Abstract
A key step in signal transduction in the visual cell is the light-induced conformational change of rhodopsin that triggers the binding and activation of the guanine nucleotide-binding protein. Site-directed mAbs against bovine rhodopsin were produced and used to detect and characterize these conformational changes upon light activation. Among several antibodies that bound exclusively to the light-activated state, an antibody (IgG subclass) with the highest affinity (Ka ≈ 6 × 10−9 M) was further purified and characterized. The epitope of this antibody was mapped to the amino acid sequence 304–311. This epitope extends from the central region to the cytoplasmic end of the seventh transmembrane helix and incorporates a part of a highly conserved NPXXY motif, a critical region for signaling and agonist-induced internalization of several biogenic amine and peptide receptors. In the dark state, no binding of the antibody to rhodopsin was detected. Accessibility of the epitope to the antibody correlated with formation of the metarhodopsin II photointermediate and was reduced significantly at the metarhodopsin III intermediate. Further, incubation of the antigen–antibody complex with 11-cis-retinal failed to regenerate the native rhodopsin chromophore. These results suggest significant and reversible conformational changes in close proximity to the cytoplasmic end of the seventh transmembrane helix of rhodopsin that might be important for folding and signaling.
Rhodopsin, a member of a large family of seven-transmembrane (TM) helix receptors (Fig. 1), triggers a light-dependent, phosphodiesterase-mediated, cyclic guanosine monophosphate-regulated, and G protein-coupled signal transduction cascade (1–7). Rhodopsin contains 11-cis-retinal covalently bound via a protonated Schiff base (PSB) to the ɛ-amino group of Lys-296 on TM7. The carboxyl group of Glu-113 on TM3, which resides well within the membrane, neutralizes the positive charge on the retinyl-PSB in the dark state (8–10). This dominant electrostatic interaction and the steric constraint imposed by the very nature of the chromophore in its immediate protein environment supports the inactive ground-state conformation. Upon light absorption, retinal isomerizes to the all-trans configuration that drives the protein through a series of transient and thermally stable intermediates (11). Charge separation between the retinyl-PSB and Glu-113 with subsequent protonation of the latter culminates in formation of metarhodopsin II (Meta II), an active intermediate that accommodates the high-affinity sites for interaction with several regulatory proteins of the visual transduction cascade.
Figure 1.
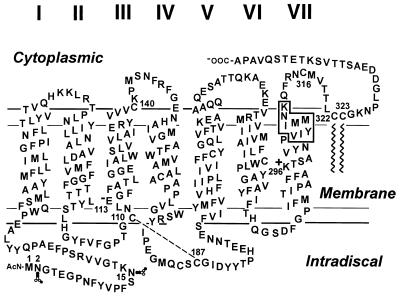
Secondary structure model of bovine rhodopsin. The transmembrane helices are numbered I–VII from the amino to the carboxyl terminus. Asn-2 and Asn-15 are N-glycosylated, Cys-110 and Cys-187 are linked by a disulfide bond, and Cys-322 and Cys-323 are palmitoylated. Exposed cysteine residues on the second (Cys-140) and fourth (Cys-316) cytoplasmic loops are indicated. Lys-296, the site of attachment of retinal, and Glu-113, the counterion to the protonated retinyl-Schiff base, also are indicated. The VIYIMMNK sequence (boxed letters) represents the epitope of the TM7C antibody and incorporates a part of one of the most conserved motifs (NPXXY) in the family of G protein-coupled receptors.
Recently, there has been considerable progress in understanding retinal–protein interactions and the structural rearrangements that the protein undergoes upon cis → trans isomerization of the chromophore (12, 13). Despite these advances, a clear understanding of how changes deep in the membrane are relayed to the surface of the protein, thereby allowing an opening of its conformation for interaction with the signaling machinery, remains largely unknown. It is evident that a comprehensive understanding of how a receptor communicates with this machinery will require the identification of light- or agonist-induced conformational changes in molecular detail. To this end, single and double cysteine-substitution mutants of rhodopsin in conjunction with site-directed spin labeling and electron paramagnetic resonance spectroscopy have provided valuable information about both the dark- and light-activated states (14–21). Further, metal-binding sites or disulfide bonds have been engineered between the TM helices to restrain possible light-induced conformational changes at specific locations in rhodopsin (22, 23). Two important conclusions to arise from these studies are that the cytoplasmic termini of TM3 and TM6 are close in proximity and that the light-induced movement of these helices relative to each other is required to adopt an active conformation.
In the present paper, we show by using an antirhodopsin mAb that light induces the exposure of an epitope that extends from the region between Lys-296 and the cytoplasmic end of TM7. Furthermore, we demonstrate that this region of the protein, which contains the highly conserved NPXXY motif implicated in signaling and agonist-induced internalization of several G protein-coupled receptors (24–26), becomes accessible to the antibody exclusively at the Meta II stage of activation.
MATERIALS AND METHODS
Materials.
Protein A and Con A Sepharose were purchased from Pharmacia. The hybridoma isotyping kit and the alkaline phosphatase-conjugated goat anti-mouse IgG were from Calbiochem. Horseradish peroxidase-conjugated goat anti-mouse IgG was from Promega. ELISA plates were from Nunc, and polyvinylidene fluoride transfer membranes were from Millipore. Peptides corresponding to the carboxyl-terminal region of TM7 were synthesized at the peptide synthesis facilities of the Max Planck Institute for Biophysics (Frankfurt). Bovine retinae were from W. L. Lawson (Lincoln, NE), and 11-cis-retinal was a gift of R. Crouch (Medical University of South Carolina and the National Eye Institute). The A5 mAb, which is specific for the amino-terminal sequence of bacteriorhodopsin, has been described (27). Other materials used in this investigation have been described (28).
Preparation of Rod Outer Segments and Disk Membranes.
Bovine rod outer segments (ROS) were prepared according to a standard procedure (29). Briefly, dark-adapted retina were shaken in an isotonic solution containing 20 mM Hepes, pH 7.0/130 mM KCl/0.5 mM MgCl2/1 mM CaCl2/1 mM DTT/1 μM 11-cis-retinal. The suspension was filtered through a nylon mesh and layered on top of a discontinuous (0.78–1.2 M) sucrose gradient. The membrane fraction was collected after centrifugation at 25,000 × g for 30 min and washed with saline. Disk membranes were prepared from this ROS preparation according to ref. 30 by using 2.5% (wt/vol) Ficoll for flotation.
Preparation of Opsin-Containing Membranes (Apomembranes).
Disk membranes containing 10 mg of rhodopsin were suspended in 5 ml of 50 mM Tris⋅HCl, pH 7.5/100 mM NaCl. An equal volume of a 200 mM solution of NH2OH (pH 7.0) was added and the sample was incubated on ice under a 150-W tungsten lamp for 15 min. The membranes were pelleted by centrifugation and washed with 50 mM Tris⋅HCl, pH 7.5/100 mM NaCl. Both washes (NH2OH followed by buffer alone) were repeated two more times. The resulting apomembranes were resuspended in 50 mM Tris⋅HCl, pH 7.5/100 mM NaCl and used immediately or stored at −80°C.
Production of Hybridomas and Purification of Antirhodopsin Antibodies.
BALB/c mice were immunized i.p. with 0.05 mg of disk membrane rhodopsin four times at 2-week intervals and then an additional three times at 1-month intervals. The first immunization was carried out in Freund’s complete adjuvant. The second, third, and fourth immunizations were done in Freund’s incomplete adjuvant and the rest were done in saline. The animals were boosted 10 days after the last immunization for 3 consecutive days by intraperitoneal injection of 0.01 mg of disk membrane rhodopsin in saline. On the fourth day, the splenocytes were fused with myeloma cells by using a standard procedure (31). The cultures were screened by solid-phase ELISA, and positives were cloned by the end-point dilution procedure. Antibodies were purified from ascites fluid by (NH4)2SO4 precipitation followed by DEAE-Sephacel chromatography using a linear gradient of NaCl (1–500 mM) in 10 mM NaH2PO4, pH 8.0. Fractions containing IgG were pooled according to the ELISA results (see below) and the combined fractions were reprecipitated in 50% (wt/vol) (NH4)2SO4. The precipitate was dissolved in 10 mM NaH2PO4, pH 8.0/150 mM NaCl and dialyzed against the same buffer at 4°C.
ELISA Assays.
Microtiter plates were coated with 100 μg/ml of disk membrane rhodopsin or apomembranes in 100 mM carbonate-buffered saline, pH 8.5. The wells then were blocked with PBS (10 mM NaH2PO4, pH 7.0/150 mM NaCl) containing 3% (wt/vol) BSA to remove any unbound rhodopsin/opsin. Antibody fractions were incubated in the rhodopsin/opsin-coated microtiter plates for 1 h at 20°C, washed with PBS containing 0.05% (vol/vol) Tween 20 to remove excess primary antibody, and then incubated with horseradish peroxidase-conjugated IgG (1:10,000 in wash buffer) for 1 h at 20°C. Excess peroxidase-conjugated IgG was washed away before adding the 3,3′,5,5′-tetramethylbenzidine peroxidase substrate. Reactions were monitored at 492 nm by using an automated ELISA reader (Dynatech).
Selection of Antibodies.
The selection of mAbs was carried out according to their light-dependent binding to rhodopsin. Briefly, samples containing 5 mg of native (unbleached) or photoactivated rhodopsin membranes and 3–5 mg of purified antibody in 10 mM NaH2PO4, pH 8.0/150 mM NaCl were incubated for 20 min at 10°C in the dark. The reaction mixtures were washed three times with the same buffer by centrifugation. Bound antibodies were eluted from the membranes with 100 mM sodium citrate, pH 4.0, and separated by centrifugation. The supernatants were neutralized with 1 M Tris⋅HCl, pH 9.0, and dialyzed against PBS at 4°C. The affinity constants of the antibodies for rhodopsin were determined according to ref. 32. A single mAb with the highest affinity for photoactivated rhodopsin and designated TM7C (directed against the carboxyl-terminal region of the seventh transmembrane helix) was selected for further experiments.
Epitope Mapping of mAb TM7C.
The epitope of the TM7C antibody was mapped by using the bovine opsin peptides in competitive inhibition ELISA assays. The competition assays were carried out by first preincubating serial concentrations of the synthetic TM7 peptides (1 × 10−3 M to 1 × 10−7 M) with 100 μg of TM7C antibody in PBS/0.2% (wt/vol) BSA. After 16 h at 4°C, the antibody/peptide mixture was added to microtiter plates coated with 100 μg of apomembranes and incubated for 2 h at room temperature. The plates were washed with PBS containing 0.05% (vol/vol) Tween 20 and incubated with the second antibody followed by the peroxidase substrate, and the reactions were monitored at 492 nm as described above.
TM7C Binding to Meta II.
Mixtures of rhodopsin (4 mg) and mAbs TM7C or A5 (3–4 mg) in 50 mM Hepes, pH 8.0/150 mM NaCl/2 mM MgCl2/0.2% (wt/vol) BSA were incubated on ice for 60 min in the dark. Equal aliquots of the suspension were transferred into several centrifuge tubes and incubated for 3–5 min at room temperature in the dark. An increasing number of flashes from an orange light (λ > 540 nm) then was applied to consecutive samples. Under these conditions, ≈0.5% of the rhodopsin is converted to Meta II (λmax ≈ 380 nm) after each flash. The samples were incubated for 5 min at room temperature in the dark, diluted with prechilled buffer, and centrifuged at 30,000 × g for 30 min. The pellets were washed two more times with the same buffer without BSA, resuspended in 100 μl of the same buffer containing 125I-protein A, and incubated for 5 min at room temperature. The reaction mixtures were washed several times by centrifugation with the incubation buffer and the pellets were analyzed for 125I radioactivity by gamma counting. Alternatively, aliquots were removed from each sample before 125I-protein A addition and subjected to SDS/PAGE followed by immunoblotting.
TM7C and Meta III Interaction.
A sample containing 2 mg of rhodopsin in 20 mM Hepes, pH 7.6/100 mM NaCl/2 mM MgCl2 was illuminated with orange light (λ > 540 nm) for 10–15 min on ice and allowed to incubate for another 2–3 h at 10°C in the dark. Under these conditions, virtually all of the Meta II (λmax ≈ 380 nm) decayed to Meta III (λmax ≈ 465 nm). Aliquots containing 200 μg of Meta III rhodopsin were mixed with increasing amounts of TM7C or A5 (100–700 μg) in the same buffer supplemented with 0.2% (wt/vol) BSA. The samples were incubated for 5 min at room temperature in the dark, washed several times by centrifugation, incubated with 125I-protein A, and analyzed by gamma counting as described above.
Effect of TM7C on Rhodopsin Regeneration.
Samples containing ≈1 mg of photoactivated rhodopsin or Meta II + TM7C in 20 mM Hepes, pH 7.6/100 mM NaCl/2 mM MgCl2 were incubated at 15°C for 90 min and then mixed with a 5- to 10-fold excess of 11-cis-retinal. After 10–20 min in the dark at room temperature, chromophore regeneration was monitored by UV/visible spectroscopy. All spectra were recorded by using antigen–antibody complexes or their mixtures as blanks before the addition of retinal. The effect of mAb A5 on rhodopsin regeneration served as a control.
SDS/PAGE Analysis and Immunoblotting.
Protein samples were analyzed by reducing SDS/PAGE (33) with 5% stacking and a 15 or 16% resolving gel and electroblotted onto poly(vinylidene difluoride) membranes (34). Rhodopsin was detected by using the rho 1D4 (35) or TM7C primary antibodies and horseradish peroxidase-conjugated goat anti-mouse IgG as the second antibody. The protein bands were visualized by chemiluminescence.
Other Methods.
UV/visible spectra were recorded at 20°C with a Perkin–Elmer λ6 spectrophotometer. Fragments of rhodopsin were generated by limited proteolysis (36) or gene expression (28, 37). Protein A was iodinated with Na125I by using the chloramine-T procedure (38).
RESULTS
Antibody Characterization and Epitope Mapping.
An mAb designated TM7C (recognizing the carboxyl-terminal portion of TM7) was isotyped as an IgG and selected for further analysis on the basis of its exclusive binding to photoactivated rhodopsin and/or apomembranes. In ELISA assays, TM7C showed a Ka ≈ 6 × 10−9 M toward photobleached disk membranes. To map the epitope of TM7C, its interaction with various opsin fragments and synthetic peptides was analyzed. Immunoblot analysis demonstrated that TM7C exclusively recognizes the 250–332 fragment (see Fig. 1) resulting from limited proteolysis of rhodopsin with Staphylococcus aureus V8 protease. Limited proteolysis of rhodopsin with trypsin or Asp-N protease, which removes the highly immunogenic sites on the carboxyl-terminal tail, resulted in essentially the same antibody-staining pattern. Furthermore, ELISA assays showed no inhibition of antibody binding to light-activated rhodopsin in the presence of the 318–348 carboxyl-terminal fragment obtained by cyanogen bromide cleavage or by two synthetic peptides, 248–252 and 312–317, which comprise the cytoplasmically exposed and membrane surface-adjacent extensions of TM6 and TM7, respectively. These findings suggested that the TM7C epitope is linear and might be located on the membrane-spanning portion of these helices or their connectivity on the intradiscal side.
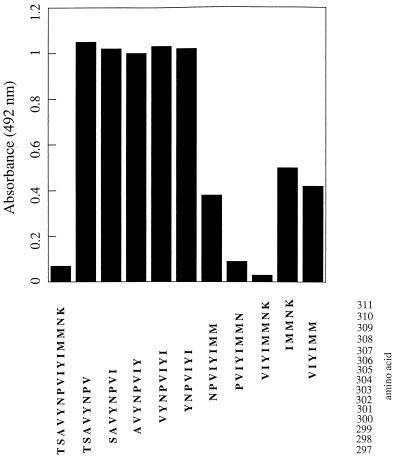
To further define the TM7C epitope, the 250–332 fragment that includes TM6 and TM7 was digested with trypsin and the resulting peptides were purified by reverse-phase HPLC. The only peptide recognized by TM7C in the ELISA assays included the sequence extending from Thr-297, adjacent to the retinal binding Lys-296, to Lys-311 located on the putative membrane/cytoplasmic border (see Fig. 1). To finally map the epitope, the binding of TM7C to apomembranes in the presence of several truncated versions of the 297–311 peptide was analyzed by competitive inhibition assays. As shown in Fig. 2, the 297–311 peptide blocked the interaction of TM7C with opsin. Similar levels of inhibition were observed when using carboxyl-terminal portions of this peptide (amino acids 303–310 or 304–311). On the contrary, there was little or no inhibition when TM7C was incubated with the 297–304 peptide. These results indicate that the epitope for TM7C is confined to the amino acid sequence 304–311 on TM7.
Figure 2.
Epitope mapping of the TM7C mAb. Peptides corresponding to various bovine opsin TM7 sequences were synthesized and tested against the TM7C antibody in competitive inhibition assays (Materials and Methods). The data shown are representative of three separate ELISA assays and were obtained at TM7 peptide concentrations of 1 mM. The Val-304 → Lys-311 peptide was found to be the shortest and most potent competitor. The A492 value obtained in the absence of any peptide was ≈1.0–1.1.
TM7C Specifically Interacts with Meta II.
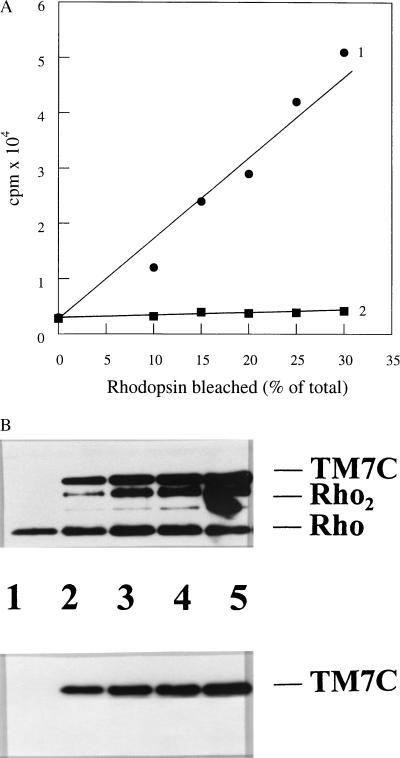
Since the primary selection for the antibodies was carried out according to their light-dependent binding to rhodopsin, the 304–311 epitope of TM7C might represent a part of the rhodopsin structure that gets exposed upon light activation. To test this hypothesis, rhodopsin was mixed with TM7C and equal aliquots of the reaction mixture were activated by an increasing number of light flashes. Bound antibody was detected and quantitated by using 125I-protein A. As shown in Fig. 3A (trace 1), the binding of TM7C to rhodopsin is directly proportional to the number of flashes imposed. Neither prolonged incubation in the dark nor bleaching under the same conditions in the presence of the antibacteriorhodopsin mAb A5 (trace 2) resulted in appreciable 125I-protein A binding. These results suggest a high specificity in the binding of TM7C to Meta II. The binding of TM7C to photoactivated rhodopsin also was tested by taking aliquots of the reaction mixture before incubation with protein A and subjecting the samples to SDS/PAGE. Consistent with the data in Fig. 3A, immunoblot analysis shows that the amount of TM7C antibody binding increases with the amount of photolyzed rhodopsin (Fig. 3B). Interestingly, the intensity of the combined signals corresponding to the rhodopsin monomers and the rhodopsin dimers formed upon photobleaching also increases with the amount of photolyzed rhodopsin (Fig. 3 B Upper). This may result from a preferential interaction or a tighter binding between the rho 1D4 antibody and photobleached rhodopsin as previously observed in solution (39).
Figure 3.
Binding of TM7C to light-activated rhodopsin. Purified disk membranes and TM7C or A5 antibodies were incubated for 60 min in the dark. Equal aliquots of the suspension then were subjected to an increasing number of the orange light (λ > 540 nm) flashes. After incubation for 5 min at room temperature, the samples were washed three times by centrifugation, and the pellets resuspended and incubated with 125I-protein A or analyzed by SDS/PAGE (Materials and Methods). (A) Binding of mAbs TM7C (trace 1) and A5 (trace 2) as a function of Meta II formation. The resuspended pellets were incubated with 125I-protein A for 5 min at room temperature, washed extensively by centrifugation, and then analyzed by gamma counting. (B) Binding of TM7C to Meta II as shown by immunoblotting. Equal amounts of antigen–antibody complex, prepared as described above, were analyzed by reducing SDS/PAGE and immunoblotting before incubation with 125I-protein A. Rhodopsin (and rhodopsin dimers) was detected by using the anti-rhodopsin rho1D4 mAb followed by incubation with the horseradish peroxidase-conjugated secondary antibody (Upper). The binding of the second antibody also results in detection of the TM7C heavy chains. The same blot was then stripped of the primary and secondary antibodies and reprobed with only the secondary antibody to detect the heavy chains of TM7C (Lower). Lane 1, dark control; lanes 2–5, correspond to ≈10, 15, 20, and 30% photobleaching of the total rhodopsin in the sample, respectively.
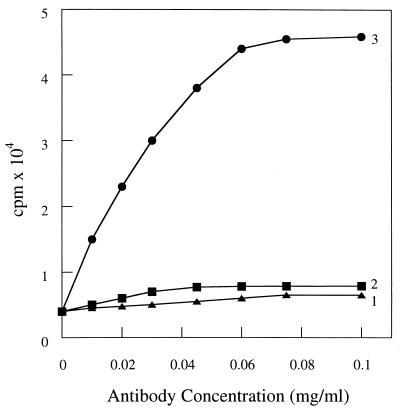
To test whether TM7C also binds to the Meta III intermediate, samples containing equivalent amounts of Meta III were incubated with increasing concentrations of TM7C or A5 antibodies. The washed antigen–antibody mixtures were incubated with 125I-protein A and the concentration–response curves for binding of the antibodies to Meta III were determined. The representative concentration–response curves show very little but significant binding of TM7C compared with that of A5 (Fig. 4, traces 2 and 1, respectively). This low level of TM7C binding to the Meta III preparation may be attributed to the presence of a minor amount of Meta II or some other form of photobleached rhodopsin in the reaction mixture. However, when compared with the saturable binding of TM7C to Meta II (Fig. 4, trace 3), these findings suggest that the TM7C epitope exposed at the Meta II intermediate is not accessible for the antibody at Meta III.
Figure 4.
Comparative analysis of TM7C binding to Meta II and Meta III. Aliquots of Meta II or Meta III were incubated with increasing amounts of TM7C or A5 antibodies for 5 min at room temperature in the dark. The samples were washed several times by centrifugation, incubated with 125I-protein A, and analyzed by gamma counting (Materials and Methods). Antibody binding is shown when Meta III was incubated with increasing concentrations of A5 (trace 1) or TM7C (trace 2). Binding of TM7C to Meta II was used as a positive control and follows saturation kinetics (trace 3).
TM7C Blocks Regeneration of Rhodopsin.
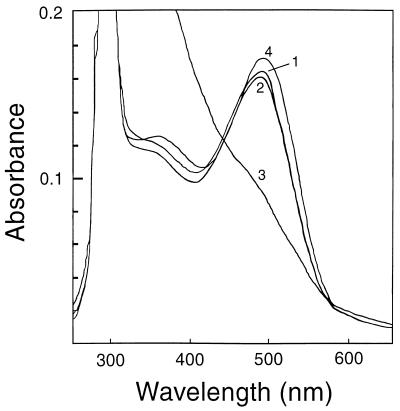
Reversible regeneration with exogenously added 11-cis-retinal is a remarkable characteristic that provides important information on rhodopsin functionality. This feature was used to investigate whether the binding of TM7C, or its presence in the regeneration mixture, affects light-activated rhodopsin’s ability to regenerate the ground-state chromophore. For these experiments, preparations of Meta II were mixed with TM7C and/or A5 antibodies and incubated with a 5- to 10-fold excess of 11-cis-retinal in the dark. As shown in Fig. 5, chromophore formation from Meta II in the absence of any antibody (trace 1) or in the presence of A5 (trace 2) gave essentially the same yield of 500-nm absorbance, ≈90–95% of that of the native (unbleached) rhodopsin control. In contrast, regeneration of rhodopsin from the Meta II-TM7C complex was almost completely blocked (trace 3). Incubating dark-adapted rhodopsin with both the TM7C and A5 antibodies had no effect on the chromophore (trace 4). Notably, the binding of TM7C to Meta II significantly accelerated its rate of decay, a process that could be prevented by preincubation of the antibody with the 304–311 peptide (data not shown).
Figure 5.
Effect of TM7C on rhodopsin regeneration. Equimolar concentrations of the antigen–antibody complexes or their mixtures were incubated with 5–10 molar excess of 11-cis-retinal for 10–20 min in the dark at room temperature. UV/visible spectra were recorded by using the antigen–antibody mixtures as blanks before the addition of retinal (Materials and Methods). Pigment regeneration from bleached rhodopsin in the absence of antibody (trace 1), in the presence of anti-bacteriorhodopsin antibody A5 (trace 2), and in the presence of TM7C (trace 3) is shown. UV/visible spectrum of 1:1:1 mixture of rhodopsin, TM7C, and A5 was incubated overnight in the dark at 4°C (trace 4).
DISCUSSION
An early search for light-induced structural changes on the cytoplasmic surface of rhodopsin has shown a small but significant increase in the susceptibility of this region to proteolytic cleavage. This sensitivity to macromolecular probes has been observed to increase at the Meta II stage of rhodopsin activation and is reversed in a time-dependent manner (40). These findings imply that structural rearrangements in the retinal-binding site are propagated into changes at the periphery of the molecule via coordinated movement of the TM helices. This conclusion also is supported by earlier observations on photoinduced conformational transitions in spin-labeled, solubilized rhodopsin (41), as elaborated more recently by using spin-labeled single and double cysteine mutants of rhodopsin (14–21). The prime objective of the present study was to further analyze these light-induced conformational changes in rhodopsin by using mAbs and to examine their dynamics and possible involvement in receptor folding and functioning.
A host of mAbs directed against different regions of rhodopsin have been isolated and characterized (35, 42–45). Our interest in the present study was stimulated by the fact that certain mAbs that fail to interact with rhodopsin in the dark recognize the photoreceptor upon light activation (46). In the experiments reported in this paper, one such mAb (TM7C) has been used to monitor conformational changes in photoactivated rhodopsin. The reported specificities of mAbs generated in different laboratories suggest three regions of rhodopsin as major immunogenic sites: the amino-terminal, the carboxyl-terminal, and the loop connecting TM2 and TM3 (see Fig. 1). The TM7C antibody displays high specificity toward the peptide fragment 304–311 (Fig. 2), which extends from the center of TM7 to its cytoplasmic end and incorporates a part of the highly conserved NPXXY motif. The immunogenicity of this fragment is consistent with earlier observations (42, 45) demonstrating the remarkable potential of this region to generate an immune response comparable to that of other major immunogenic sites.
Detection of light-induced TM7C binding and a demonstrated correlation of this binding with Meta II formation (Fig. 3) raise the possibility that the observed changes on TM7 actually reflect a shift in the positions of TM3 and/or TM6 that occur upon photoactivation of rhodopsin (15, 20). This explanation would be consistent with the results of an earlier electron paramagnetic resonance study (14) demonstrating little or no light-induced conformational change at Cys-316 in spin-labeled rhodopsin in the disk membrane. Although the TM7C antibody shows high specificity toward Meta II, it practically fails to interact with Meta III (Fig. 4). The small increase in TM7C binding to Meta III compared with dark-adapted rhodopsin and A5 antibody binding can be attributed to residual Meta II or other photoproducts of rhodopsin bleaching. These results are consistent with those obtained by kinetic analysis of Fourier-transform infrared difference spectra (47, 48) and time-resolved electron paramagnetic resonance spectra (14) showing that the Meta II → Meta III transition is accompanied by protein refolding to a conformational state very similar, but not identical, to that of native rhodopsin. Thus, it seems that the reversal of the conformational changes detected by these techniques is part of the same process that exposes the TM7C epitope at Meta II and buries it at Meta III.
The lack of pigment regeneration from Meta II in the presence of TM7C (Fig. 5) suggests an important role for the epitope in “shaping up” the appropriate conformation critical for rhodopsin folding. Given the importance of the highly conserved NPXXY motif in the function of several biogenic amine and peptide receptors (24–26), light-induced exposure of this epitope may be relevant to a general mechanism for receptor activation. As indicated above, whether the accessibility of the distal portion of TM7 to the antibody reflects light-induced movement of TM3 and/or TM6, or is a separate and independent process, deserves further study. It is conceivable that light-induced displacement of the TM helices exposes a small portion of the epitope that is sufficient to provide for the loose binding of TM7C. High-affinity binding resulting in tight antigen–antibody complex formation may trigger a further amplification of the structural changes that account for the observed acceleration in retinyl-Schiff base hydrolysis. Elucidation of this light-induced accessibility change mechanism and its functional importance is the goal of future work.
Acknowledgments
This work was supported by Grant EY11112 from the National Eye Institute, National Institutes of Health. N.G.A. acknowledges the generous hospitality of many colleagues at the Max Planck Institute for Biophysics, where a portion of this work was performed under an Alexander von Humboldt Research Prize.
ABBREVIATIONS
- TM
transmembrane
- Meta II
metarhodopsin II
- Meta III
metarhodopsin III
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1. Ovchinnikov Y A, Abdulaev N G, Feigina M Y, Artamonov I D, Zolotarev A S, Kostina M B, Bogachuk A S, Miroshnikov A I, Martinov V I, Kudelin A B. Bioorg Khim. 1982;8:1011–1014. [Google Scholar]
- 2.Nathans J, Hogness D. Cell. 1982;34:807–814. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- 3.Keirns J J, Miki N, Bitensky M W, Keirns M. Biochemistry. 1975;14:2760–2772. doi: 10.1021/bi00683a032. [DOI] [PubMed] [Google Scholar]
- 4.Fesenko E E, Kolesnikov S S, Lyubarsky A L. Nature (London) 1985;313:310–313. doi: 10.1038/313310a0. [DOI] [PubMed] [Google Scholar]
- 5.Wheeler G, Bitensky M W. Proc Natl Acad Sci USA. 1977;74:4238–4242. doi: 10.1073/pnas.74.10.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hargrave P A, McDowell J H. FASEB J. 1992;6:2323–2331. doi: 10.1096/fasebj.6.6.1544542. [DOI] [PubMed] [Google Scholar]
- 7.Pugh E N, Lamb T D. Biochim Biophys Acta. 1993;1141:111–149. doi: 10.1016/0005-2728(93)90038-h. [DOI] [PubMed] [Google Scholar]
- 8.Sakmar T P, Franke R R, Khorana H G. Proc Natl Acad Sci USA. 1989;96:8309–8312. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhukovsky E A, Oprian D D. Science. 1989;246:928–930. doi: 10.1126/science.2573154. [DOI] [PubMed] [Google Scholar]
- 10.Nathans J. Biochemistry. 1990;29:937–942. doi: 10.1021/bi00456a013. [DOI] [PubMed] [Google Scholar]
- 11.Yoshizawa T, Wald G. Nature (London) 1963;197:1279–1286. doi: 10.1038/1971279a0. [DOI] [PubMed] [Google Scholar]
- 12.Sheih T, Han M, Sakmar T P, Smith S O. J Mol Biol. 1997;269:373–384. doi: 10.1006/jmbi.1997.1035. [DOI] [PubMed] [Google Scholar]
- 13.Jager S, Lewis J W, Zvyaga T A, Szundi I, Sakmar T P, Kliger D S. Proc Natl Acad Sci USA. 1997;94:8557–8562. doi: 10.1073/pnas.94.16.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farahbakhsh Z T, Hideg K, Hubbell W L. Science. 1993;262:1416–1419. doi: 10.1126/science.8248781. [DOI] [PubMed] [Google Scholar]
- 15.Resek J F, Farahbakhsh Z T, Hubbell W L, Khorana H G. Biochemistry. 1993;32:12025–12031. doi: 10.1021/bi00096a012. [DOI] [PubMed] [Google Scholar]
- 16.Farahbakhsh Z T, Ridge K D, Khorana H G, Hubbell W L. Biochemistry. 1995;34:8812–8819. doi: 10.1021/bi00027a033. [DOI] [PubMed] [Google Scholar]
- 17.Altenbach C, Yang K, Farrens D L, Khorana H G, Hubbell W L. Biochemistry. 1996;35:12470–12478. doi: 10.1021/bi960849l. [DOI] [PubMed] [Google Scholar]
- 18.Yang K, Farrens D L, Altenbach C, Farahbakhsh Z T, Hubbell W L, Khorana H G. Biochemistry. 1996;35:14040–14046. doi: 10.1021/bi962113u. [DOI] [PubMed] [Google Scholar]
- 19.Yang K, Farrens D L, Hubbell W L, Khorana H G. Biochemistry. 1996;35:12464–12469. doi: 10.1021/bi960848t. [DOI] [PubMed] [Google Scholar]
- 20.Cai K, Langen R, Hubbell W L, Khorana H G. Proc Natl Acad Sci USA. 1997;94:14267–14272. doi: 10.1073/pnas.94.26.14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J M, Altenbach C, Thurmond R L, Khorana H G, Hubbell W L. Proc Natl Acad Sci USA. 1997;94:14273–14278. doi: 10.1073/pnas.94.26.14273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheikh L, Zvyaga T A, Lichtarge O, Sakmar T P, Bourne H R. Nature (London) 1996;383:347–349. doi: 10.1038/383347a0. [DOI] [PubMed] [Google Scholar]
- 23.Farrens D L, Altenbach C, Yang K, Hubbell W L, Khorana H G. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 24.Barak L S, Menard L, Ferguson S S-G, Colapietro A-M, Caron M G. Biochemistry. 1995;34:15407–15414. doi: 10.1021/bi00047a003. [DOI] [PubMed] [Google Scholar]
- 25.Hunyady L, Bar M, Bauka A, Catt K. J Biol Chem. 1995;270:16602–16609. doi: 10.1074/jbc.270.28.16602. [DOI] [PubMed] [Google Scholar]
- 26.Bohm S K, Khitin L M, Smeekens S P, Grady E F, Payan D G, Bunnett N W. J Biol Chem. 1997;272:2363–2372. doi: 10.1074/jbc.272.4.2363. [DOI] [PubMed] [Google Scholar]
- 27.Pashkov V S, Balashova T A, Zhemaeva L V, Sikilinda N N, Kutuzov M A, Abdulaev N G, Arseniev A S. FEBS Lett. 1996;381:119–122. doi: 10.1016/0014-5793(96)00094-4. [DOI] [PubMed] [Google Scholar]
- 28.Ridge K D, Lee S S J, Abdulaev N G. J Biol Chem. 1996;271:7860–7867. doi: 10.1074/jbc.271.13.7860. [DOI] [PubMed] [Google Scholar]
- 29.Wilden U, Kuhn H. Biochemistry. 1982;21:3014–3022. doi: 10.1021/bi00541a032. [DOI] [PubMed] [Google Scholar]
- 30.Smith H G, Stubbs G W, Litman B J. Exp Eye Res. 1975;20:211–217. doi: 10.1016/0014-4835(75)90134-7. [DOI] [PubMed] [Google Scholar]
- 31.Kohler G, Milstein C. Nature (London) 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 32.Blatty J D, Blatty B, Vlahos W. Immunol Methods. 1987;100:173–179. doi: 10.1016/0022-1759(87)90187-6. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli U K. Nature (London) 1970;270:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Matsudaira P. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 35.Molday R S, McKenzie D. Biochemistry. 1983;22:653–660. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- 36.Martynov V I, Kostina M B, Feigina M Y, Miroshnikov A I. Bioorg Khim. 1983;9:934–945. [PubMed] [Google Scholar]
- 37.Ridge K D, Lee S S J, Yao L L. Proc Natl Acad Sci USA. 1995;92:3204–3208. doi: 10.1073/pnas.92.8.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freychet P, Roth J, Neville J M. Biochem Biophys Res Commun. 1971;43:400–408. doi: 10.1016/0006-291x(71)90767-4. [DOI] [PubMed] [Google Scholar]
- 39.Ridge K D, Lu Z, Liu X, Khorana H G. Biochemistry. 1995;34:3261–3267. doi: 10.1021/bi00010a016. [DOI] [PubMed] [Google Scholar]
- 40.Kuhn H, Mommertz O, Hargrave P. Biochim Biophys Acta. 1982;679:95–100. [Google Scholar]
- 41.Kalamkarov G R, Grigorian G L, Fedorovich I B, Krylova V I, Ostrovskii M A. Dokl Akad Nauk SSSR. 1976;231:736–738. [PubMed] [Google Scholar]
- 42.Ovchinnikov Y A, Eganyan E R, Abdulaev N G. Biol Membr (Russia) 1988;3:1189–1196. [Google Scholar]
- 43.Molday R S. Prog Ret Res. 1986;8:173–203. [Google Scholar]
- 44.Adamus G, Zam S, Arendt A, Palczewski K, McDowell J H, Hargrave P A. Vision Res. 1991;31:17–31. doi: 10.1016/0042-6989(91)90069-h. [DOI] [PubMed] [Google Scholar]
- 45.Adamus G, Arendt A, Hargrave P A. J Neuroimmunol. 1991;34:89–97. doi: 10.1016/0165-5728(91)90118-q. [DOI] [PubMed] [Google Scholar]
- 46.Ovchinnikov Y A. Photochem Photobiol. 1987;45:909–914. doi: 10.1111/j.1751-1097.1987.tb07902.x. [DOI] [PubMed] [Google Scholar]
- 47.Rothschild E J, Gillespie J, DeGrip W J. Biophys J. 1997;51:345–350. doi: 10.1016/S0006-3495(87)83341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klinger A L, Braiman M S. Biophys J. 1992;63:1244–1255. doi: 10.1016/S0006-3495(92)81700-9. [DOI] [PMC free article] [PubMed] [Google Scholar]