Abstract
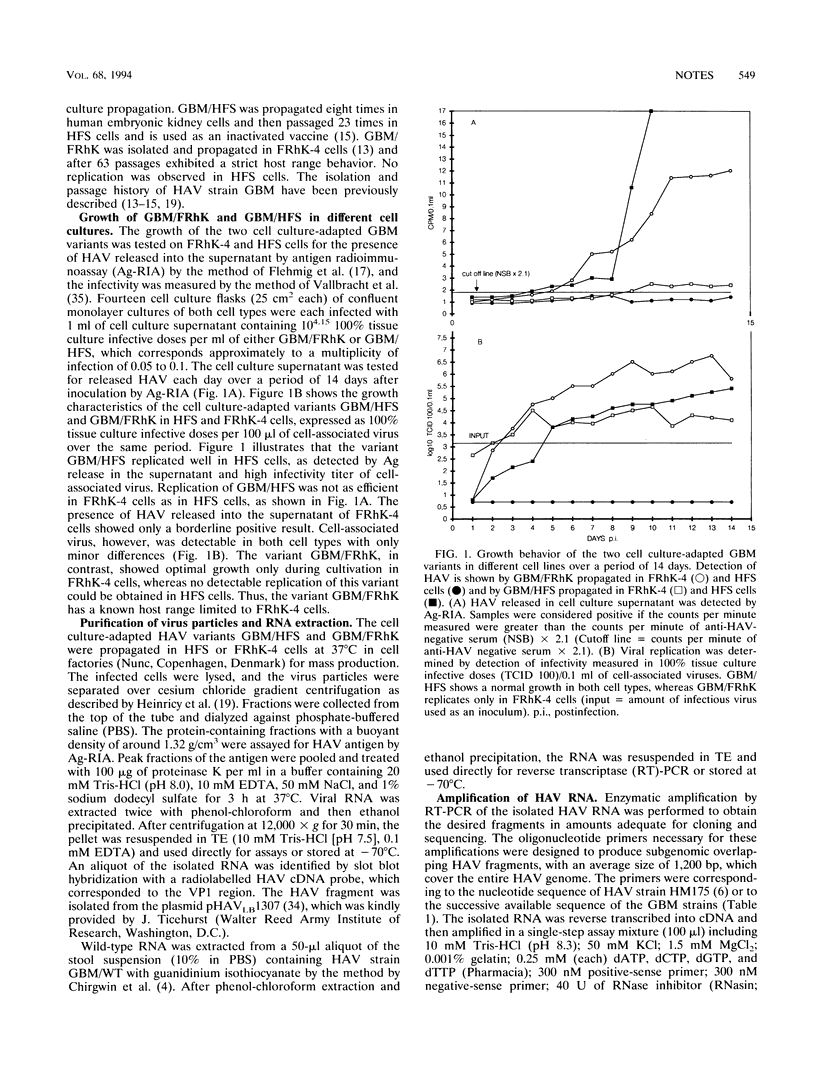
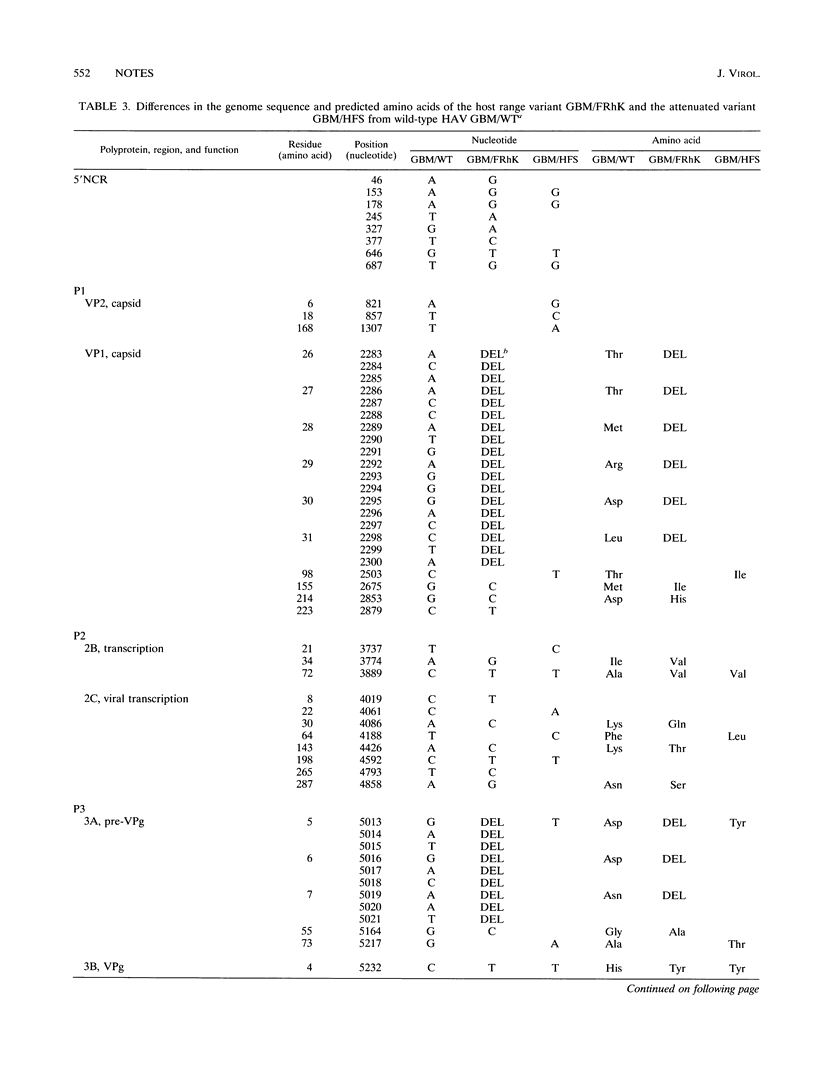
In order to study cell tropism and attenuation of hepatitis A virus (HAV), the genome of HAV wild-type GBM and two cell culture-adapted variants, GBM/FRhK and GBM/HFS, were cloned and sequenced after amplification by reverse transcriptase-PCR. During virus cultivation, the HAV variant GBM/FRhK had a strict host range for FRhK-4 cells, in contrast to GBM/HFS, which can be grown in HFS and FRhK-4 cells. The HAV variant GBM/HFS was shown to be attenuated when inoculated into chimpanzees (B. Flehmig, R. F. Mauler, G. Noll, E. Weinmann, and J. P. Gregerson, p. 87-90, in A. Zuckerman, ed., Viral Hepatitis and Liver Disease, 1988). On the basis of this biological background, the comparison of the nucleotide sequences of these three HAV GBM variants should elucidate differences which may be of importance for cell tropism and attenuation. The comparison of the genome between the GBM wild type and HAV wild types HM175 (J. I. Cohen, J. R. Ticehurst, R. H. Purcell, A. Buckler-White, and B. M. Baroudy, J. Virol. 61:50-59, 1987) and HAV-LA (R. Najarian, O. Caput, W. Gee, S. J. Potter, A. Renard, J. Merryweather, G. Van Nest, and D. Dina, Proc. Natl. Acad. Sci. USA 82:2627-2631, 1985) showed a 92 to 96.3% identity, whereas the identity was 99.3 to 99.6% between the GBM variants. Nucleotide differences between the wild-type and the cell culture-adapted variants, which were identical in both cell culture-adapted GBM variants, were localized in the 5' noncoding region; in 2B, 3B, and 3D; and in the 3' noncoding region. Our result concerning the 2B/2C region confirms a mutation at position 3889 (C-->T, alanine to valine), which had been shown to be of importance for cell culture adaptation (S. U. Emerson, C. McRill, B. Rosenblum, S. M. Feinstone, and R. H. Purcell, J. Virol. 65:4882-4886, 1991; S. U. Emerson, Y. K. Huang, C. McRill, M. Lewis, and R. H. Purcell, J. Virol. 66:650-654, 1992), whereas other mutations differ from published HAV sequence data and may be cell specific. Further comparison of the two cell culture-adapted GBM variants showed cell-specific mutations resulting in deletions of six amino acids in the VP1 region and three amino acids in the 3A region of the GBM variant GBM/FRhK.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. A. Cytopathology, plaque assay, and heat inactivation of hepatitis A virus strain HM175. J Med Virol. 1987 May;22(1):35–44. doi: 10.1002/jmv.1890220106. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. A., Day S. P., Jansen R. W., Lemon S. M. The 5' nontranslated region of hepatitis A virus RNA: secondary structure and elements required for translation in vitro. J Virol. 1991 Nov;65(11):5828–5838. doi: 10.1128/jvi.65.11.5828-5838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cohen J. I., Rosenblum B., Ticehurst J. R., Daemer R. J., Feinstone S. M., Purcell R. H. Complete nucleotide sequence of an attenuated hepatitis A virus: comparison with wild-type virus. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2497–2501. doi: 10.1073/pnas.84.8.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. I., Ticehurst J. R., Purcell R. H., Buckler-White A., Baroudy B. M. Complete nucleotide sequence of wild-type hepatitis A virus: comparison with different strains of hepatitis A virus and other picornaviruses. J Virol. 1987 Jan;61(1):50–59. doi: 10.1128/jvi.61.1.50-59.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromeans T., Fields H. A., Sobsey M. D. Replication kinetics and cytopathic effect of hepatitis A virus. J Gen Virol. 1989 Aug;70(Pt 8):2051–2062. doi: 10.1099/0022-1317-70-8-2051. [DOI] [PubMed] [Google Scholar]
- Day S. P., Murphy P., Brown E. A., Lemon S. M. Mutations within the 5' nontranslated region of hepatitis A virus RNA which enhance replication in BS-C-1 cells. J Virol. 1992 Nov;66(11):6533–6540. doi: 10.1128/jvi.66.11.6533-6540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Chastonay J., Siegl G. Replicative events in hepatitis A virus-infected MRC-5 cells. Virology. 1987 Apr;157(2):268–275. doi: 10.1016/0042-6822(87)90269-8. [DOI] [PubMed] [Google Scholar]
- Emerson S. U., Huang Y. K., McRill C., Lewis M., Purcell R. H. Mutations in both the 2B and 2C genes of hepatitis A virus are involved in adaptation to growth in cell culture. J Virol. 1992 Feb;66(2):650–654. doi: 10.1128/jvi.66.2.650-654.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S. U., Huang Y. K., Purcell R. H. 2B and 2C mutations are essential but mutations throughout the genome of HAV contribute to adaptation to cell culture. Virology. 1993 Jun;194(2):475–480. doi: 10.1006/viro.1993.1286. [DOI] [PubMed] [Google Scholar]
- Emerson S. U., McRill C., Rosenblum B., Feinstone S., Purcell R. H. Mutations responsible for adaptation of hepatitis A virus to efficient growth in cell culture. J Virol. 1991 Sep;65(9):4882–4886. doi: 10.1128/jvi.65.9.4882-4886.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flehmig B., Frank H., Frösner G. G., Gerth H. J. Hepatitis A-virus particles in stools of patients from a natural hepatitis outbreak in Germany. Med Microbiol Immunol. 1977 Oct 7;163(3):209–214. doi: 10.1007/BF02126679. [DOI] [PubMed] [Google Scholar]
- Flehmig B., Heinricy U., Pfisterer M. Immunogenicity of a killed hepatitis A vaccine in seronegative volunteers. Lancet. 1989 May 13;1(8646):1039–1041. doi: 10.1016/s0140-6736(89)92443-4. [DOI] [PubMed] [Google Scholar]
- Flehmig B. Hepatitis A virus in cell culture. II. Growth characteristics of hepatitis A virus in Frhk-4/R cells. Med Microbiol Immunol. 1981;170(2):73–81. doi: 10.1007/BF02122671. [DOI] [PubMed] [Google Scholar]
- Flehmig B., Ranke M., Frank H., Gerth H. J. Application of a solid-phase radioimmunoassay and immune electron microscopy for hepatitis A in diagnosis and research. Med Microbiol Immunol. 1978 Nov 17;166(1-4):187–194. doi: 10.1007/BF02121149. [DOI] [PubMed] [Google Scholar]
- Glass M. J., Summers D. F. A cis-acting element within the hepatitis A virus 5'-non-coding region required for in vitro translation. Virus Res. 1992 Oct;26(1):15–31. doi: 10.1016/0168-1702(92)90143-w. [DOI] [PubMed] [Google Scholar]
- Heinricy U., Stierhof Y. D., Pfisterer M., Flehmig B. Properties of a hepatitis A virus candidate vaccine strain. J Gen Virol. 1987 Sep;68(Pt 9):2487–2493. doi: 10.1099/0022-1317-68-9-2487. [DOI] [PubMed] [Google Scholar]
- Jansen R. W., Newbold J. E., Lemon S. M. Complete nucleotide sequence of a cell culture-adapted variant of hepatitis A virus: comparison with wild-type virus with restricted capacity for in vitro replication. Virology. 1988 Apr;163(2):299–307. doi: 10.1016/0042-6822(88)90270-x. [DOI] [PubMed] [Google Scholar]
- Lemon S. M., Murphy P. C., Shields P. A., Ping L. H., Feinstone S. M., Cromeans T., Jansen R. W. Antigenic and genetic variation in cytopathic hepatitis A virus variants arising during persistent infection: evidence for genetic recombination. J Virol. 1991 Apr;65(4):2056–2065. doi: 10.1128/jvi.65.4.2056-2065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locarnini S. A., Coulepis A. G., Westaway E. G., Gust I. D. Restricted replication of human hepatitis A virus in cell culture: intracellular biochemical studies. J Virol. 1981 Jan;37(1):216–225. doi: 10.1128/jvi.37.1.216-225.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morace G., Pisani G., Beneduce F., Divizia M., Panà A. Mutations in the 3A genomic region of two cytopathic strains of hepatitis A virus isolated in Italy. Virus Res. 1993 May;28(2):187–194. doi: 10.1016/0168-1702(93)90135-a. [DOI] [PubMed] [Google Scholar]
- Najarian R., Caput D., Gee W., Potter S. J., Renard A., Merryweather J., Van Nest G., Dina D. Primary structure and gene organization of human hepatitis A virus. Proc Natl Acad Sci U S A. 1985 May;82(9):2627–2631. doi: 10.1073/pnas.82.9.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A. V., Tada H., von der Helm K., Wissel T., Kiehn R., Wimmer E., Deinhardt F. The entire nucleotide sequence of the genome of human hepatitis A virus (isolate MBB). Virus Res. 1987 Aug;8(2):153–171. doi: 10.1016/0168-1702(87)90026-8. [DOI] [PubMed] [Google Scholar]
- Provost P. J., Hilleman M. R. Propagation of human hepatitis A virus in cell culture in vitro. Proc Soc Exp Biol Med. 1979 Feb;160(2):213–221. doi: 10.3181/00379727-160-40422. [DOI] [PubMed] [Google Scholar]
- Robertson B. H., Jansen R. W., Khanna B., Totsuka A., Nainan O. V., Siegl G., Widell A., Margolis H. S., Isomura S., Ito K. Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J Gen Virol. 1992 Jun;73(Pt 6):1365–1377. doi: 10.1099/0022-1317-73-6-1365. [DOI] [PubMed] [Google Scholar]
- Ross B. C., Anderson B. N., Edwards P. C., Gust I. D. Nucleotide sequence of high-passage hepatitis A virus strain HM175: comparison with wild-type and cell culture-adapted strains. J Gen Virol. 1989 Oct;70(Pt 10):2805–2810. doi: 10.1099/0022-1317-70-10-2805. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticehurst J. R., Racaniello V. R., Baroudy B. M., Baltimore D., Purcell R. H., Feinstone S. M. Molecular cloning and characterization of hepatitis A virus cDNA. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5885–5889. doi: 10.1073/pnas.80.19.5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallbracht A., Hofmann L., Wurster K. G., Flehmig B. Persistent infection of human fibroblasts by hepatitis A virus. J Gen Virol. 1984 Mar;65(Pt 3):609–615. doi: 10.1099/0022-1317-65-3-609. [DOI] [PubMed] [Google Scholar]
- Venuti A., Di Russo C., del Grosso N., Patti A. M., Ruggeri F., De Stasio P. R., Martiniello M. G., Pagnotti P., Degener A. M., Midulla M. Isolation and molecular cloning of a fast-growing strain of human hepatitis A virus from its double-stranded replicative form. J Virol. 1985 Nov;56(2):579–588. doi: 10.1128/jvi.56.2.579-588.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler C. M., Fields H. A., Schable C. A., Meinke W. J., Maynard J. E. Adsorption, purification, and growth characteristics of hepatitis A virus strain HAS-15 propagated in fetal rhesus monkey kidney cells. J Clin Microbiol. 1986 Mar;23(3):434–440. doi: 10.1128/jcm.23.3.434-440.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]


