of a solution of 20% (wt/vol) hexyl-β-d-glucopyranoside to determine the protein concentration (23) by using Coomassie Plus Protein Assay Reagent (Pierce). Equal amounts of protein (42–54 μg) were incubated in the absence or presence of 1 μg of trypsin in a total volume of 58 μl for 30 min at 30°C. Reactions were stopped by addition of 2 μl of soybean trypsin inhibitor (400 units).
Glycosidase Treatments.
Cells were harvested, and membrane fractions were prepared and treated with trypsin as described above. For subsequent treatment with endo H, individual samples received 10 μl of solution containing 3.5% (wt/vol) SDS and 7% (vol/vol) 2-mercaptoethanol. After heating at 100°C for 10 min, each sample received sequential additions of 9 μl of 0.5 M sodium citrate (pH 5.5), 5 μl of solution containing 17× protease inhibitors (a concentration of 1× corresponds to 10 μg/ml of leupeptin, 5 μg/ml of pepstatin A, and 2 μg/ml of aprotinin), followed by 1 μl of endo H (0.05 units). For treatment with PNGase F, trypsin-treated samples were denatured in the presence of SDS and 2-mercaptoethanol as described above and then received sequential additions of 7 μl of 0.5 M sodium phosphate (pH 7.5), 7 μl of solution containing 10% (vol/vol) Nonidet P-40 and 12× protease inhibitors, followed by 1 μl of PNGase F (7.7 × 10−3 units). All reactions were incubated overnight at 37°C and stopped by addition of 20 μl of buffer G containing 0.25 M Tris⋅HCl at pH 6.8, 2% SDS, 4% 2-mercaptoethanol, 10% (vol/vol) glycerol, and 0.05% (wt/vol) bromophenol blue. The mixtures then were heated at 100°C for 5 min and subjected to SDS/PAGE (5–12% gradient gels).
Immunoblot Analysis.
mAbs IgG-2A4 directed against the basic-helix-loop-helix domain of human SREBP-1 (24), IgG-7D4 directed against amino acids 32–250 of hamster SREBP-2 (25), and IgG-9D5 directed against amino acids 540–707 of hamster SCAP (7) were prepared as described. mAb IgG-HSV-Tag was obtained from Novagen. Gels were calibrated with molecular weight markers (Amersham or Bio-Rad). After SDS/PAGE, proteins were transferred to Hybond-C extra nitrocellulose sheets (Amersham) and incubated with antibodies at the indicated concentration. Bound antibodies were visualized by chemiluminescence with horseradish peroxidase-conjugated, affinity-purified donkey anti-mouse IgG (Jackson ImmunoResearch) using the Supersignal substrate system (Pierce). Filters were exposed to Kodak X-Omat Blue XB-1 film (NEN) at room temperature for the indicated time.
RESULTS
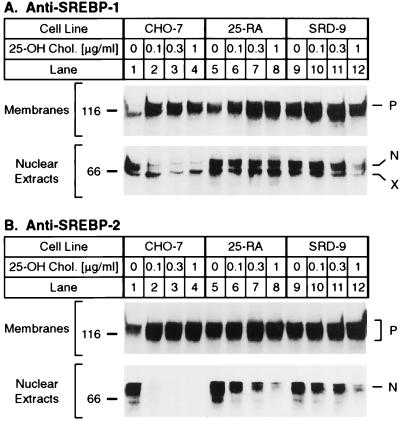
Fig. 1 shows immunoblots of the two isoforms of SREBP, SREBP-1 and SREBP-2, in SDS/PAGE gels of membrane and nuclear extracts from wild-type CHO-7 cells and from two lines of 25-hydroxycholesterol-resistant mutant CHO cell lines. The resistant cells were obtained by chemical mutagenesis followed by selection for growth in the presence of 25-hydroxycholesterol and the absence of cholesterol. The previously described 25-RA cells are resistant to 25-hydroxycholesterol because of a D443N substitution in SCAP (2). The SRD-9 cells have not been described previously. When wild-type CHO-7 cells were incubated with increasing concentrations of 25-hydroxycholesterol, the amounts of SREBP-1 and SREBP-2 in nuclear extracts declined by more than 90% at the lowest concentration tested (0.1 mg/ml) (Fig. 1 A and B). The 25-RA cells and SRD-9 cells were markedly resistant to 25-hydroxycholesterol. The amounts of SREBP-1 and SREBP-2 in the nuclear extracts declined only by about 50% at the highest concentration tested (1 μg/ml).
Figure 1.
Immunoblot analysis of SREBP-1 and SREBP-2 in wild-type CHO-7 and mutant 25-hydroxycholesterol-resistant cells. On day 0, the indicated cell line was set up in medium A supplemented with 5% lipoprotein-deficient serum. On day 2, the cells were switched to medium A containing 5% lipoprotein-deficient serum, 50 μM compactin, 50 μM sodium mevalonate, and 0.1% (vol/vol) ethanol containing the indicated final concentration of 25-hydroxycholesterol (25-OH Chol.). On day 3, the cells were harvested and fractionated into nuclear extract and 105 g membrane fraction as described (21, 29). Aliquots of the fractions (40 μg protein) were subjected to 7% SDS/PAGE and transferred to nitrocellulose. Immunoblot analysis was carried out with 10 μg/ml of IgG-2A4 (A) or 2 μg/ml of IgG-7D4 (B). Filters were exposed to film for 3 min (A, Upper), 45 s (A, Lower), 2 s (B, Upper), or 10 s (B, Lower). N and P denote the nuclear and precursor forms of SREBPs, respectively. X denotes a cross-reacting protein of unknown identity.
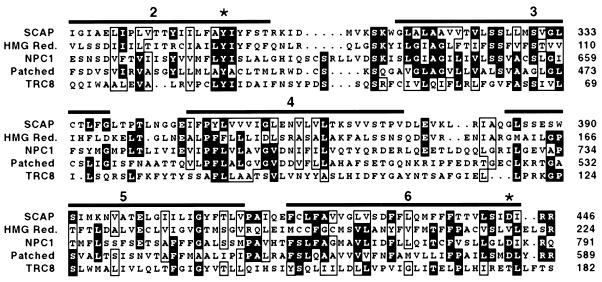
To determine whether the SRD-9 cells had a point mutation in the sterol-sensing domain of SCAP, we used reverse-transcription–PCR to amplify this region of the SCAP mRNA. Single-strand conformational polymorphism analysis of one amplified fragment revealed an abnormality (see Materials and Methods). Accordingly, we cloned the PCR-amplified segment of the SCAP mRNA and determined its nucleotide sequence. The data revealed that the SRD-9 cells had undergone an A to G substitution in codon 298 of SCAP, changing the codon from tyrosine (TAC) to cysteine (TGC). This change was observed in six of 10 independent clones, suggesting that SRD-9 cells were heterozygous for the Y298C mutation. Fig. 2 shows that Tyr-298 is conserved in the sterol-sensing domains of all five proteins that are known to possess this domain. Tyr-298 lies within the first of the five membrane-spanning sequences that are found in the sterol-sensing domain. Fig. 2 also shows the location of Asp-443, which is the site of the D443N mutation in the 25-RA cells as well as in the SRD-4 and SRD-8 cells, two other 25-hydroxycholesterol-resistant cell lines harboring a D443N mutation in SCAP (9). This residue lies at the end of the fifth membrane-spanning sequence of the sterol-sensing domain. Asp-443 is conserved in three of the five proteins.
Figure 2.
Comparison of sterol-sensing domains in SCAP, HMG CoA reductase, NPC1, Patched, and TRC8. Sequences are from Chinese hamster SCAP (amino acids 280–446), Chinese hamster HMG CoA reductase (amino acids 57–224), mouse NPC1 (amino acids 617–791), mouse Patched (amino acids 420–589), and human TRC8 (amino acids 16–182). GenBank accession numbers are U67060, L00165, AF003348, U46155, and AF064801, respectively. Sequences were aligned with pileup from the Genetics Computer Group Sequence Analysis Software Package, version 8.0. Residues identical in at least three of the five proteins are highlighted in black. Residues chemically similar in at least four of the five proteins are boxed. Overbars denote positions of transmembrane domains 2–6 in HMG CoA reductase (10). ∗ denote positions of point mutations in hamster SCAP (Y298C and D443N).
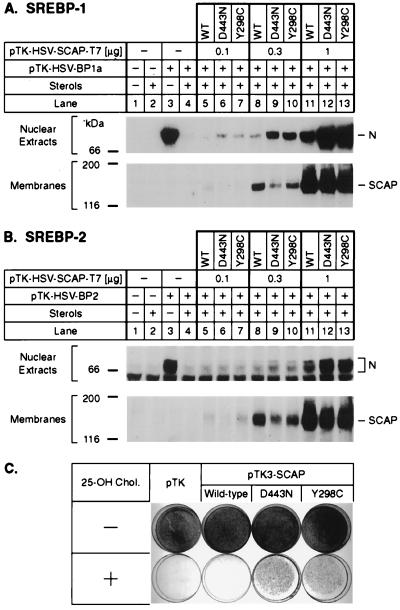
Fig. 3 shows experiments designed to confirm that the Y298C mutation is capable of causing a sterol-resistant phenotype. In the first experiment, human kidney 293 cells were transfected with a cDNA encoding either wild-type SCAP or one of the two mutant versions (D443N or Y298C). Expression was driven by the weak HSV thymidine kinase promoter, which produces a relatively low level of expression. The cells also were transfected with cDNAs encoding HSV-tagged SREBP-1a (Fig. 3A) or SREBP-2 (Fig. 3B). The cells were incubated overnight in the presence of a mixture of 25-hydroxycholesterol and cholesterol as indicated, after which nuclear extracts were subjected to SDS/PAGE and blotted with an antibody to the HSV tag. In the absence of cotransfected SCAP, the 293 cells showed abundant SREBP-1a and SREBP-2 in the nucleus when incubated in the absence of sterols (Fig. 3 A and B, lanes 3). In the presence of the sterol mixture the amounts of nuclear SREBP-1a and SREBP-2 were reduced by more than 95% (lane 4). Transfection of 0.1 or 0.3 μg of cDNA encoding wild-type SCAP did not increase the amount of nuclear SREBP-1a or SREBP-2 appreciably (lanes 5 and 8). At these concentrations both of the mutant forms of SCAP produced a clear increase in nuclear SREBP-1a, and a distinct, but lesser, increase in nuclear SREBP-2 (lanes 6, 7, 9, and 10). When a higher amount of SCAP cDNA was transfected, the wild-type SCAP caused an increase in nuclear SREBPs (lane 11), but the mutant forms of SCAP had a more pronounced effect (lanes 12 and 13). These results are consistent with previous experiments showing that overexpression of wild-type SCAP stimulates the cleavage of SREBPs in the presence of sterols, but that the D443N mutant is much more potent in this regard (2).
Figure 3.
Activity of mutant SCAP(Y298C) in transfected cells. (A and B) Stimulation of cleavage of SREBPs by mutant SCAP(Y298C) in transfected 293 cells grown in the presence of sterols. On day 2 of growth, 293 cells were cotransfected with 9 μg of either pTK-HSV-BP1a encoding HSV epitope-tagged human SREBP-1a (A, lanes 3–13) or pTK-HSV-BP2 (B, lanes 3–13) together with the indicated amount of epitope-tagged wild-type SCAP (lanes 5, 8, and 11), SCAP(D443N) (lanes 6, 9, and 12), or SCAP(Y298C) (lanes 7, 10, and 13). The total amount of DNA in each dish was adjusted to 10 μg by addition of pTK empty vector. Three hours after transfection, cells were switched to medium B containing 10% lipoprotein-deficient serum, 50 μM compactin, 50 μM sodium mevalonate, and 0.2% ethanol in the absence (− sterols) or presence (+ sterols) of 1 μg/ml of 25-hydroxycholesterol plus 10 μg/ml of cholesterol as indicated. On day 3, 25 μg/ml of N-acetyl-leucinol-leucinol-norleucinol (ALLN) was added directly to the medium 4 h before cell harvesting and fractionation, which was carried out as described in Fig. 1. Aliquots of nuclear extract (A, 80 μg and B, 55 μg) and membranes (A, 100 μg and B, 110 μg) were subjected to 7% SDS/PAGE, transferred to nitrocellulose, and immunoblotted with either 0.5 μg/ml of IgG-HSV-Tag antibody (nuclear extracts) or 10 μg/ml of IgG-9D5 (membranes). Filters were exposed to film for 5 min (A), 30 s (B, Upper), and 50 s (B, Lower). N denotes the nuclear cleaved form of SREBPs. (C) Growth of CHO-7 cells. On day 0, CHO-7 cells were set up (5 × 105 cells/100-mm dish) in medium C (medium A containing 5% lipoprotein-deficient serum). On day 2, cells were transfected with either no plasmid, empty vector pTK, pTK3-SCAP encoding wild-type SCAP, pTK3-SCAP(D443N), or pTK3-SCAP(Y298C). All plasmids contained the G418-resistance gene neo. Three hours after transfection, cells were washed twice with PBS and refed with medium C. On day 3, cells were switched to medium D (medium C containing 0.75 mg/ml of G418) and refed every 2–3 days. No cells survived in dishes mock-transfected without any plasmid. On day 14, all surviving cells from duplicate, vector-transfected dishes were trypsinized, pooled, and set up in duplicate dishes in medium D (1.5 × 105 cells/60-mm dish). On day 15, cells were refed with medium D (Upper) or medium D containing 0.3 μg/ml of 25-hydroxycholesterol (Lower), and refed every 2–3 days. On day 18 (Upper) and day 22 (Lower), cells were fixed in 1% (vol/vol) glutaraldehyde, stained with crystal violet, and photographed.
Fig. 3C shows a growth experiment in which wild-type CHO cells were transfected with cDNAs encoding wild-type SCAP or one of the two mutant versions. The cells then were grown in the presence of 25-hydroxycholesterol and the absence of cholesterol. Transfection with wild-type SCAP did not permit the cells to grow under these conditions, but the two mutant forms of SCAP restored growth.
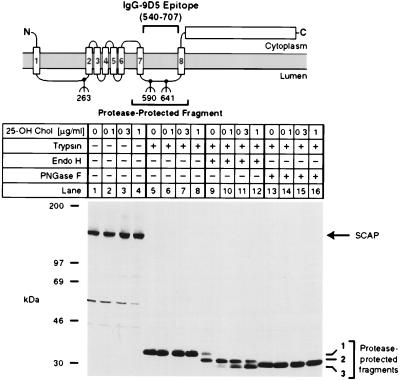
We previously reported that SCAP contains a glycosylated luminal loop that is protected from proteolysis when intact membrane vesicles are digested with trypsin (5) (see diagram in Fig. 4). The protected fragment can be visualized by SDS/PAGE and immunoblotting with IgG-9D5, a mAb whose epitope lies within the protected fragment. The previous studies showed that this fragment contains two N-linked sugar chains that can be released with PNGase F, a nonspecific glycosidase that digests high mannose sugars of the type found in the ER as well as mannose-trimmed carbohydrates that are generated by Golgi-specific enzymes (26, 27). We now sought to determine the sensitivity of these carbohydrate chains to endo H, a glycosidase that cleaves N-linked sugars with a high mannose or hybrid structure, but is inactive against N-linked sugars with a complex structure (26).
Figure 4.
Sterols alter endo H sensitivity of SCAP. The diagram shows a schematic of the domain structure of SCAP, denoting the approximate position of the protease-resistant fragment recognized by mAb IgG-9D5. Numbers below the diagram denote sites of N-linked glycosylation (5). On day 0, CHO-7 cells were set up in medium A supplemented with 10% fetal calf serum and 50 μg protein/ml of low density lipoprotein. On day 2, cells were switched to medium A containing 10% lipoprotein-deficient serum, 50 μM compactin, 50 μM sodium mevalonate, and 0.1% ethanol containing the indicated final concentration of 25-hydroxycholesterol (25-OH Chol.). After incubation for 16 h, cells were harvested, and membrane fractions were prepared as described in Materials and Methods. Aliquots of the membrane fraction (42 μg protein) were incubated in the absence (lanes 1–4) or presence (lanes 5–16) of 17 μg/ml of trypsin. Proteolysis was stopped, and the samples were incubated for 16 h at 37°C in the absence (lanes 1–8) or presence (lanes 9–16) of the indicated glycosidase, subjected to SDS/PAGE, transferred to nitrocellulose, and immunoblotted with 10 μg/ml of IgG-9D5. The filter was exposed to film for 1.5 min. Numbers 1–3 on the right denote differentially glycosylated forms of the protease-resistant SCAP fragment containing either two (band 1), one (band 2), or no (band 3) N-linked oligosaccharides (5).
In the experiment of Fig. 4, CHO-7 cells were incubated in inducing medium in the absence of sterols or in the presence of increasing concentrations of 25-hydroxycholesterol. After 16 h membrane vesicles were digested with trypsin, and the protected fragments were further digested with either endo H or PNGase F. In the absence of trypsin, intact SCAP migrated at 150 kDa, and there was no effect of 25-hydroxycholesterol on the amount or migration of the protein (Fig. 4, lanes 1–4). Trypsin generated a protected fragment of 36 kDa (designated fragment 1), which was not affected by 25-hydroxycholesterol (lanes 5–8). Treatment of fragment 1 with PNGase F reduced its apparent size to 32 kDa (designated fragment 3), and again there was no effect of 25-hydroxycholesterol (lanes 13–16). A different result was obtained with endo H. In the sterol-deprived cells, endo H treatment only partially removed the N-linked sugar chains (lane 9). A portion of fragment 1 remained intact, and another portion was reduced to an intermediate fragment of 34 kDa (designated fragment 2). With the addition of increasing amounts of 25-hydroxycholesterol, fragment 1 disappeared, and we observed a gradual increase in fragment 3 (lanes 10–12). We interpret this finding to indicate that in the presence of 25-hydroxycholesterol the N-linked sugars on SCAP exist in a high mannose form, whereas in the absence of sterols the N-linked sugars are modified to a complex structure.
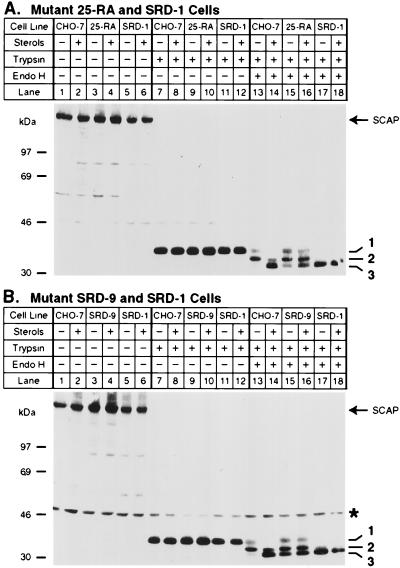
To determine whether the 25-hydroxycholesterol-resistant forms of SCAP would show the same changes in endo H sensitivity we repeated this experiment with 25-RA cells (Fig. 5A). As a control we used SRD-1 cells. These cells have normal SCAP, but they are resistant to 25-hydroxycholesterol by virtue of the production of a truncated SREBP that enters the nucleus without a requirement for proteolysis (18). Membranes from sterol-deprived CHO-7 cells showed a persistence of fragment 1 and an absence of fragment 3 when treated with trypsin followed by endo H (Fig. 5A, lane 13). Fragment 1 declined and fragment 3 was generated when the cells had been incubated with sterols (lane 14). In sterol-deprived 25-RA cells (lane 15), we found a mixture of fully endo H-resistant SCAP (fragment 1) and fully endo H-sensitive SCAP (fragment 3). When sterols were added, there was a slight decrease in the amount of fragment 1, but it remained much more prominent than it was in sterol-treated CHO-7 cells (compare lanes 16 and 14). There was also a slight increase in the amount of fragment 3, but it remained relatively less abundant than it was in the CHO-7 cells. In the SRD-1 cells, all of the SCAP was in the fully endo H-sensitive form even in the absence of added sterols (lane 17), and there was no change when 25-hydroxycholesterol was added (lane 18). Virtually identical results were obtained when SRD-9 cells were compared with SRD-1 cells (Fig. 5B). These data indicate that the mutant forms of SCAP are resistant to the effects of sterols on endo H sensitivity.
Figure 5.
Regulated endo H sensitivity of SCAP is perturbed in 25-hydroxycholesterol-resistant CHO cell lines that harbor mutations in SCAP or SREBP-2. On day 0, the indicated cells were set up in medium A supplemented with 10% fetal calf serum. On day 2, cells were switched to medium A containing 10% lipoprotein-deficient serum, 50 μM compactin, 50 μM sodium mevalonate, and 0.2% ethanol in either the absence (− sterols) or presence (+ sterols) of 1 μg/ml of 25-hydroxycholesterol plus 10 μg/ml of cholesterol as indicated. After incubation for 16 h, cells were harvested, and membrane fractions were prepared as described in Materials and Methods. Aliquots of membranes (A, 50 μg and B, 54 μg) were incubated in the absence or presence of 17 μg/ml of trypsin as indicated. Proteolysis was stopped, and the samples were incubated at 37°C for 16 h either in the absence (lanes 1–12) or presence (lanes 13–18) of endo H, subjected to SDS/PAGE, and transferred to nitrocellulose. Filters were blotted with 10 μg/ml of IgG-9D5 and exposed to film for 2 min (A) and 20 sec (B). ∗ denotes a cross-reacting protein of unknown identity. Numbers 1–3 on the right denote differentially N-glycosylated forms of the protease-resistant SCAP fragment as described in the legend to Fig. 4.
DISCUSSION
In this paper we report the identification of a point mutation in the sterol-sensing domain of SCAP that creates a dominant 25-hydroxycholesterol-resistant phenotype in CHO cells. The mutation changes Tyr-298 to Cys, thereby replacing a residue that is conserved in all five known sterol-sensing domains. A cDNA encoding the Y298C mutant form of SCAP was able to confer 25-hydroxycholesterol resistance when introduced into human kidney 293 cells or wild-type CHO cells (Fig. 3). In experiments not shown, we found that the phenotype is attributable to the loss of a tyrosine and not to the acquisition of a cysteine. Replacement of Tyr-298 with Ala produced the same 25-hydroxycholesterol-resistant phenotype.
Considered together with previous data, the current results identify two residues in SCAP that are crucial for regulation by 25-hydroxycholesterol, namely Asp-443 and Tyr-298. Inasmuch as both of these residues lie within the putative sterol-sensing domain, these data provide strong support for the hypothesis that this domain serves as a sterol sensor in SCAP and, by analogy, in the other proteins that contain sterol-sensing domains (Fig. 2).
Additional support for a sterol-sensing function of SCAP comes from the finding that 25-hydroxycholesterol affected the processing of the N-linked carbohydrate chains of wild-type SCAP, but not mutant SCAP, as indicated by susceptibility to endo H digestion. These studies were made possible by the observation of a trypsin-resistant fragment of SCAP that contains two N-linked carbohydrate chains (5). When SCAP from sterol-deprived cells was digested with trypsin, a high proportion of these chains were endo H resistant. Addition of 25-hydroxycholesterol led to the elimination of all peptide fragments carrying two endo H-resistant chains (fragment 1 in Figs. 4 and 5) and to the appearance of fragments of SCAP in which both N-linked chains were endo H sensitive (fragment 3).
A different pattern of carbohydrate processing was seen when we studied the 25-RA and SRD-9 cells, which produce 25-hydroxycholesterol resistant variants of SCAP (Fig. 5). These cells are heterozygous for the D443N mutation or the Y298C mutation, respectively. Half of their SCAP is wild type, and the other half is mutant. In the absence of sterols these cells showed one population of SCAP that was endo H resistant (fragment 1) and another population that was endo H sensitive (fragment 3). We believe that the endo H-resistant form represents the mutant SCAP and the endo H-sensitive form represents the wild-type protein. Why is the wild-type protein endo H sensitive in these cells, even in the absence of exogenous sterols? The answer may lie in the fact that these mutant cells overproduce cholesterol as a result of their defect in sterol-mediated feedback regulation. Two additional findings support this hypothesis. First, the addition of 25-hydroxycholesterol had little effect on the carbohydrate chains of these cells, which continued to show evidence of endo H-sensitive and resistant forms. Second, in the SRD-1 cells SCAP was fully endo H sensitive, even in the absence of sterols. The SRD-1 cells produce a truncated form of SREBP-2 that enters the nucleus without proteolysis, and it cannot be down-regulated even though SCAP is normal (18). These cells are 25-hydroxycholesterol resistant, and they overproduce cholesterol (17) just as do the 25-RA and SRD-9 cells, but all of the SCAP in the SRD-1 cells is wild type and therefore behaves as though it has been exposed to sterols, i.e., it is endo H sensitive.
In the classic pathway of glycoprotein biosynthesis, high mannose N-linked carbohydrate chains are added to asparagine residues in the ER (27). These chains contain two N-acetylglucosamine residues, nine mannose residues, and three glucose moieties. One mannose and three glucoses are removed while the protein is in the ER. During this period the chains are susceptible to removal by endo H. When the protein reaches the cis/medial Golgi apparatus, the N-linked sugars are successively modified by a series of three Golgi-specific glycosidases and glycosyltransferases, at which point the chains are no longer susceptible to endo H. Considered in this context, the current data raise the possibility that in sterol-deprived wild-type cells SCAP moves from the ER to the cis/medial Golgi complex where its N-linked sugars are rendered endo H resistant. When 25-hydroxycholesterol is present, SCAP remains in a pre-Golgi compartment, and its sugars remain endo H sensitive. We cannot rule out the possibility that SCAP moves to the cis/medial Golgi under all conditions and that in the presence of sterols the protein undergoes a conformational change that renders it resistant to the carbohydrate-modifying enzymes that it encounters in the Golgi.
In addition to the fully endo H-resistant fragment 1 and the fully sensitive fragment 3, SCAP contains fragment 2, which is partially endo H sensitive (see Figs. 4 and 5). According to the scheme presented above, SCAP molecules bearing fragment 2 must have entered the Golgi, but only one of the two carbohydrate chains was modified to an endo H-resistant form. It is well known that some carbohydrate chains can pass through the Golgi and remain in an endo H-sensitive form (28). We attempted to use in vitro mutagenesis to eliminate one of the carbohydrate chains, but these experiments were unsuccessful because overexpressed SCAP in transfected cells remains in an endo H-sensitive form, even under maximal inducing conditions.
Considered together, the current data suggest that SCAP moves from the ER to the cis/medial Golgi whenever it is active, i.e., in sterol-deprived wild-type cells or in sterol-treated mutant cells that express the Y298C or D443N allele. A crucial question is whether the movement of SCAP from ER to Golgi is required for SCAP to stimulate cleavage of SREBPs, or whether this movement occurs after SREBP cleavage. At the time of cleavage, SCAP is in a complex with the COOH-terminal half of SREBPs (7, 8), and it is possible that SCAP moves with the COOH-terminal domain to the Golgi after cleavage has taken place.
Acknowledgments
We thank Jianxin Yang for help in isolation of SRD-9 cells; Kara Robinson for excellent technical assistance; Lisa Beatty for invaluable help with cultured cells; and Michelle Laremore and Jeff Cormier for DNA sequencing. This work was supported by research funds from the National Institutes of Health (HL20948) and the Perot Family Foundation.
ABBREVIATIONS
- CHO
Chinese hamster ovary
- endo
endoglycosidase
- ER
endoplasmic reticulum
- HSV
herpes simplex virus
- HMG
3-hydroxy-3-methylglutaryl
- PNGase F
peptide N-glycosidase F
- SREBP
sterol regulatory element binding protein
- SCAP
SREBP cleavage-activating protein
- SRD-9 cells
a mutant cell line expressing a sterol regulatory-defective phenotype.
References
- 1. Brown M S, Goldstein J L. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 2.Hua X, Nohturfft A, Goldstein J L, Brown M S. Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- 3.Duncan E A, Brown M S, Goldstein J L, Sakai J. J Biol Chem. 1997;272:12778–12785. doi: 10.1074/jbc.272.19.12778. [DOI] [PubMed] [Google Scholar]
- 4.Duncan E A, Dave U P, Sakai J, Goldstein J L, Brown M S. J Biol Chem. 1998;273:17801–17809. doi: 10.1074/jbc.273.28.17801. [DOI] [PubMed] [Google Scholar]
- 5.Nohturfft A, Brown M S, Goldstein J L. J Biol Chem. 1998;273:17243–17250. doi: 10.1074/jbc.273.27.17243. [DOI] [PubMed] [Google Scholar]
- 6.Neer E J, Schmidt C J, Nambudripad R, Smith T F. Nature (London) 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 7.Sakai J, Nohturfft A, Cheng D, Ho Y K, Brown M S, Goldstein J L. J Biol Chem. 1997;272:20213–20221. doi: 10.1074/jbc.272.32.20213. [DOI] [PubMed] [Google Scholar]
- 8.Sakai J, Nohturfft A, Goldstein J L, Brown M S. J Biol Chem. 1998;273:5785–5793. doi: 10.1074/jbc.273.10.5785. [DOI] [PubMed] [Google Scholar]
- 9.Nohturfft A, Hua X, Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1996;93:13709–13714. doi: 10.1073/pnas.93.24.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olender E H, Simoni R D. J Biol Chem. 1992;267:4223–4235. [PubMed] [Google Scholar]
- 11.Loftus S K, Morris J A, Carstea E D, Gu J Z, Cummings C, Brown A, Ellison J, Ohno K, Rosenfeld M A, Tagle D A, et al. Science. 1997;277:232–235. doi: 10.1126/science.277.5323.232. [DOI] [PubMed] [Google Scholar]
- 12.Tabin C J, McMahon A P. Trends Cell Biol. 1997;7:442–446. doi: 10.1016/S0962-8924(97)01159-8. [DOI] [PubMed] [Google Scholar]
- 13.Porter J A, Young K E, Beachy P A. Science. 1996;274:255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- 14.Gemmill R M, West J D, Boldog F, Tanaka N, Robinson L J, Smith D I, Li F, Drabkin H A. Proc Natl Acad Sci USA. 1998;95:9572–9577. doi: 10.1073/pnas.95.16.9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang T-Y, Limanek J S. J Biol Chem. 1980;255:7787–7795. [PubMed] [Google Scholar]
- 16.Goldstein J L, Basu S K, Brown M S. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 17.Metherall J E, Goldstein J L, Luskey K L, Brown M S. J Biol Chem. 1989;264:15634–15641. [PubMed] [Google Scholar]
- 18.Yang J, Sato R, Goldstein J L, Brown M S. Gene Dev. 1994;8:1910–1919. doi: 10.1101/gad.8.16.1910. [DOI] [PubMed] [Google Scholar]
- 19.Hua X, Sakai J, Ho Y K, Goldstein J L, Brown M S. J Biol Chem. 1995;270:29422–29427. doi: 10.1074/jbc.270.49.29422. [DOI] [PubMed] [Google Scholar]
- 20.Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Proc Natl Acad Sci USA. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hua X, Sakai J, Brown M S, Goldstein J L. J Biol Chem. 1996;271:10379–10384. doi: 10.1074/jbc.271.17.10379. [DOI] [PubMed] [Google Scholar]
- 22.Kozak M. Microbiol Rev. 1983;47:1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fanger B O. Anal Biochem. 1987;162:11–17. doi: 10.1016/0003-2697(87)90004-2. [DOI] [PubMed] [Google Scholar]
- 24.Sato R, Yang J, Wang X, Evans M J, Ho Y K, Goldstein J L, Brown M S. J Biol Chem. 1994;269:17267–17273. [PubMed] [Google Scholar]
- 25.Yang J, Brown M S, Ho Y K, Goldstein J L. J Biol Chem. 1995;270:12152–12161. doi: 10.1074/jbc.270.20.12152. [DOI] [PubMed] [Google Scholar]
- 26.Maley F, Trimble R B, Tarentino A L, Plummer T H., Jr Anal Biochem. 1989;180:195–204. doi: 10.1016/0003-2697(89)90115-2. [DOI] [PubMed] [Google Scholar]
- 27.Kornfeld R, Kornfeld S. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 28.Li E, Kornfeld S. J Biol Chem. 1979;254:1600–1605. [PubMed] [Google Scholar]
- 29.Sakai J, Duncan E A, Rawson R B, Hua X, Brown M S, Goldstein J L. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]