Abstract
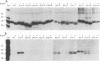
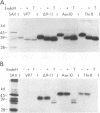
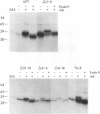
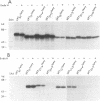
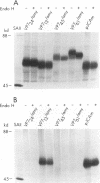
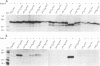
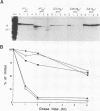
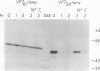
The rotavirus outer capsid glycoprotein, VP7, is an endoplasmic reticulum (ER) membrane-associated glycoprotein in both infected and transfected cells. It was previously demonstrated in this laboratory and by others that both the cleaved signal sequence (H2) and the first NH2-terminal 61 amino acids of VP7 are sufficient and necessary for ER retention of this molecule. Using site-specific mutagenesis and transfection techniques, we show that residues Ile-9, Thr-10, and Gly-11 were specifically necessary for ER retention. These results further define the ER retention sequence of VP7 and demonstrate that conservative changes, apparently innocuous in only three adjacent amino acids, can lead to major solubility and compartmentalization changes. It was found that placement of the first 31 mature NH2-terminal residues of VP7, in addition to the cleaved ER translocation signal sequence, was sufficient to retain the enzymatically active chimeric alpha-amylase in the ER; this enzyme is normally secreted. Deletions of the residues Ile-9, Thr-10, and Gly-11 within the amylase chimera containing 31 VP7 amino acids resulted in secretion of enzymatically active protein. It was also observed that the residues of VP7 presented in certain chimeras were able to abolish alpha-amylase enzymatic activity. These chimeras are presumably misfolded since it was demonstrated by pulse-chase experiments that these molecules are degraded in the ER. We surmise that a favorable conformation is necessary for retention since ER retention and activity of the chimeras depend on the primary sequence context.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bellamy A. R., Both G. W. Molecular biology of rotaviruses. Adv Virus Res. 1990;38:1–43. doi: 10.1016/s0065-3527(08)60858-1. [DOI] [PubMed] [Google Scholar]
- Both G. W., Mattick J. S., Bellamy A. R. Serotype-specific glycoprotein of simian 11 rotavirus: coding assignment and gene sequence. Proc Natl Acad Sci U S A. 1983 May;80(10):3091–3095. doi: 10.1073/pnas.80.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson G., Duée E., Haser R., Payan F. Three dimensional structure of porcine pancreatic alpha-amylase at 2.9 A resolution. Role of calcium in structure and activity. EMBO J. 1987 Dec 20;6(13):3909–3916. doi: 10.1002/j.1460-2075.1987.tb02731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Bonifacino J. S., Yuan L. C., Klausner R. D. Selective degradation of T cell antigen receptor chains retained in a pre-Golgi compartment. J Cell Biol. 1988 Dec;107(6 Pt 1):2149–2161. doi: 10.1083/jcb.107.6.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahllöf B., Wallin M., Kvist S. The endoplasmic reticulum retention signal of the E3/19K protein of adenovirus-2 is microtubule binding. J Biol Chem. 1991 Jan 25;266(3):1804–1808. [PubMed] [Google Scholar]
- Estes M. K., Cohen J. Rotavirus gene structure and function. Microbiol Rev. 1989 Dec;53(4):410–449. doi: 10.1128/mr.53.4.410-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Jackson M. R., Nilsson T., Peterson P. A. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 1990 Oct;9(10):3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabcenell A. K., Atkinson P. H. Processing of the rough endoplasmic reticulum membrane glycoproteins of rotavirus SA11. J Cell Biol. 1985 Oct;101(4):1270–1280. doi: 10.1083/jcb.101.4.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabcenell A. K., Poruchynsky M. S., Bellamy A. R., Greenberg H. B., Atkinson P. H. Two forms of VP7 are involved in assembly of SA11 rotavirus in endoplasmic reticulum. J Virol. 1988 Aug;62(8):2929–2941. doi: 10.1128/jvi.62.8.2929-2941.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausner R. D., Sitia R. Protein degradation in the endoplasmic reticulum. Cell. 1990 Aug 24;62(4):611–614. doi: 10.1016/0092-8674(90)90104-m. [DOI] [PubMed] [Google Scholar]
- Kornfeld R., Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lewis M. J., Pelham H. R. A human homologue of the yeast HDEL receptor. Nature. 1990 Nov 8;348(6297):162–163. doi: 10.1038/348162a0. [DOI] [PubMed] [Google Scholar]
- Lewis M. J., Sweet D. J., Pelham H. R. The ERD2 gene determines the specificity of the luminal ER protein retention system. Cell. 1990 Jun 29;61(7):1359–1363. doi: 10.1016/0092-8674(90)90699-f. [DOI] [PubMed] [Google Scholar]
- Maass D. R., Atkinson P. H. Rotavirus proteins VP7, NS28, and VP4 form oligomeric structures. J Virol. 1990 Jun;64(6):2632–2641. doi: 10.1128/jvi.64.6.2632-2641.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor E. A., Svensson B. A super-secondary structure predicted to be common to several alpha-1,4-D-glucan-cleaving enzymes. Biochem J. 1989 Apr 1;259(1):145–152. doi: 10.1042/bj2590145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer C. E., Doms R. W., Bole D. G., Helenius A., Rose J. K. Heavy chain binding protein recognizes incompletely disulfide-bonded forms of vesicular stomatitis virus G protein. J Biol Chem. 1990 Apr 25;265(12):6879–6883. [PubMed] [Google Scholar]
- Munro S., Pelham H. R. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987 Mar 13;48(5):899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- Nilsson T., Jackson M., Peterson P. A. Short cytoplasmic sequences serve as retention signals for transmembrane proteins in the endoplasmic reticulum. Cell. 1989 Aug 25;58(4):707–718. doi: 10.1016/0092-8674(89)90105-0. [DOI] [PubMed] [Google Scholar]
- Ohkuma S., Poole B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3327–3331. doi: 10.1073/pnas.75.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R. Control of protein exit from the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:1–23. doi: 10.1146/annurev.cb.05.110189.000245. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Hardwick K. G., Lewis M. J. Sorting of soluble ER proteins in yeast. EMBO J. 1988 Jun;7(6):1757–1762. doi: 10.1002/j.1460-2075.1988.tb03005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R., Jackson R. J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem. 1976 Aug 1;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- Petrie B. L., Greenberg H. B., Graham D. Y., Estes M. K. Ultrastructural localization of rotavirus antigens using colloidal gold. Virus Res. 1984;1(2):133–152. doi: 10.1016/0168-1702(84)90069-8. [DOI] [PubMed] [Google Scholar]
- Poruchynsky M. S., Atkinson P. H. Primary sequence domains required for the retention of rotavirus VP7 in the endoplasmic reticulum. J Cell Biol. 1988 Nov;107(5):1697–1706. doi: 10.1083/jcb.107.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pytela R., Pierschbacher M. D., Argraves S., Suzuki S., Ruoslahti E. Arginine-glycine-aspartic acid adhesion receptors. Methods Enzymol. 1987;144:475–489. doi: 10.1016/0076-6879(87)44196-7. [DOI] [PubMed] [Google Scholar]
- Päbo S., Bhat B. M., Wold W. S., Peterson P. A. A short sequence in the COOH-terminus makes an adenovirus membrane glycoprotein a resident of the endoplasmic reticulum. Cell. 1987 Jul 17;50(2):311–317. doi: 10.1016/0092-8674(87)90226-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J. K., Doms R. W. Regulation of protein export from the endoplasmic reticulum. Annu Rev Cell Biol. 1988;4:257–288. doi: 10.1146/annurev.cb.04.110188.001353. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste J., Kuismanen E. Pre- and post-Golgi vacuoles operate in the transport of Semliki Forest virus membrane glycoproteins to the cell surface. Cell. 1984 Sep;38(2):535–549. doi: 10.1016/0092-8674(84)90508-7. [DOI] [PubMed] [Google Scholar]
- Schneewind O., Jones K. F., Fischetti V. A. Sequence and structural characteristics of the trypsin-resistant T6 surface protein of group A streptococci. J Bacteriol. 1990 Jun;172(6):3310–3317. doi: 10.1128/jb.172.6.3310-3317.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza J. C., Hardwick K. G., Dean N., Pelham H. R. ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell. 1990 Jun 29;61(7):1349–1357. doi: 10.1016/0092-8674(90)90698-e. [DOI] [PubMed] [Google Scholar]
- Shields D., Blobel G. Efficient cleavage and segregation of nascent presecretory proteins in a reticulocyte lysate supplemented with microsomal membranes. J Biol Chem. 1978 Jun 10;253(11):3753–3756. [PubMed] [Google Scholar]
- Stirzaker S. C., Both G. W. The signal peptide of the rotavirus glycoprotein VP7 is essential for its retention in the ER as an integral membrane protein. Cell. 1989 Mar 10;56(5):741–747. doi: 10.1016/0092-8674(89)90677-6. [DOI] [PubMed] [Google Scholar]
- Stirzaker S. C., Poncet D., Both G. W. Sequences in rotavirus glycoprotein VP7 that mediate delayed translocation and retention of the protein in the endoplasmic reticulum. J Cell Biol. 1990 Oct;111(4):1343–1350. doi: 10.1083/jcb.111.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirzaker S. C., Whitfeld P. L., Christie D. L., Bellamy A. R., Both G. W. Processing of rotavirus glycoprotein VP7: implications for the retention of the protein in the endoplasmic reticulum. J Cell Biol. 1987 Dec;105(6 Pt 2):2897–2903. doi: 10.1083/jcb.105.6.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K., Hoshino Y., Nishikawa K., Green K. Y., Maloy W. L., Morita Y., Urasawa S., Kapikian A. Z., Chanock R. M., Gorziglia M. Cross-reactive and serotype-specific neutralization epitopes on VP7 of human rotavirus: nucleotide sequence analysis of antigenic mutants selected with monoclonal antibodies. J Virol. 1988 Jun;62(6):1870–1874. doi: 10.1128/jvi.62.6.1870-1874.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarentino A. L., Maley F. Purification and properties of an endo-beta-N-acetylglucosaminidase from Streptomyces griseus. J Biol Chem. 1974 Feb 10;249(3):811–817. [PubMed] [Google Scholar]
- Tartakoff A. M. Temperature and energy dependence of secretory protein transport in the exocrine pancreas. EMBO J. 1986 Jul;5(7):1477–1482. doi: 10.1002/j.1460-2075.1986.tb04385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartakoff A. M., Vassalli P. Lectin-binding sites as markers of Golgi subcompartments: proximal-to-distal maturation of oligosaccharides. J Cell Biol. 1983 Oct;97(4):1243–1248. doi: 10.1083/jcb.97.4.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkacz J. S., Lampen O. Tunicamycin inhibition of polyisoprenyl N-acetylglucosaminyl pyrophosphate formation in calf-liver microsomes. Biochem Biophys Res Commun. 1975 Jul 8;65(1):248–257. doi: 10.1016/s0006-291x(75)80086-6. [DOI] [PubMed] [Google Scholar]
- Vaux D., Tooze J., Fuller S. Identification by anti-idiotype antibodies of an intracellular membrane protein that recognizes a mammalian endoplasmic reticulum retention signal. Nature. 1990 Jun 7;345(6275):495–502. doi: 10.1038/345495a0. [DOI] [PubMed] [Google Scholar]
- Whitfeld P. L., Tyndall C., Stirzaker S. C., Bellamy A. R., Both G. W. Location of sequences within rotavirus SA11 glycoprotein VP7 which direct it to the endoplasmic reticulum. Mol Cell Biol. 1987 Jul;7(7):2491–2497. doi: 10.1128/mcb.7.7.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibo M., Poole B. Protein degradation in cultured cells. II. The uptake of chloroquine by rat fibroblasts and the inhibition of cellular protein degradation and cathepsin B1. J Cell Biol. 1974 Nov;63(2 Pt 1):430–440. doi: 10.1083/jcb.63.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagouras P., Rose J. K. Carboxy-terminal SEKDEL sequences retard but do not retain two secretory proteins in the endoplasmic reticulum. J Cell Biol. 1989 Dec;109(6 Pt 1):2633–2640. doi: 10.1083/jcb.109.6.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982 Oct 25;10(20):6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva A. M., Balch W. E., Helenius A. Quality control in the endoplasmic reticulum: folding and misfolding of vesicular stomatitis virus G protein in cells and in vitro. J Cell Biol. 1990 Sep;111(3):857–866. doi: 10.1083/jcb.111.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]