Abstract
The immediate early (IE) proteins of human cytomegalovirus (hCMV) have diverse roles in directing viral and host cell transcription. Among these is the ability of IE2 to induce transcription of the IL1B gene that codes for IL-1β in monocytes. This function is partially explained by interaction between IE2 and the host cell transcription factor Spi-1/PU.1 (Spi-1). We now show that maximal IE2 function also depends on productive interactions localizing to two C/EBP sites on the IL1B promoter suggesting either bi- or tri-molecular interactions between IE2, Spi-1 and C/EBPβ at two different locations on the promoter. The IE2 interaction region on Spi-1 was previously mapped to the DNA-binding ETS domain and overlaps the region of Spi-1 that interacts with the transcription factor C/EBPβ, a factor known to be critical for the induction of IL1B in response to Toll/IL-1 receptor (TIR) family signal transduction. The Spi-1 interacting region of IE2 maps to amino acids 315–328, a sequence that also interacts with the bZIP domain of C/EBPβ. An expression vector coding for amino acids 291–364 of IE2 can suppress LPS induction of a cotransfected IL1B enhancer-promoter fragment in a monocyte cell line. This inhibition is likely the result of competition between Spi-1 and C/EBPβ, thus blunting gene induction.
Keywords: Interleukin 1β, Cytomegalovirus, IE2, Transcription factors, peptide inhibitor
1 Introduction
Interleukin 1β (IL-1β) is a potent cytokine with diverse roles in inflammation including induction of the acute phase response, activation of the cellular components of acquired immunity, and promotion of chronic inflammation (Auron, 1998). Monocytes express large quantities of IL-1β via induction of its gene (IL1B) after contact with a variety of exogenous and endogenous stimulants typically associated with infection or injury (Fig 1a). LPS, a classic inducer of IL1B, is a gram negative bacterial cell wall component that binds to Toll-like receptor 4 (TLR4) on monocytes (O’Neill and Dinarello, 2000). The TLR/IL-1β system appears to have evolved under selective pressure as an early warning system to allow defensive maneuvers by the host’s immune system in order to combat infection.
Fig 1. Regulation of IL1B expression.
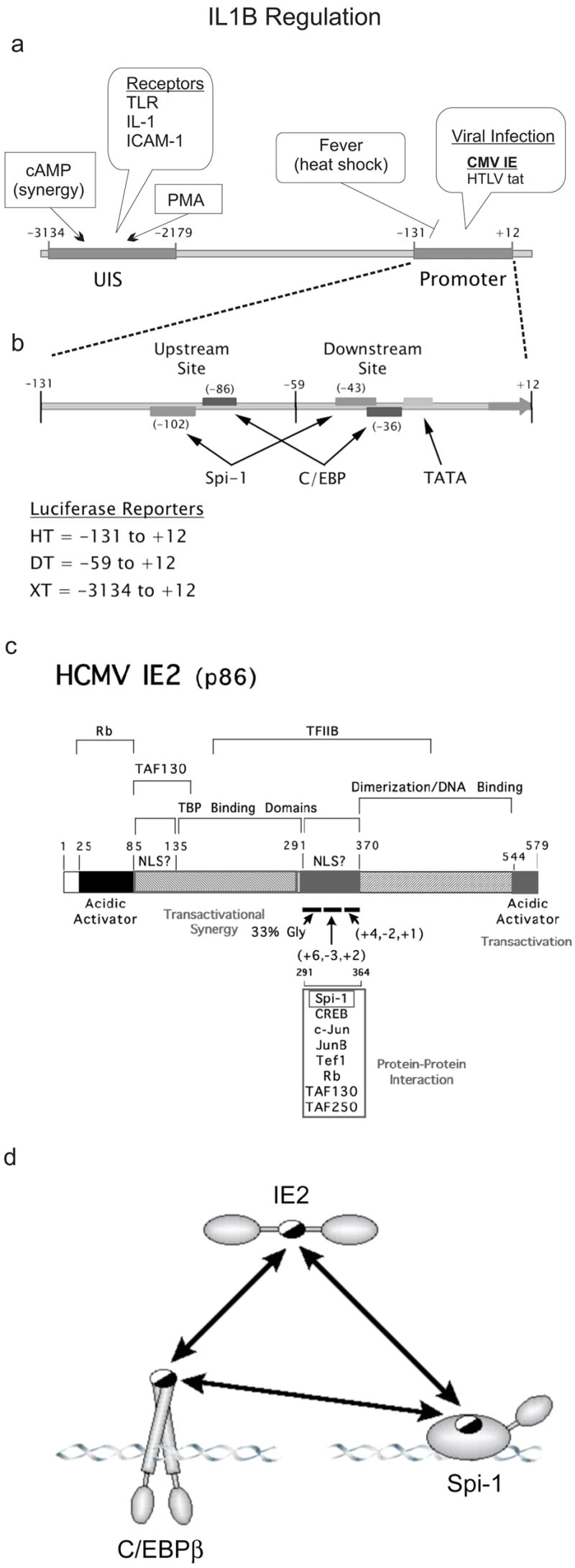
a) Schematic of IL1B regulatory region. An upstream induction sequence (UIS) is located at −3134 to− 2179. This region is acted on by PMA and cAMP and receptor engagement by TLR, IL-1, and ICAM-1 (Koyama et al., 1996; Shirakawa et al., 1993; Tsukada et al., 1994). The promoter sequence, −131 to +12 (HT) is stimulated by the viral transactivators CMV IE and HTLV tax (Tsukada et al., 1997; Wara-aswapati et al., 1999). The promoter also contains a heat shock factor repressor site (Cahill et al., 1996). b) Exploded diagram of the IL1B promoter region. The relative Spi-1, C/EBP and TATA sites are noted. Also depicted are the IL1B sequences used for the luciferase reporters (HT, DT, XT). c) Schematic of IE2 protein. Shown are three independent TBP-binding domains and two possible NLS sites. Also shown are several proteins known to interact at the central portion of the molecule as well as the general amino acid composition (gly-rich and charge) of the amino, central, and carboxy regions of the 291–364 peptide. d) Model of tri-molecular interactions between IE2, Spi-1 and C/EBPβ. Mapping experiments from previous reports have indicated that amino acid sequence 291–364 of IE2 interacts with the DBD of Spi-1 over a region that includes one of two arginines required for DNA binding. This arginine is critical for interaction with the last 15 amino acids of the C/EBPβ bZIP domain (c-terminus) (Listman et al.). This c-terminus also interacts with IE2. Half-filled ovals denote mutual interactions sites.
LPS induction of IL1B requires interaction between two independent elements, a promoter that contains a TATA box and an upstream LPS-responsive enhancer, also known as the upstream inducible sequence (UIS) (Fig 1a) (Auron and Webb, 1994). The UIS functions to promote transcription most strongly when bound by a heterodimeric complex composed of C/EBPβ and either CREB or ATF1 (Chandra et al., 1995; Tsukada et al., 1994), both members of the bZIP family of transcription factors. The IL1B promoter requires binding by Spi-1/PU.1 (Spi-1), a winged-helix-turn-helix protein primarily restricted to myeloid lineage cells (Klemsz and Maki, 1996; Pahl et al., 1993; Paul et al., 1991; Ray et al., 1990) and C/EBPβ, a bZIP protein with broader tissue expression (Lekstrom-Himes and Xanthopoulos, 1998). Spi-1 is necessary, but insufficient for strong IL1B transcription in monocytes (Kominato et al., 1995). Gene induction depends upon a mutual interaction between Spi-1 and C/EBPβ that is supported by binding to sites on the IL1B promoter (Fig 1b) (Listman et al., 2005; Wara-aswapati et al., 1999; Yang et al., 2000). A NF-κB site is also present upstream of the core promoter (Hiscott et al., 1993). Although NF-κB is important for maximum IL1B gene expression, it is not the only factor responsible for induction events (Baldassare et al., 1999; Park et al., 2005; Tsukada et al., 1994). The promoter functionally couples to a far-upstream enhancer via a mechanism that likely involves a cooperative long-range interaction between Spi-1 and C/EBPβ (Kominato et al., 1995; Listman et al., 2005; Yang et al., 2000). Chromatin remodeling has also been implicated in IL1B regulation as others have shown an open promoter conformation in myeloid cell lines (Liang et al., 2006).
A variety of stimuli besides LPS can induce IL1B gene expression in monocytes. Among them is the human cytomegalovirus (hCMV) (Crump et al., 1992; Dudding et al., 1989; Iwamoto et al., 1990). In contrast to the rapid and transient response to LPS stimulation (lasting several hours) (Fenton et al., 1987), human peripheral blood monocytes infected with hCMV display a rapid but sustained level of IL1B expression that is measured in days (Dudding et al., 1989). Two mechanisms can explain this response. The first mechanism is mediated by the binding of the viral coat proteins bG and gH to the monocyte TLR2 that leads to NF-κB, and likely C/EBPβ, activation via p38 dependent phosphorylation events (Baldassare et al., 1999; Compton, 2004; Yurochko and Huang, 1999). This is presumably a transient response that parallels the monocytic response to other invading organisms. The second mechanism likely relies upon direct transactivation of the IL1B promoter by the immediate-early (IE) proteins encoded within the viral genome (Mocarski, 1996; Stinski et al., 1982). The IE locus produces several splice products including IE1 and IE2 (Mocarski, 1996). Expression of this locus in transient transfection assays of THP-1 cells results in potent activation of an IL1B promoter reporter at levels comparable to those seen with an enhancer/promoter reporter (Iwamoto et al., 1990; Wara-aswapati et al., 1999). IE1 appears to have a more permissive role and at higher levels of expression, inhibits transactivation (Yang et al., 2002). The larger IE2 isoform was shown to provide a powerful transactivation signal, even in the absence of an enhancer, that was dependant on a direct interaction with the DNA binding domain (DBD) of Spi-1 (Wara-aswapati et al., 1999). This same domain also supports a cooperative interaction with a few other proteins including IRF-4 and C/EBPβ (Escalante et al., 2002; Listman et al., 2005), highlighting its importance for functions other than DNA binding. The molecular basis of these cooperative protein interactions might be manipulated to inhibit gene transcription making them important targets for study.
The molecular details of the IE2 interaction with Spi-1 are only partially understood. We previously mapped the IE2 interaction to the ETS domain (DBD) of Spi-1 in biochemical assays and the complementary binding site on IE2 was inferred to be located between residues 291–364, because deletion constructs of IE2 missing this sequence resulted in loss of IE2 transactivation potential in a gene reporter assay (Wara-aswapati et al., 1999). This report demonstrated that the deleted sequence is necessary, but did not determine whether it is sufficient for interaction with Spi-1. This same region was shown by others to contain one of two nuclear localization sequences (NLS), one of three TBP binding regions, and is required for IE2 interaction with at least seven other transcription factors (Fig 1c) (Ahn et al., 1998; Caswell et al., 1993; Fortunato et al., 1997; Jupp et al., 1993; Kim et al., 2000; Lukac et al., 1997; Lukac et al., 1994; Pizzorno et al., 1991; Scully et al., 1995). Recently, we reported the existence of a mutually exclusive three-way interaction involving associations between any two of the proteins: IE2 ; C/EBPβ; and Spi-1 (Fig 1d) (Listman et al., 2005). Therefore, we hypothesized that IE2 functions by tethering to either Spi-1 or C/EBPβ bound to their cognate sites on IL1B. The TADs of C/EBPβ and IE2 can each strongly transactivate the low-level TAD activity of Spi-1 (Wara-aswapati et al., 1999; Yang et al., 2000). However, the functional relevance of the IE2–C/EBPβ interaction is unknown. Consequently, we further hypothesized that expression of TAD-defective IE2 should repress IL1B transcription by competing with both C/EBPβ and wt IE2.
Herein, we investigate the mechanism of IE2 function by refining a map of the Spi-1 and C/EBPβ interaction sites, revealing that as few as 14 amino acids of the original 74 are necessary for association with both Spi-1 and C/EBPβ. This sequence encodes one of the two NLS for IE2 and is surprising because of our recent determination that the NLS for Spi-1 may include amino acid residues critical for DNA binding and support interaction with the bZIP domain of C/EBPβ and IE2 (Listman et al., 2005). These results have implications for the development of novel anti-inflammatory agents and the growing recognition of the multifunctional roles played by nuclear localization sequences.
2. Methods and Materials
2.1. Cell Culture
HeLa S3 cells (gift of Phillip A. Sharp) were cultured as previously described (Kominato et al., 1995). Raw 264.7 cells were obtained from the American Type Culture Collection (ATCC). The cells were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% heat inactivated FBS (Hyclone), 0.5% penicillin-streptomycin and 2mM L-Glutamine (Cellgro). For LPS stimulation, RAW 264.7 cells were treated with E. coli serotype 055:B5 LPS (Sigma) dissolved in endotoxin free PBS at a concentration of 10μg/ml.
2.2. Reporter Constructs and Expression Vectors
The wild type human IL1B promoter-luciferase reporters (−131/+12 and –59/+12) were constructed previously (Wara-aswapati et al., 1999). The mutant promoters were generated by PCR and inserted into pGL3-Basic vector (Promega) using primers 5’cccctttcctttaacttgattgggacctcaggtattcaac3’ and 5’gttgaatacctgaggtcccaatcaagttaaaggaaagggg3’ for downstream mutant and primers 5’ctcagcctcctacaactgcttttaaccactataaaaacagag3’ and 5’cgctgtttttatagtggttaaaagcagaagtaggaggctgag3’ for the upstream mutant. The IE expression vectors pEQ276, pEQ273 and pEQ326, gifts from Adam Geballe (Fred Hutchinson Cancer Research Center, Seattle, WA), contain hCMV IE1+2, IE1, and IE2 genomic DNA, respectively. The plasmids expressing full-length C/EBPβ and a truncated version (C/EBPβ SPl), which has an internal deletion between the two SplI restriction sites, were constructed previously in expression vectors pcDNA3.1 or pcDNA1 (Invitrogen) (Tsukada et al., 1994). bZIP pcDNA3.1 expression vector (containing aa 269–345 of C/EBPβ) was constructed by PCR (Yang et al., 2000). The construction of the murine Spi 1 pRC/CMV expression vector was previously described (Galson et al., 1993). Spi1 cDNA was also PCR amplified and inserted into pGEX2T (Amersham) for use in GST pull down assays (Wara-aswapati et al., 1999). Also for GST pull down assays, hCMV IE1 and IE2 cDNA was previously constructed within pGEX 3X (Klucher et al., 1993). IE2 291–364delA pGEX 2T was PCR amplified using four primers: an internal pair to create a 14 aa deletion 5’ctgcggccatcagagcagcatctccgagttggacaacg3’ and 5’cgttgtccaactcggagatgctgctctgatggccgcag3’, and external primers: 5’ctcctcggatccagccaccatgggcgcggcgg3’, and 5’ctcgaattcctcgtcaatcttgacgcgaccccg3’. IE2 291–343 and IE2 291–364 pGEX 2T were PCR amplified using primers 5’ctcctcggatccagccaccatgggcgcggcgg3’, and 5’ggaattcatctttcatgatattgcgcaccttctc3’, and primers 5’ctcctcggatccagccaccatgggcgcggcgg3’, and 5’ctcgaattcctcgtcaatcttgacgcgaccccg3’, respectively. The IE2 291–364 pCDNA3.1V5/His vector was constructed by PCR using primers: 5’ctcctcggatccgccaccatgagccaccatgggcgcggcgg3’ and 5’ctcgaattcctcgtcaatcttgacgcgaccccg3’. The IE2 291–264 pEGFPC3 expression vector was constructed by ligation cloning using restriction enzymes Hind III and Eco RI from a pCDNA3.1V5/HisA intermediate vector constructed with primers 5’ctcctcggatccagccaccatgggcgcggcgg3’ and 5’ctcgaattcctcgtcaatcttgacgcgaccccg3’.
2.3. Transfections and Reporter Assays
HeLa S3 cells were transfected using Effectene (Qiagen) according to the manufacturer’s instructions and as previously described (Listman et al., 2005). Raw 264.7 cells were transfected using Calcium Phosphate reagent from the Mammalian Transfection Kit (Stratagene) as described previously (Unlu et al., 2007). Total amount of transfected DNA per well was kept constant within each experiment by adding empty parental vectors. Twenty-four or forty-eight hours after transfection, cells were lysed in Cell Culture Lysis buffer (Promega) and luciferase activities were determined using the Luciferase Assay System (Promega). β-galactosidase activity was also measured and used for transfection efficiency normalization in HeLa experiments. Error bars represent the standard error of the mean for triplicate samples. Experiments were repeated a minimum of three times.
2.4. GST Pull Down Assays
GST fusion proteins were harvested from E. coli BL21pLysS (Promega) using previously described methods (Wara-aswapati et al., 1999). Briefly, cultures were induced with 0.5 mM IPTG for 3–4 hours, pellets were suspended in NETN buffer (20 mM Tris, 100 mM NaCl, 1mM EDTA, 0.005 % NP-40) with 1mM DTT, PefaBloc (Roche, Indianapolis, IN), and one CompleteTM Protease Inhibitor Cocktail (Roche) tablet/50 ml. Suspensions were sonicated on ice and supernatants containing fusion proteins were incubated overnight with pre-washed Glutathione–Sepharose beads at 4oC. After washing with NETN, beads were incubated with in vitro translated proteins prepared using TNT T7 Quick Coupled Reticulocyte Lysate System (Promega) and labeled with fresh 35S methionine (Amersham) according to the manufacturer’s instructions. After washes with NETN, beads were separated by SDS-PAGE, and analyzed for binding using autoradiography.
2.5. Western Blot
Lysates from transient transfections were separated by SDS-PAGE and transferred to a PVDF membrane (Millipore). The membranes were incubated with either anti-C/EBPβ antibody (sc-150 or sc-746 as indicated, Santa Cruz), anti-PU.1 (Spi-1) antibody (sc-352, Santa Cruz), or anti-IE antibody (MAB810, Chemicon) that reacts with the common amminoterminus of IE1 and IE2, followed by incubation with a horseradish peroxidase-labeled secondary antibody. Bands were detected by chemiluminescence (National Diagnostics).
3. Results
3.1. hCMV IE proteins cooperatively transactivate the IL1B promoter with Spi-1 and C/EBPβ
IE-dependent transactivation of an IL1B promoter (HT) luciferase reporter was evaluated following transfection into HeLa S3 cells. This reporter does not contain the IL1B upstream enhancer (UIS), but can exhibit constitutive activity under some conditions. HeLa S3 cells, unlike monocytes, do not require LPS to support IL1B promoter activity and are ideal for these experiments because the reporters have very low background expression in the absence of ectopic Spi-1 and C/EBPβ (Yang et al., 2000). The inability to test the UIS is not likely a major issue, since we have previously reported that the UIS is dispensable in monocytes for IE activation of the IL1B promoter (Wara-aswapati et al., 1999). An IE genomic expression vector coding for both IE1 and IE2 was co-transfected alone or in the indicated combinations with cDNA expression vectors encoding Spi-1 and C/EBPβ for 48 hours (Fig 2a). The IE expression vector alone resulted in several fold induction of the reporter at levels comparable to either Spi-1 or C/EBPβ alone (not readily apparent due to the y-axis scale). However, IE more actively induced the reporter when co-expressed with Spi-1 or C/EBPβ, to a level comparable with that of Spi-1 plus C/EBPβ in the absence of IE. Maximal activity resulted when all three molecules were co-expressed. The results were not confounded by IE effects on the C/EBPβ and Spi-1 expression vectors, because the cis-repression site is lacking in these vectors’ CMV promoter. Furthermore, Western blotting did not show enhancement of Spi-1 or C/EBPβ expression in samples containing IE (Fig 2b). Interestingly, the C/EBPβ antibody detects a pair of slightly higher molecular weight proteins that closely correspond to the more abundant forms found after transfection with the C/EBPβ expression vector. These bands may represent endogenous isoforms of C/EBP that are cross-reacting with the antibody.
Fig 2. Transactivation of the IL1B promoter by IE Protein.
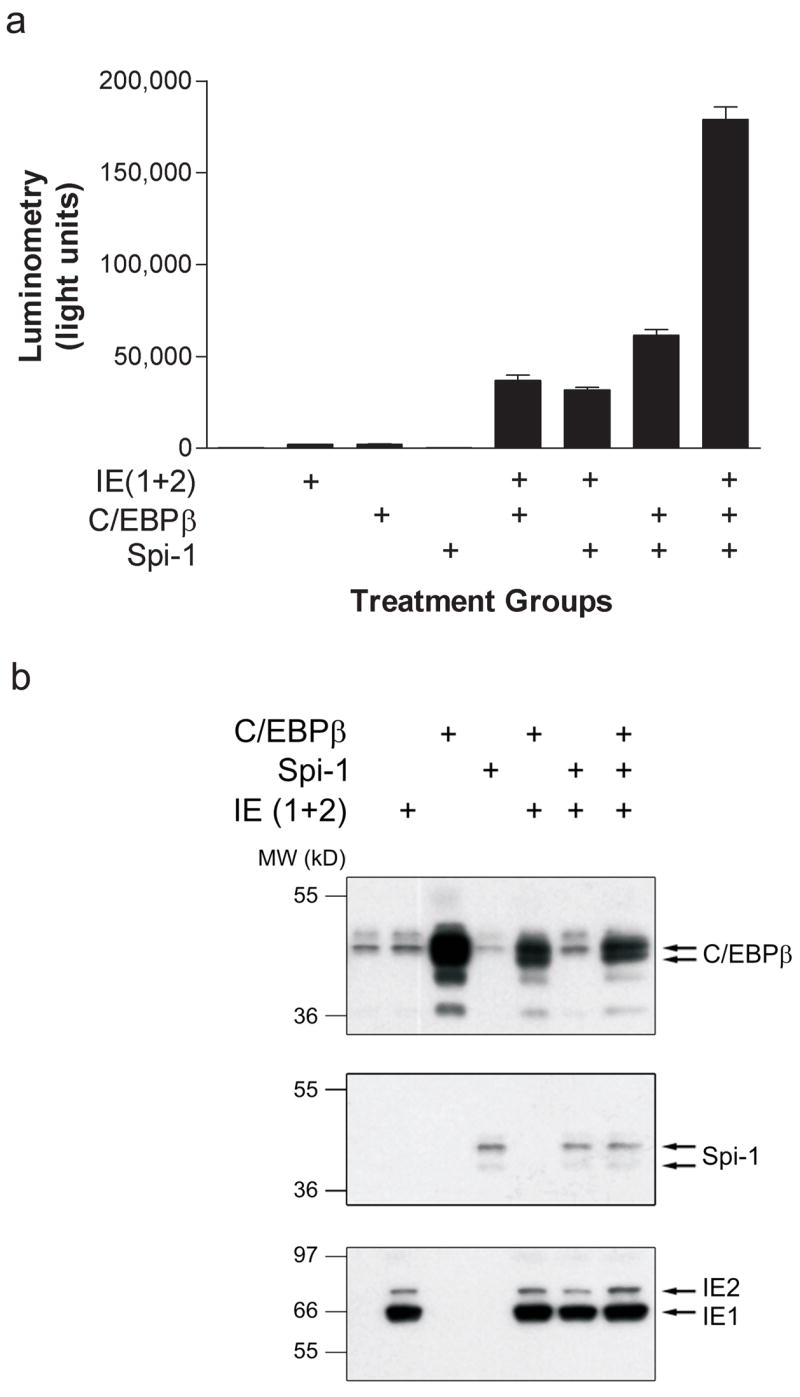
a) Hela S3 cells were transiently transfected with the IL1B luciferase reporter HT and co-transfected with expression vectors for C/EBPβ, Spi-1 and pEQ273, a plasmid that expresses both isoforms of IE (1+2). Cells were harvested at 48 hrs and cell lysates analyzed by luminometry. Results are displayed as mean light units ± SEM of triplicate samples and are representative of at least three experiments. b) Lysates from (a) were analyzed by Western blot using a rabbit polyclonal antibodies to C/EBPβ (sc-150, Santa Cruz Biotechnology), Spi-1 (Santa Cruz Biotechnology), and IE (Chemicon) as indicated. The IE antibody recognizes a common epitope at the N-terminus of IE1 and IE2. Secondary antibodies were labeled with horseradish peroxidase and detected using enhanced chemiluminescence.
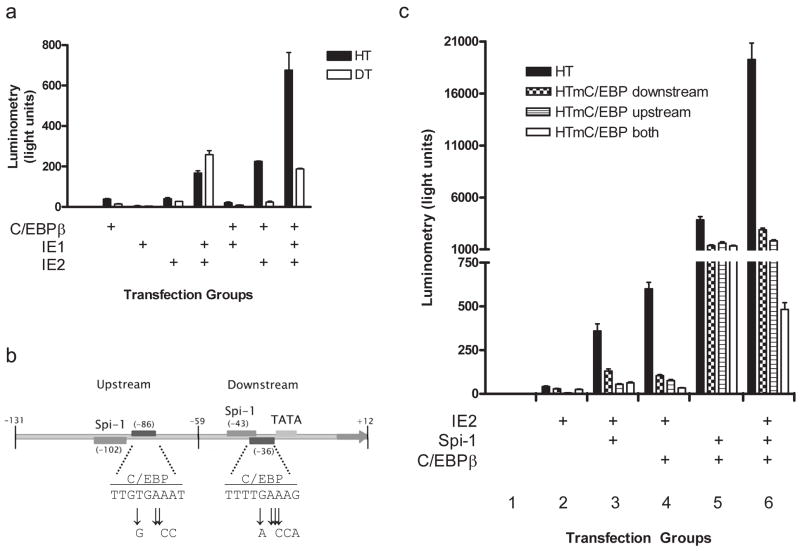
3.2. IE-C/EBPβ cooperative function has differential requirements for IE isoforms and C/EBP sites within the IL1B promoter
To investigate the nature of IE-C/EBPβ based effects on the IL1B promoter, we hypothesized that IE would functionally depend upon at least one of the two C/EBP sites. We hypothesized that the upstream C/EBP site was most logical because it binds C/EBPβ more avidly than the downstream C/EBP site in gel shift assays (unpublished observation). To test this, we compared the responsiveness of the HT and shorter DT (containing only the TATA proximal site) reporters to IE and C/EBPβ (Fig 3a). In contrast to above, where one vector expressed both IE isoforms, here we used vectors that express the individual isoforms. While DT did respond to IE1 + IE2 alone, the response was not augmented by the addition of C/EBPβ. However, the HT reporter did show a several fold induction when IE and C/EBPβ were cotransfected. These data also show that the cooperative interaction depends on IE2 because the IE1 isoform functioned only when IE2 was co-expressed. Clarification of the relative roles for each C/EBP site was further investigated by making mutations in base pairs previously shown to interfere with C/EBPβ binding (Kominato et al., 1995) (Fig 3b). IE2 again cooperatively functioned with Spi-1 or C/EBPβ using wild type HT, although, much more vigorously when both were co-expressed (Fig 3c, black bars in groups 3, 4, and 6, respectively). Coexpression of Spi-1 with C/EBPβ in the absence of IE2 yielded intermediate activity (Fig 3c, black bar in group 5). In contrast, ablation of C/EBPβ binding at either site reduced cooperative function with Spi-1 and C/EBPβ by approximately 50% and there was no additive effect by simultaneous mutations at both sites (Fig 3c, group 5). However, when IE2 was introduced, there was a more dramatic reduction in reporter activity (much greater than 50%) and this effect was additive when both sites were simultaneously mutated and C/EBPβ was coexpressed (Fig 3c, groups 4, and 6). Unexpectedly, a similar pattern was observed in cells co-transfected with Spi-1 and IE2, in the absence of co-transfected C/EBPβ (Fig 3c, group 3). This may be due to the low level of C/EBP expression in HeLa S3 cells, as revealed by the Western blot data above.
Fig 3. Promoter analysis of IE function.
a) Effects of C/EBPβ and the individual isoforms of IE on HT and DT reporters. Transfections were carried out in HeLa S3 cells as described in Fig 2 with the exception that Spi-1 expression vector was not used and the IE (1+2) vector (pEQ276) was substituted by vectors that express either IE1 (pEQ276) or IE2 (pEQ326), as indicated. Results are displayed as mean light units ± SEM of triplicate samples and are representative of at least three experiments. b) Schematic of mutations made at C/EBP sites in HT reporters used in (c). c) Gene reporter analysis of mutated CEBP sites on IL1B promoter. HeLa S3 cells were transiently transfected with mutant or wt IL1B promoter reporters and cotransfected with IE2 (pEQ326), C/EBPβ, and Spi-1 in six groups, as indicated. Results are displayed as mean light units ± SEM of triplicate samples and are representative of at least three experiments.
3.3. IE2 physically interacts with C/EBPβ and replaces the function of a portion of the C/EBPβ transactivation domain (TAD)
A physical interaction between C/EBPβ and IE2 could account for the IE function observed. We tested the capacity of the C/EBPβ bZIP domain to physically interact with either IE1 or IE2 in a GST pull-down assay because we previously reported (Listman et al., 2005) that this domain was crucial for Spi-1–C/EBPβ cooperativity (Fig 4a). The radiolabled bZIP probe revealed binding to IE2 and the Spi-1 control, but not to IE1 or the GST control. We had previously shown that the cooperativity between Spi-1 and IE on the IL1B promoter did not absolutely require the Spi-1 TAD. To determine if the same is true for C/EBPβ, a deletion construct missing sequence coding for amino acids 41–245 ( spl) that was previously reported to be essential for IL1B enhancer activity (Tsukada et al., 1994) was tested in the reporter assay. This deletion removes most of the functional C/EBPβ TAD but retains the N-terminus important for chromatin remodeling in some systems (Fig 4b) (Kowenz-Leutz and Leutz, 1999). Surprisingly, the absence of a majority of the C/EBPβ TAD had no effect on the synergistic activation of the HT reporter by IE, while the C/EBPβ bZIP domain (amino acids 259–345) was insufficient for synergy (Fig 4c).
Fig 4. Biochemical and structural requirements of IE and C/EBPβ function.
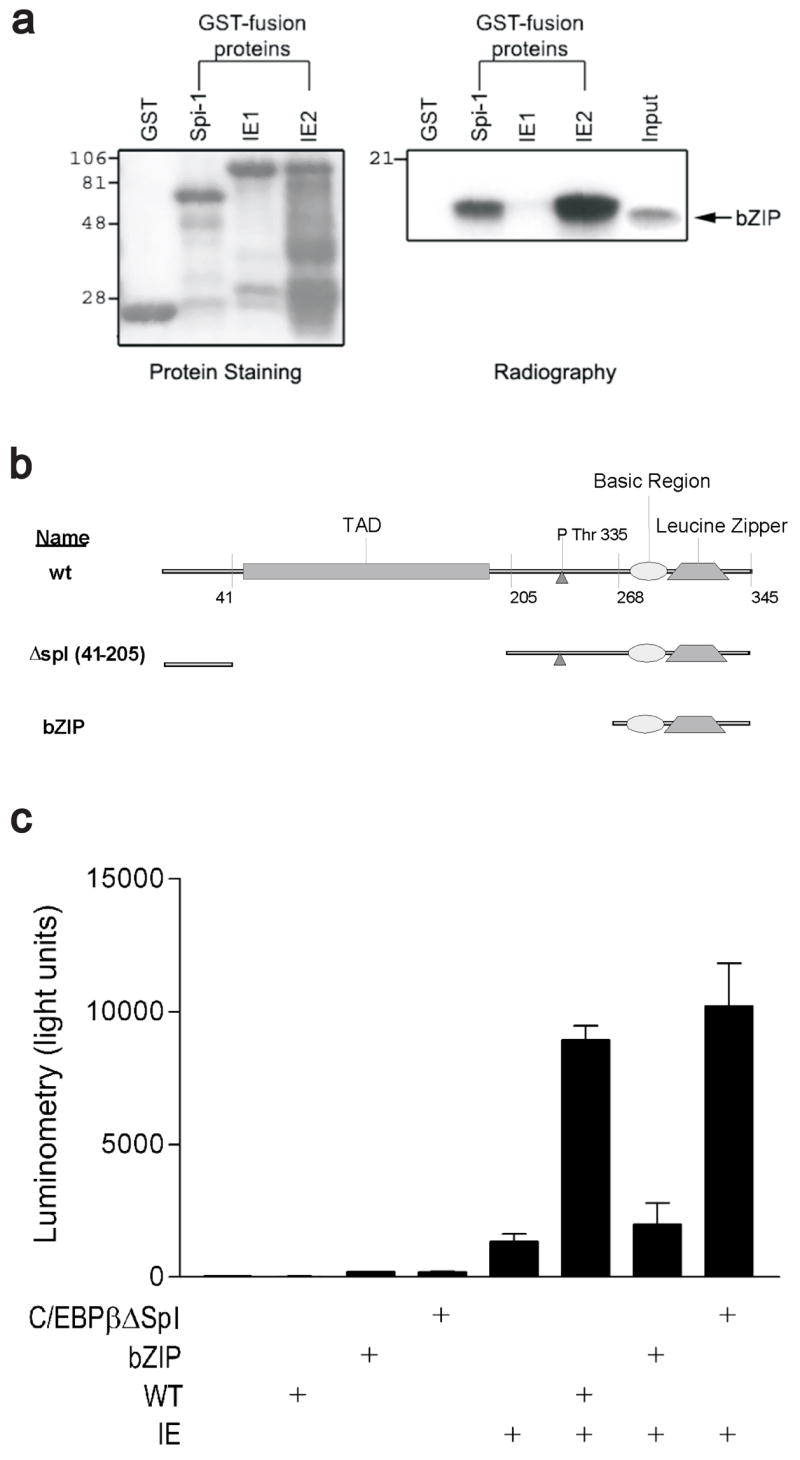
a) Capacity of IE1 and IE2 to interact with C/EBPβ was determined using a GST pulldown assay. The indicated GST fusion products were incubated with an in vitro translated radiolabeled probe of the C/EBPβ bZIP domain and washed to remove nonspecific binding. The retained protein complexes were isolated by denaturing PAGE. The GST fusion proteins were resolved by staining (left panel) and bound probe detected by autoradiography (right panel). b) Schematic of structure/function relationships in primary sequence of C/EBPβ and regions deleted for functional analysis in (c). c) Analysis of C/EBPβ deletion constructs shown in (b). Transient transfection in HeLa S3 showing functional activation of the HT reporter by IE with indicated WT or truncated forms of C/EBPβ. Results are displayed as mean light units ± SEM of triplicate samples and are representative of at least three experiments.
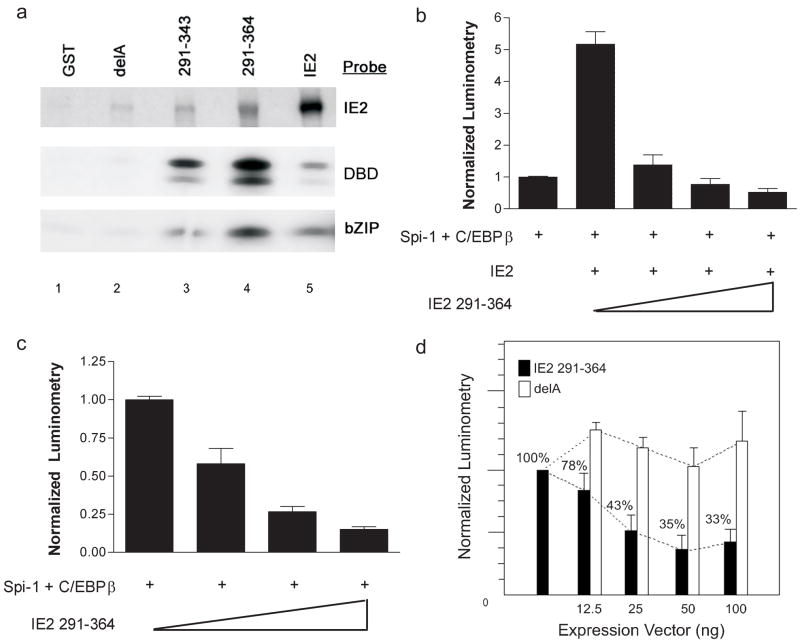
3.4. An internal fragment of IE2 binds to Spi-1 DBD and C/EBPβ bZIP domains and possesses dominant negative function
Previous works suggested that the IE2 sequence 291–364 was a critical determinant for a cooperative protein interaction on the IL1B gene (Wara-aswapati et al., 1999) (Listman et al., 2005). This notion was first tested by using a GST – IE2 291–364 fusion protein in a pulldown assay to evaluate the capacity of this sequence to interact with the DBD’s of Spi-1 and C/EBPβ (Fig 5a). The results showed that IE2 self-associates, as previously reported (Chiou et al., 1993), as well as effectively interacts with the DBD of both Spi-1 and C/EBPβ (lane 5). In contrast with the full-length wt IE2, the GST 291–364 peptide interacted more strongly with the DBDs than with wt IE2 , suggesting a distinct contact geometry (lane 4). Two other sub-fragments with deletions of the more highly charged regions of IE 291–364 were also tested in this assay. GST 291–343 peptide (deletion of last 20 residues) still retained binding, though less well when compared to the longer GST 291–364 peptide (lane 3). A fragment (delA) containing an internal deletion of residues 315–328, and a spuriously introduced deletion at residues 295–308 along with a C309R substitution, revealed minimal interaction of the fragment to all three probes (lane 2). The binding specificity was localized to the intended deletion sequence 315–328 after another peptide (sequence 310–364) missing the unintended deletion retained interaction with Spi-1 and C/EBPβ in our GST pulldown (not shown).
Fig 5. Structural and functional analysis of IE2.
a) IE2 peptide fragments (indicated across the top of panel) were expressed as GST fusion products and binding to the indicated probes carried out in a GST pulldown assay. Construct labeled delA encompasses IE2 sequences 291–364 but contains a deletion of sequences 315–328. Probes labeled DBD = Spi-1 ETS domain and bZIP = C/EBPβ bZIP domain. b) Functional analysis of peptide fragment IE2 291–364 in IL1B gene reporter assay using transient transfection of HeLa S3 cells. Increasing amounts of IE2 291–364 expression vector were added to constant amounts of Spi-1, C/EBPβ and IE2 as indicated. Luminometry was normalized to HT reporter activity when co-transfected with Spi-1 + C/EBPβ alone. c) Similar to (b) except full length IE2 expression vector was not used in the transfections. d) Inhibitory capacity of the IE2 291–364 peptide was tested using the full-length IL1B reporter XT in transient transfections of RAW 264.7 cells. Increasing amounts of expression vector for IE2 291–364 or the delA construct described in (a) were cotransfected with the XT reporter using CaPO4 precipitation in RAW 264.7 cells. After overnight incubations, the cells were stimulated by LPS (10μg/ml) for 6 hours and cells harvested for luminometry. The data were normalized to the response of empty vector and replicated three times before combining the results to yield the above graph. Results shown represent the mean percentage of normalized value ± SEM.
Co-transfecting an IE2 291–364 His-tagged expression vector with C/EBPβ and Spi-1 resulted in a dose-dependent decrease in reporter activity in both the presence (Fig 5b) and absence (Fig 5c) of full-length IE2. The reporter was tested in the absence of IE2, because our previous studies (Listman et al., 2005) argue for an overlap between IE2 and C/EBPβ binding to Spi-1. The fact that both C/EBP and IE2-dependent activities are inhibited supports this model. In order to test the inhibitory capabilities of the IE2 peptides on IL1B gene expression in the presence of the UIS, a complete monocyte-specific 3.8 kb regulatory region of the IL1B gene containing both the LPS-responsive enhancer and the promoter (Shirakawa et al., 1993; Unlu et al., 2007) was transfected as a luciferase reporter into LPS-treated RAW264.7 monocyte cells (Fig 5d). This cell line expresses abundant levels of Spi-1 (Kominato et al., 1995) and C/EBPβ and has been demonstrated to support UIS function and respond to LPS induction of both endogenous and transfected IL1B genes (Shirakawa et al., 1993) (Tsukada et al., 1994). The results show that co-transfection of the 3.8 kbp complete IL1B regulatory region with the IE2 291–364 peptide expression vector containing a His tag resulted in dose-dependent inhibition of activity down to 33% compared to the delA control peptide.
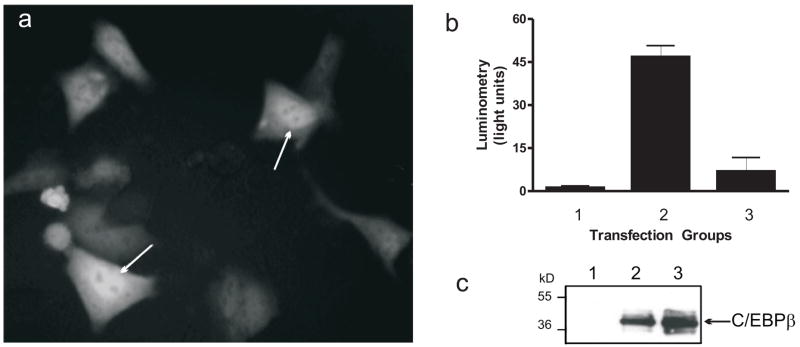
Various mechanisms were examined in order to explain the inhibition by the IE2 291–364. Because the IE2 291–364 region is missing one of two consensus NLS, we could not be sure whether a positive function was lost due to a lack of nuclear localization. Therefore, we transferred the sequence into a GFP expression vector to create a fusion product to verify nuclear localization of the peptide. At 24 h after transient transfection into HeLa S3 cells, GFP-fused 291–364 peptide resulted in both nuclear and cytoplasmic localization (Fig 6a). The GFP IE2 291–364 fusion product also retained inhibitory activity when compared to the empty GFP expression vector (Fig 6b, bar 3 vs. 2, respectively). Another possible confounding effect could involve inhibition of IE2 interaction with TBP because sequence 291–364 was previously mapped as one of three TBP binding sites on IE2. As the C/EBPβ expression vector used for these transfections contains a classic TATA box-dependent promoter, this possibility was simply investigated by quantifying C/EBPβ expression from the lysates used in Fig 6b by Western blot (Fig 6c). The results showed no decrease in C/EBPβ expression when co-transfected with the GFP IE2 291–364 expression vector (lane 3) relative to the GFP control (lane 2). Absence of detectable signal in the negative control (lane 1) compared with the blot shown in Fig 2b is due to use of a different antibody with reduced sensitivity.
Fig 6. Localization and specificity of the IE2 291–364 peptide.
a) Cellular localization of GFP–IE2 291–364 fusion product. HeLa S3 cells were transiently transfected with IE2 291–364pEGFP. After 24 hr, plates were visualized by fluorescence microscopy. b) Functional analysis of the GFP–IE2 291–364 fusion product described in (a). HeLa S3 cells were transfected with the HT reporter and cotransfected with the following expression vectors: Group 1 = reporter only; group 2 = Spi-1 + C/EBPβ + GFP; group 3 = Spi-1 + C/EBPβ + GFP-IE2 291–364. 48 hours following transfection, cells were lifted and lysates analyzed by luminometry and displayed as the mean light units ± SEM. c) C/EBPβ expression from the pCDNA3.1 expression vector used in (b) was detected by Western blot ( 198, Santa Cruz). The lane numbers correspond to the transfection groups in (b).
4. Discussion
HCMV induction of IL1B appears to be a complex process that is mediated by both the host cell response (via TLR signaling) and the viral encoded IE proteins. We have hypothesized that the later mechanism explains the robustness of the IL1B response because we previously reported that co-expression of the CMV IE proteins can vigorously transactivate the IL1B promoter even in the absence of the inducible UIS enhancer (Wara-aswapati et al., 1999). Bypassing enhancer may allow IE to function on the promoter and to escape the usual desensitization of the TLR response resulting from loss of an effective interaction between the common signaling components MyD88 and IRAK (Kramer et al., 1995; Lehner et al., 2001). This remarkable event may not be the only example of this kind of activation in the absence of an endogenous gene enhancer. HTLV1 virus can transactivate the IL1B gene by the viral Tax protein that also functions by interacting with Spi-1 (Tsukada et al., 1997). Others have shown that the EBV protein EBVNA2 can transactivate its cis response element in the presence of Spi-1 in B cells (Laux et al., 1994).
The magnitude of the IL1B promoter response to IE2 probably relates to its ability to interact with multiple components of the transcriptional machinery including TBP and Spi-1, among others, at amino acids 291–364 (Ahn et al., 1998; Caswell et al., 1993; Fortunato et al., 1997; Jupp et al., 1993; Kim et al., 2000; Lukac et al., 1997; Lukac et al., 1994; Pizzorno et al., 1991; Scully et al., 1995). Here the list of interacting partners is expanded with the addition of C/EBPβ. In this regard, the ability of IE2 to physically interact with the bZIP domain of C/EBPβ may serve as another docking point for IE2 on the IL1B promoter. This is an interesting finding because we have previously reported that cooperative function between Spi-1 and C/EBPβ also depends on an interaction mediated by the last 15 residues of the C/EBPβ bZIP domain and a surface on Spi-1 that includes arginine 232, the most critical arginine for DNA binding by this molecule (Listman et al., 2005). This interaction was proposed as a mechanism of stabilizing binding by these molecules and as a coupling mechanism between either both promoter sites or the far-upstream UIS and the promoter. Arginine 232 may also be important for IE2 interaction with Spi-1, although less so than for C/EBPβ (Listman et al., 2005).
The potency of IE2 depends upon its strong acidic transactivation domains (Wara-aswapati et al., 1999). This functionality probably explains why IE2 can compensate for the loss of sequence 41–245 of the C/EBPβ TAD. However, the N-terminus of C/EBPβ, which has been associated with chromatin remodeling, is also required. This function is necessary for maximal activity of some chromatin embedded genes (Kowenz-Leutz and Leutz, 1999). The role for chromatin remodeling for IL1B is interesting because monocytic cell types appear to have an open chromatin at the promoter with Spi-1 bound and poised for action (Liang et al., 2006). However, in HeLa S3 cells, endogenous IL1B is inactive, suggesting it has a more compact structure. Therefore, it is possible that the C/EBPβ amino terminus is necessary requisite for induction by the IE2.
What remains less well defined is the stoichiometry and precise location of the IE2 interactions because of the multiple binding sites for Spi-1 and C/EBPβ on the promoter. The mechanism of IE function at the IL1B promoter was previously shown to depend primarily on a cooperative interaction between Spi-1 and IE2 at the downstream Spi-1 site (Wara-aswapati et al., 1999). However, the role of C/EBPβ was not thoroughly evaluated in our earlier works. Herein, we observed low level reporter activity by C/EBPβ in the absence of ectopic Spi-1 (Fig 2a). This weak activity was not previously appreciated and may have been overlooked due either to the increased sensitivity of the luciferase vs. CAT assay or to the low level expression of a C/EBP isoform observed in the HeLa S3 cells used for some of the reporter assays (Fig 2b). A critical role for C/EBPβ is further highlighted by the significantly impaired IE2 response observed when the C/EBP sites found on the ILI1B promoter were mutated. Interestingly, these mutations exert more negative influence on IE cooperativity in this system, than on C/EBPβ cooperativity with Spi-1 in the absence of IE. We previously reported that the downstream C/EBP site is dispensable for cooperative function with Spi-1 and is supported by tethering between the DBD of Spi-1 and C/EBPβ (Listman et al., 2005; Yang et al., 2000). In fact, as shown here, C/EBPβ can function without any DNA binding site on the IL1B promoter when Spi-1 is present (function only reduced by approximately 50%). Taken together, these findings indicate that the IE2 effect on the IL1B promoter has more stringent spatial requirements for C/EBPβ than that required for cooperative function between Spi-1 and C/EBPβ alone. One plausible explanation could be that IE2 interaction with Spi-1 at the downstream site displaces C/EBPβ from Spi-1 to the weak C/EBP downstream DNA binding site. In this situation, mutation of the downstream site relinquishes all C/EBPβ interaction at this location on the promoter. In contrast, at the upstream location, the C/EBP site is relatively strong compared with the downstream site (unpublished observations) and IE2 function may depend more on a direct interaction with DNA bound C/EBPβ.
An alternate explanation for this stringency could be that IE2 interacts directly with the DNA overlapping one or both of the C/EBP sites. This seems unlikely at least for the upstream site, based on our current understanding of IE2 DNA binding (Waheed et al.). The situation differs at the downstream C/EBP site, which is in close proximity to the TATA box. IE2 has been shown to bind over certain TATA box sequences (Wang et al., 2000). This hypothesis is supported by the relatively simple, but specific, requirements for IE2 promoter function described by others (Lukac et al., 1994). IE2 could also couple with the enhancer via a long-range interaction with heterodimeric C/EBPβ bound to its site in the UIS. Therefore, it is possible that IE2 could replace C/EBPβ and TBP at this location. Binding by IE2 to this site would also explain why it does not function cooperatively with C/EBPβ using the DT reporter (in the absence of Spi-1), while we previously showed that this same reporter is very responsive to Spi-1 and C/EBPβ, even with the same mutated C/EBP site (Yang et al., 2000). However, we have been unable to reliably detect IE2 binding to the IL1B promoter. Therefore, this issue remains unresolved. Regardless of the mechanisms involved, be it direct interaction with C/EBPβ or to DNA, IE function is strongly dependent on both C/EBP sites for its activity.
The promiscuity of binding between these molecules probably explains the robust dominant negative effect of the IE2 291–364 peptide on reporter activity, particularly in the heterologous assays in HeLa S3 cells where the IL1B UIS is not particularly responsive compared to the myeloid lineage RAW 264.7 line. Because our biochemical assays localized a Spi-1 and C/EBPβ interaction region to depend upon only 14 residues of IE2 (amino acids 315–328), we propose that the interaction is mutually exclusive on IE2. As predicted by our model (Fig 1d), small peptides encompassing this region, but excluding other portions of the IE2 transactivation domain, could competitively antagonize productive interactions between Spi-1, C/EBPβ and IE2 (Fig 7). Moreover, these interactions explain why the peptide also inhibits promoter activity even in the absence of IE expression.
Fig 7. Model of competitive antagonism by IE2 291–364.
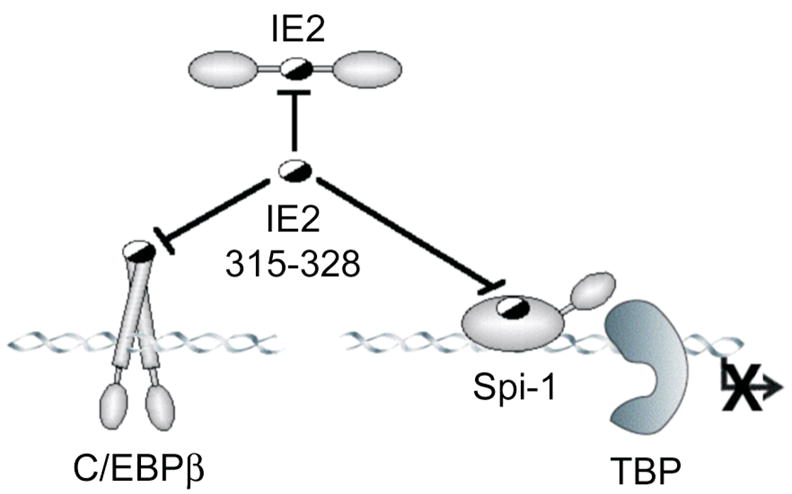
The normal cooperative interactions between Spi-1, C/EBPβ and IE2 on the IL1B promoter (shown in Fig 1d) are blocked by the 291–364 fragment of IE2 because it encompasses region 315–328, which binds to Spi-1 and C/EBPβ, but lacks other transactivation elements and the DNA binding domain of IE2 (shown in Fig 1c). Half-filled ovals denote mutual interactions sites.
The teleology of the IL1B promoter response to IE2 is somewhat counterintuitive, but may serve the bidding of both virus and host. For example, during primary infection by hCMV, cytokine production is essential for a robust immune response in order to halt the spread of infection. However, like other herpes family viruses, hCMV can evade the host’s immune system by transitioning from viral replication to a poorly understood state of viral latency where viral DNA remains in the host cell, but no viral replication is apparent. In this regard, the major cytokines of the innate immune system may have a role. There is some evidence in models of solid organ transplantation that TNFα can reverse latency back to viral replication (Docke et al., 1994; Hummel et al., 2001). On the other hand, IL-1β has been shown in bone marrow stromal cells to inhibit viral replication and perhaps induce latency (Iwata et al., 1999). Whether the redundant mechanisms used to induce cytokines is more beneficial to the host or the virus is unknown. Nevertheless, given the current interest in the role of latent viral infections in a variety of chronic inflammatory diseases in humans, defining the structural and functional components of IE mediated transactivation of the above cytokines might lead to valuable insights for the development of novel anti-inflammatory agents that block IL-1β production at the earliest phase of gene expression.
Acknowledgments
This work was supported by grants from the NIH to J.A.L (K08DK002788) and P.E.A (CA068544).
Abbreviations
- hCMV
human Cytomegalovirus
- DBD
DNA binding domain
- IL1B
gene encoding IL-1β
- TLR
Toll-like receptor
- bZIP
basic leucine zipper domain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn JH, Chiou CJ, Hayward GS. Evaluation and mapping of the DNA binding and oligomerization domains of the IE2 regulatory protein of human cytomegalovirus using yeast one and two hybrid interaction assays. Gene. 1998;210:25–36. doi: 10.1016/s0378-1119(98)00056-0. [DOI] [PubMed] [Google Scholar]
- Auron PE. The interleukin 1 receptor: ligand interactions and signal transduction. Cytokine Growth Factor Rev. 1998;9:221–37. doi: 10.1016/s1359-6101(98)00018-5. [DOI] [PubMed] [Google Scholar]
- Auron PE, Webb AC. Interleukin-1: a gene expression system regulated at multiple levels. Eur Cytokine Netw. 1994;5:573–92. [PubMed] [Google Scholar]
- Baldassare JJ, Bi Y, Bellone CJ. The role of p38 mitogen-activated protein kinase in IL-1 beta transcription. J Immunol. 1999;162:5367–73. [PubMed] [Google Scholar]
- Cahill CM, Waterman WR, Xie Y, Auron PE, Calderwood SK. Transcriptional repression of the prointerleukin 1beta gene by heat shock factor 1.PG - 24874–9. J Biol Chem. 1996;271 [PubMed] [Google Scholar]
- Caswell R, Hagemeier C, Chiou CJ, Hayward G, Kouzarides T, Sinclair J. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J Gen Virol. 1993;74(Pt 12):2691–8. doi: 10.1099/0022-1317-74-12-2691. [DOI] [PubMed] [Google Scholar]
- Chandra G, Cogswell JP, Miller LR, Godlevski MM, Stinnett SW, Noel SL, Kadwell SH, Kost TA, Gray JG. Cyclic AMP signaling pathways are important in IL-1 beta transcriptional regulation. J Immunol. 1995;155:4535–43. [PubMed] [Google Scholar]
- Chiou CJ, Zong J, Waheed I, Hayward GS. Identification and mapping of dimerization and DNA-binding domains in the C terminus of the IE2 regulatory protein of human cytomegalovirus. J Virol. 1993;67:6201–14. doi: 10.1128/jvi.67.10.6201-6214.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton T. Receptors and immune sensors: the complex entry path of human cytomegalovirus. Trends Cell Biol. 2004;14:5–8. doi: 10.1016/j.tcb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Crump JW, Geist LJ, Auron PE, Webb AC, Stinski MF, Hunninghake GW. The immediate early genes of human cytomegalovirus require only proximal promoter elements to upregulate expression of interleukin-1 beta. Am J Respir Cell Mol Biol. 1992;6:674–7. doi: 10.1165/ajrcmb/6.6.674. [DOI] [PubMed] [Google Scholar]
- Docke WD, Prosch S, Fietze E, Kimel V, Zuckermann H, Klug C, Syrbe U, Kruger DH, von Baehr R, Volk HD. Cytomegalovirus reactivation and tumour necrosis factor. Lancet. 1994;343:268–9. doi: 10.1016/s0140-6736(94)91116-9. [DOI] [PubMed] [Google Scholar]
- Dudding L, Haskill S, Clark BD, Auron PE, Sporn S, Huang ES. Cytomegalovirus infection stimulates expression of monocyte-associated mediator genes. J Immunol. 1989;143:3343–52. [PubMed] [Google Scholar]
- Escalante CR, Brass AL, Pongubala JM, Shatova E, Shen L, Singh H, Aggarwal AK. Crystal structure of PU.1/IRF-4/DNA ternary complex. Mol Cell. 2002;10:1097–105. doi: 10.1016/s1097-2765(02)00703-7. [DOI] [PubMed] [Google Scholar]
- Fenton MJ, Clark BD, Collins KL, Webb AC, Rich A, Auron PE. Transcriptional regulation of the human prointerleukin 1 beta gene. J Immunol. 1987;138:3972–9. [PubMed] [Google Scholar]
- Fortunato EA, Sommer MH, Yoder K, Spector DH. Identification of domains within the human cytomegalovirus major immediate-early 86-kilodalton protein and the retinoblastoma protein required for physical and functional interaction with each other. J Virol. 1997;71:8176–8185. doi: 10.1128/jvi.71.11.8176-8185.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galson DL, Hensold JO, Bishop TR, Schalling M, D’Andrea AD, Jones C, Auron PE, Housman DE. Mouse beta-globin DNA-binding protein B1 is identical to a proto- oncogene, the transcription factor Spi-1/PU.1, and is restricted in expression to hematopoietic cells and the testis. Mol Cell Biol. 1993;13:2929–41. doi: 10.1128/mcb.13.5.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J, Marois J, Garoufalis J, D’Addario M, Roulston A, Kwan I, Pepin N, Lacoste J, Nguyen H, Bensi G, et al. Characterization of a functional NF-kappa B site in the human interleukin 1 beta promoter: evidence for a positive autoregulatory loop. Mol Cell Biol. 1993;13:6231–40. doi: 10.1128/mcb.13.10.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel M, Zhang Z, Yan S, DePlaen I, Golia P, Varghese T, Thomas G, Abecassis MI. Allogeneic transplantation induces expression of cytomegalovirus immediate-early genes in vivo: a model for reactivation from latency. J Virol. 2001;75:4814–22. doi: 10.1128/JVI.75.10.4814-4822.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto GK, Monick MM, Clark BD, Auron PE, Stinski MF, Hunninghake GW. Modulation of interleukin 1 beta gene expression by the immediate early genes of human cytomegalovirus. J Clin Invest. 1990;85:1853–7. doi: 10.1172/JCI114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata M, Vieira J, Byrne M, Horton H, Torok-Storb B. Interleukin-1 (IL-1) inhibits growth of cytomegalovirus in human marrow stromal cells: inhibition is reversed upon removal of IL-1. Blood. 1999;94:572–8. [PubMed] [Google Scholar]
- Jupp R, Hoffmann S, Stenberg RM, Nelson JA, Ghazal P. Human cytomegalovirus IE86 protein interacts with promoter-bound TATA-binding protein via a specific region distinct from the autorepression domain. J Virol. 1993;67:7539–46. doi: 10.1128/jvi.67.12.7539-7546.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-M, Hong Y, Jeang KT, Kim S. Transactivation activity of the human cytomegalovirus IE2 protein occurs at steps subsequent to TATA box-binding protein recruitment. J Gen Virol. 2000;81:37–46. doi: 10.1099/0022-1317-81-1-37. [DOI] [PubMed] [Google Scholar]
- Klemsz MJ, Maki RA. Activation of transcription by PU.1 requires both acidic and glutamine domains. Mol Cell Biol. 1996;16:390–7. doi: 10.1128/mcb.16.1.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher KM, Sommer M, Kadonaga JT, Spector DH. In vivo and in vitro analysis of transcriptional activation mediated by the human cytomegalovirus major immediate-early proteins. Mol Cell Biol. 1993;13:1238–50. doi: 10.1128/mcb.13.2.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominato Y, Galson D, Waterman WR, Webb AC, Auron PE. Monocyte expression of the human prointerleukin 1 beta gene (IL1B) is dependent on promoter sequences which bind the hematopoietic transcription factor Spi-1/PU.1. Mol Cell Biol. 1995;15:59–68. [PMC free article] [PubMed] [Google Scholar]
- Kowenz-Leutz E, Leutz A. A C/EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol Cell. 1999;4:735–43. doi: 10.1016/s1097-2765(00)80384-6. [DOI] [PubMed] [Google Scholar]
- Koyama Y, Tanaka Y, Saito K, Abe M, Nakatsuka K, Morimoto I, Auron PE, Eto S. Cross-linking of intercellular adhesion molecule 1 (CD54) induces AP-1 activation and IL-1beta transcription. J Immunol. 1996;157:5097–103. [PubMed] [Google Scholar]
- Kramer B, Wiegmann K, Kronke M. Regulation of the human TNF promoter by the transcription factor Ets. J Biol Chem. 1995;270:6577–83. doi: 10.1074/jbc.270.12.6577. [DOI] [PubMed] [Google Scholar]
- Laux G, Adam B, Strobl LJ, Moreau-Gachelin F. The Spi-1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-J kappa interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. Embo J. 1994;13:5624–32. doi: 10.1002/j.1460-2075.1994.tb06900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner MD, Morath S, Michelsen KS, Schumann RR, Hartung T. Induction of cross-tolerance by lipopolysaccharide and highly purified lipoteichoic acid via different Toll-like receptors independent of paracrine mediators. J Immunol. 2001;166:5161–7. doi: 10.4049/jimmunol.166.8.5161. [DOI] [PubMed] [Google Scholar]
- Lekstrom-Himes J, Xanthopoulos KG. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J Biol Chem. 1998;273:28545–8. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- Liang MD, Zhang Y, McDevit D, Marecki S, Nikolajczyk BS. The IL-1beta gene is transcribed from a poised promoter architecture in monocytes. J Biol Chem. 2006:M510700200. doi: 10.1074/jbc.M510700200. [DOI] [PubMed] [Google Scholar]
- Listman JA, Wara-aswapati N, Race JE, Blystone LW, Walker-Kopp N, Yang Z, Auron PE. Conserved ETS Domain Arginines Mediate DNA Binding, Nuclear Localization, and a Novel Mode of bZIP Interaction. J Biol Chem. 2005;280:41421–41428. doi: 10.1074/jbc.M509143200. [DOI] [PubMed] [Google Scholar]
- Lukac DM, Harel NY, Tanese N, Alwine JC. TAF-like functions of human cytomegalovirus immediate-early proteins. J Virol. 1997;71:7227–39. doi: 10.1128/jvi.71.10.7227-7239.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac DM, Manuppello JR, Alwine JC. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J Virol. 1994;68:5184–93. doi: 10.1128/jvi.68.8.5184-5193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski ES. Cytomegalorviruses and Their Replication. In: Fields BN, Knipe DM, Holey PM, Chanock RM, Melnick JL, Monath TP, Roizman B, Straus SE, editors. Fields Virology. Lipincott-Raven Publishers; Philadelphia: 1996. [Google Scholar]
- O’Neill LA, Dinarello CA. The IL-1 receptor/toll-like receptor superfamily: crucial receptors for inflammation and host defense. Immunol Today. 2000;21:206–9. doi: 10.1016/s0167-5699(00)01611-x. [DOI] [PubMed] [Google Scholar]
- Pahl HL, Scheibe RJ, Zhang DE, Chen HM, Galson DL, Maki RA, Tenen DG. The proto-oncogene PU.1 regulates expression of the myeloid-specific CD11b promoter. J Biol Chem. 1993;268:5014–20. [PubMed] [Google Scholar]
- Park JM, Greten FR, Wong A, Westrick RJ, Arthur JS, Otsu K, Hoffmann A, Montminy M, Karin M. Signaling pathways and genes that inhibit pathogen-induced macrophage apoptosis--CREB and NF-kappaB as key regulators. Immunity. 2005;23:319–29. doi: 10.1016/j.immuni.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Paul R, Schuetze S, Kozak SL, Kozak CA, Kabat D. The Sfpi-1 proviral integration site of Friend erythroleukemia encodes the ets-related transcription factor Pu.1. J Virol. 1991;65:464–7. doi: 10.1128/jvi.65.1.464-467.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzorno MC, Mullen MA, Chang YN, Hayward GS. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J Virol. 1991;65:3839–52. doi: 10.1128/jvi.65.7.3839-3852.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Culine S, Tavitain A, Moreau-Gachelin F. The human homologue of the putative proto-oncogene Spi-1: characterization and expression in tumors. Oncogene. 1990;5:663–8. [PubMed] [Google Scholar]
- Scully AL, Sommer MH, Schwartz R, Spector DH. The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J Virol. 1995;69:6533–6540. doi: 10.1128/jvi.69.10.6533-6540.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa F, Saito K, Bonagura CA, Galson DL, Fenton MJ, Webb AC, Auron PE. The human prointerleukin 1 beta gene requires DNA sequences both proximal and distal to the transcription start site for tissue-specific induction. Mol Cell Biol. 1993;13:1332–44. doi: 10.1128/mcb.13.3.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinski M, Thomsen D, Stenberg R, Goldstein L. Organization and expression of the immediate early genes of human cytomegalovirus. J Virol. 1982;46:1–14. doi: 10.1128/jvi.46.1.1-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada J, Misago M, Serino Y, Ogawa R, Murakami S, Nakanishi M, Tonai S, Kominato Y, Morimoto I, Auron PE, Eto S. Human T-cell leukemia virus type I Tax transactivates the promoter of human prointerleukin-1beta gene through association with two transcription factors, nuclear factor-interleukin-6 and Spi-1. Blood. 1997;90:3142–53. [PubMed] [Google Scholar]
- Tsukada J, Saito K, Waterman WR, Webb AC, Auron PE. Transcription factors NF-IL6 and CREB recognize a common essential site in the human prointerleukin 1 beta gene. Mol Cell Biol. 1994;14:7285–97. doi: 10.1128/mcb.14.11.7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unlu S, Kumar A, Waterman WR, Tsukada J, Wang KZ, Galson DL, Auron PE. Phosphorylation of IRF8 in a pre-associated complex with Spi-1/PU.1 and non-phosphorylated Stat1 is critical for LPS induction of the IL1B gene. Mol Immunol. 2007 doi: 10.1016/j.molimm.2007.02.016. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waheed I, Chiou CJ, Ahn JH, Hayward GS. Binding of the human cytomegalovirus 80-kDa immediate-early protein (IE2) to minor groove A/T-rich sequences bounded by CG dinucleotides is regulated by protein oligomerization and phosphorylation. Virology. 1998;252:235–57. doi: 10.1006/viro.1998.9448. [DOI] [PubMed] [Google Scholar]
- Wang YC, Huang CF, Tung SF, Lin YS. Competition with TATA box-binding protein for binding to the TATA box implicated in human cytomegalovirus IE2-mediated transcriptional repression of cellular promoters. DNA Cell Biol. 2000;19:613–9. doi: 10.1089/104454900750019371. [DOI] [PubMed] [Google Scholar]
- Wara-aswapati N, Yang Z, Waterman WR, Koyama Y, Tetradis S, Choy BK, Webb AC, Auron PE. Cytomegalovirus IE2 protein stimulates interleukin 1beta gene transcription via tethering to Spi-1/PU.1. Mol Cell Biol. 1999;19:6803–14. doi: 10.1128/mcb.19.10.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wara-Aswapati N, Chen C, Tsukada J, Auron PE. NF-IL6 (C/EBPbeta) vigorously activates il1β gene expression via a Spi- 1 (PU.1) protein-protein tether. J Biol Chem. 2000;275:21272–7. doi: 10.1074/jbc.M000145200. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wara-aswapati N, Yoshida Y, Walker N, Galson DL, Listman J, Auron PE. Dual regulatory role of human cytomegalovirus immediate-early protein in IL1B transcription is dependent upon Spi-1/PU.1. Biochem Biophys Res Commun. 2002;294:854–63. doi: 10.1016/S0006-291X(02)00562-4. [DOI] [PubMed] [Google Scholar]
- Yurochko AD, Huang ES. Human cytomegalovirus binding to human monocytes induces immunoregulatory gene expression. J Immunol. 1999;162:4806–16. [PubMed] [Google Scholar]