Abstract
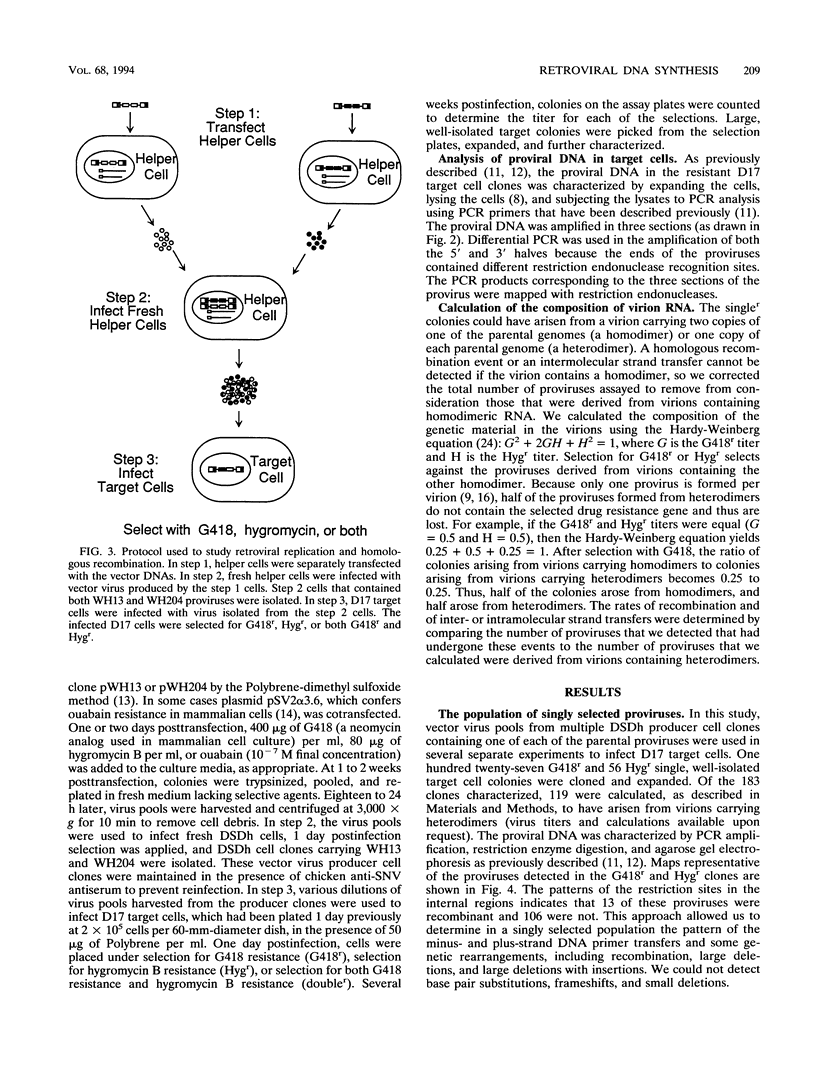
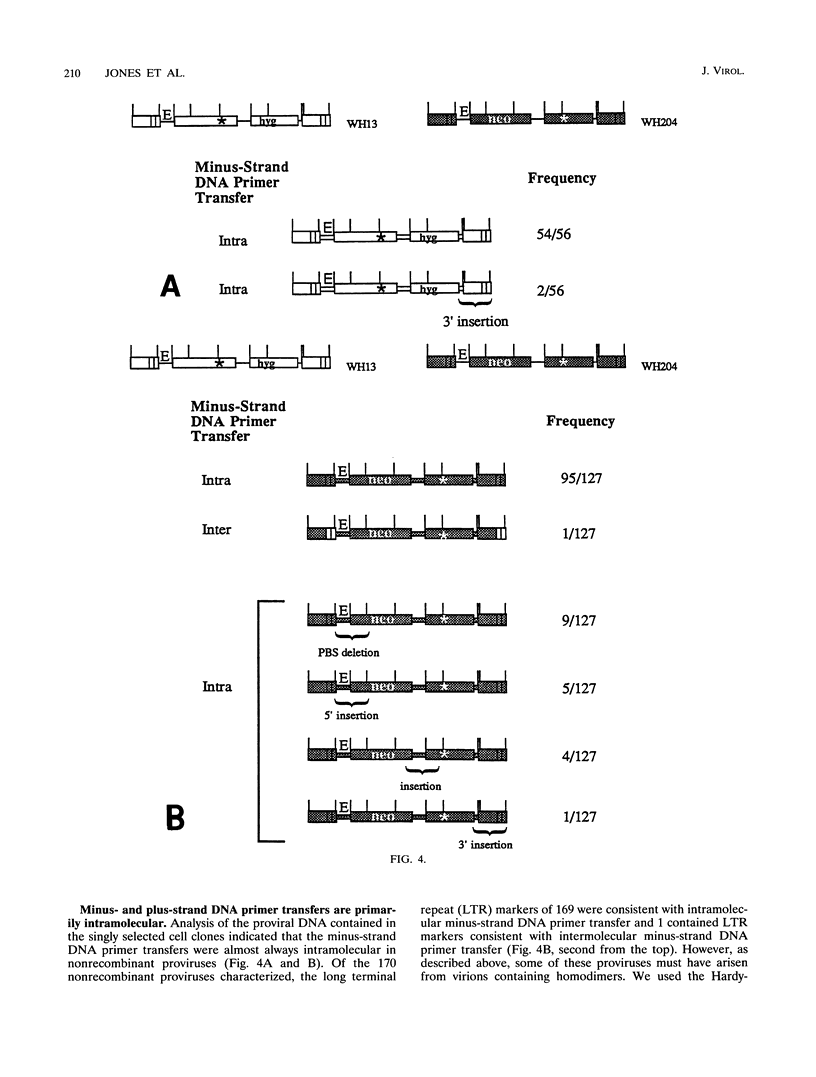
We used previously characterized spleen necrosis virus-based retroviral vectors and helper cells to study the strand transfers that occur during the reverse-transcription phase of a single cycle of retroviral replication. The conditions used selected only for formation of an active provirus rather than for expression of multiple drug resistance markers. In nonrecombinant proviruses the minus- and plus-strand DNA primer transfers were almost completely intramolecular. However, as previously reported, recombinant proviruses contained approximately equal proportions of inter- and intramolecular minus-strand DNA primer transfers. Thus, we conclude that in the absence of recombination, one molecule of retroviral RNA is sufficient for viral DNA synthesis. Large deletions and deletions with insertions were detected primarily at a limited number of positions which appear to be hot spots for such events, the primer binding site and regions containing multiple inverted repeats. At these hot spots, the rate of deletions and deletions with insertions visible with PCR was about 10% per genome per replication cycle. Other deletions and deletions with insertions (detectable with PCR) occurred at a rate of about 0.5%/kb per replication cycle. Crossovers occurred at a rate of about 6%/kb per replication cycle under single-selection conditions. This rate is comparable to the rate that we reported previously under double-selection conditions, indicating that retroviral homologous recombination is not highly error prone. The combined rates of deletions and deletions with insertions at hot spots (10% per genome per replication cycle) and other sites (0.5%/kb per replication cycle) and the rate of crossovers (6%/kb per replication cycle) indicate that on average, full-size (10-kb) type C retroviruses undergo an additional or aberrant strand transfer about once per cycle of infection.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Coffin J. M. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979 Jan;42(1):1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- Dougherty J. P., Temin H. M. High mutation rate of a spleen necrosis virus-based retrovirus vector. Mol Cell Biol. 1986 Dec;6(12):4387–4395. doi: 10.1128/mcb.6.12.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty J. P., Wisniewski R., Yang S. L., Rhode B. W., Temin H. M. New retrovirus helper cells with almost no nucleotide sequence homology to retrovirus vectors. J Virol. 1989 Jul;63(7):3209–3212. doi: 10.1128/jvi.63.7.3209-3212.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa E., Mitra S. W., Goff S., Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979 Sep;18(1):93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- Hajjar A. M., Linial M. L. A model system for nonhomologous recombination between retroviral and cellular RNA. J Virol. 1993 Jul;67(7):3845–3853. doi: 10.1128/jvi.67.7.3845-3853.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. S., Temin H. M. Effect of gamma radiation on retroviral recombination. J Virol. 1992 Jul;66(7):4457–4463. doi: 10.1128/jvi.66.7.4457-4463.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. S., Temin H. M. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci U S A. 1990 Feb;87(4):1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. S., Temin H. M. Retroviral recombination and reverse transcription. Science. 1990 Nov 30;250(4985):1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- Jones J. S., Allan R. W., Temin H. M. Alteration of location of dimer linkage sequence in retroviral RNA: little effect on replication or homologous recombination. J Virol. 1993 Jun;67(6):3151–3158. doi: 10.1128/jvi.67.6.3151-3158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai S., Nishizawa M. New procedure for DNA transfection with polycation and dimethyl sulfoxide. Mol Cell Biol. 1984 Jun;4(6):1172–1174. doi: 10.1128/mcb.4.6.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent R. B., Emanuel J. R., Ben Neriah Y., Levenson R., Housman D. E. Ouabain resistance conferred by expression of the cDNA for a murine Na+, K+-ATPase alpha subunit. Science. 1987 Aug 21;237(4817):901–903. doi: 10.1126/science.3039660. [DOI] [PubMed] [Google Scholar]
- O'Rear J. J., Temin H. M. Spontaneous changes in nucleotide sequence in proviruses of spleen necrosis virus, an avian retrovirus. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1230–1234. doi: 10.1073/pnas.79.4.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban A. T., Fiore D. Ordered interstrand and intrastrand DNA transfer during reverse transcription. Science. 1988 Aug 26;241(4869):1064–1069. doi: 10.1126/science.2457948. [DOI] [PubMed] [Google Scholar]
- Pathak V. K., Temin H. M. 5-Azacytidine and RNA secondary structure increase the retrovirus mutation rate. J Virol. 1992 May;66(5):3093–3100. doi: 10.1128/jvi.66.5.3093-3100.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak V. K., Temin H. M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6024–6028. doi: 10.1073/pnas.87.16.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peliska J. A., Benkovic S. J. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992 Nov 13;258(5085):1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- Pulsinelli G. A., Temin H. M. Characterization of large deletions occurring during a single round of retrovirus vector replication: novel deletion mechanism involving errors in strand transfer. J Virol. 1991 Sep;65(9):4786–4797. doi: 10.1128/jvi.65.9.4786-4797.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields A., Witte W. N., Rothenberg E., Baltimore D. High frequency of aberrant expression of Moloney murine leukemia virus in clonal infections. Cell. 1978 Jul;14(3):601–609. doi: 10.1016/0092-8674(78)90245-3. [DOI] [PubMed] [Google Scholar]
- Stuhlmann H., Berg P. Homologous recombination of copackaged retrovirus RNAs during reverse transcription. J Virol. 1992 Apr;66(4):2378–2388. doi: 10.1128/jvi.66.4.2378-2388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain A., Coffin J. M. Mechanism of transduction by retroviruses. Science. 1992 Feb 14;255(5046):841–845. doi: 10.1126/science.1371365. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin H. M. Sex and recombination in retroviruses. Trends Genet. 1991 Mar;7(3):71–74. doi: 10.1016/0168-9525(91)90272-R. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Structure, variation and synthesis of retrovirus long terminal repeat. Cell. 1981 Nov;27(1 Pt 2):1–3. doi: 10.1016/0092-8674(81)90353-6. [DOI] [PubMed] [Google Scholar]
- Watanabe S., Temin H. M. Construction of a helper cell line for avian reticuloendotheliosis virus cloning vectors. Mol Cell Biol. 1983 Dec;3(12):2241–2249. doi: 10.1128/mcb.3.12.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Temin H. M. 3' junctions of oncogene-virus sequences and the mechanisms for formation of highly oncogenic retroviruses. J Virol. 1993 Apr;67(4):1747–1751. doi: 10.1128/jvi.67.4.1747-1751.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Temin H. M. Rate and mechanism of nonhomologous recombination during a single cycle of retroviral replication. Science. 1993 Jan 8;259(5092):234–238. doi: 10.1126/science.8421784. [DOI] [PubMed] [Google Scholar]



