Abstract
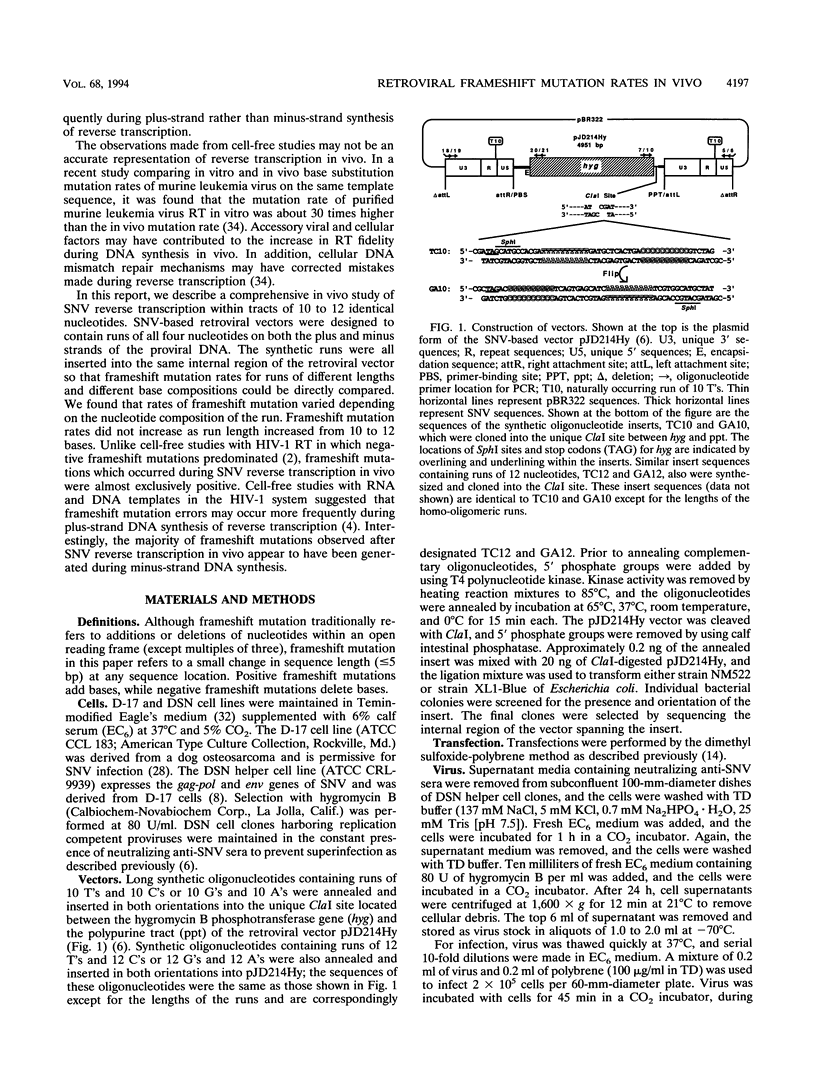
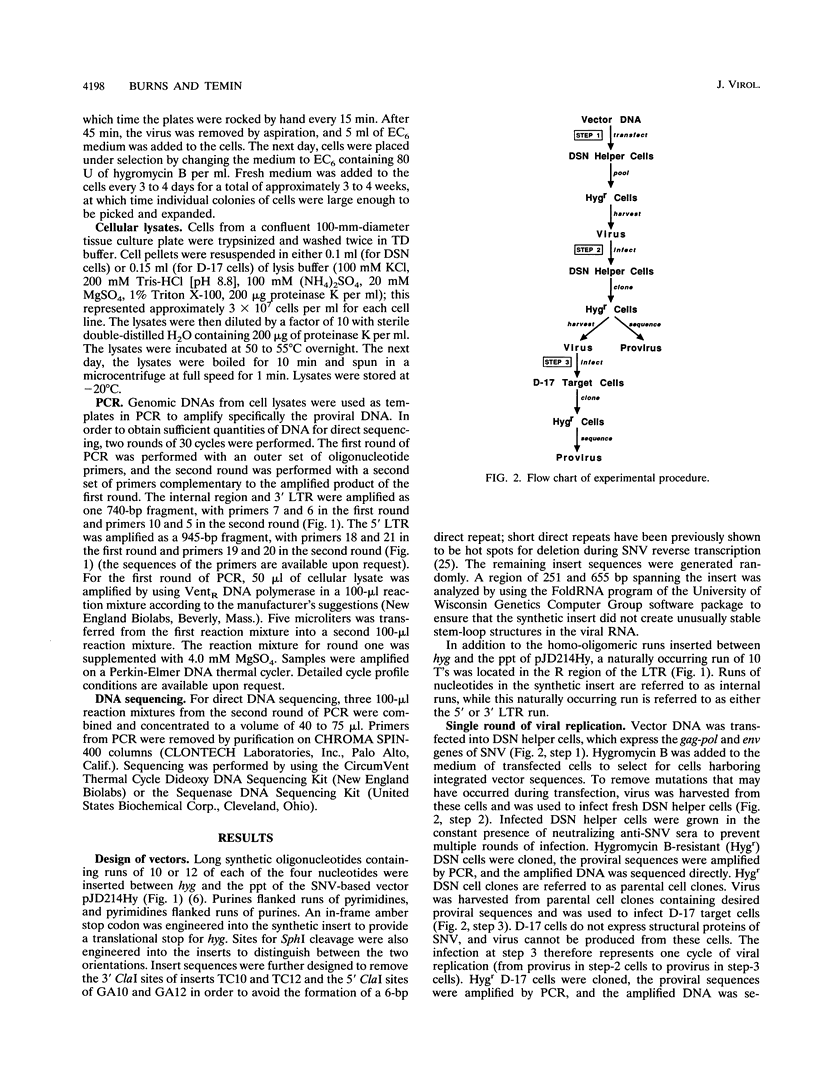
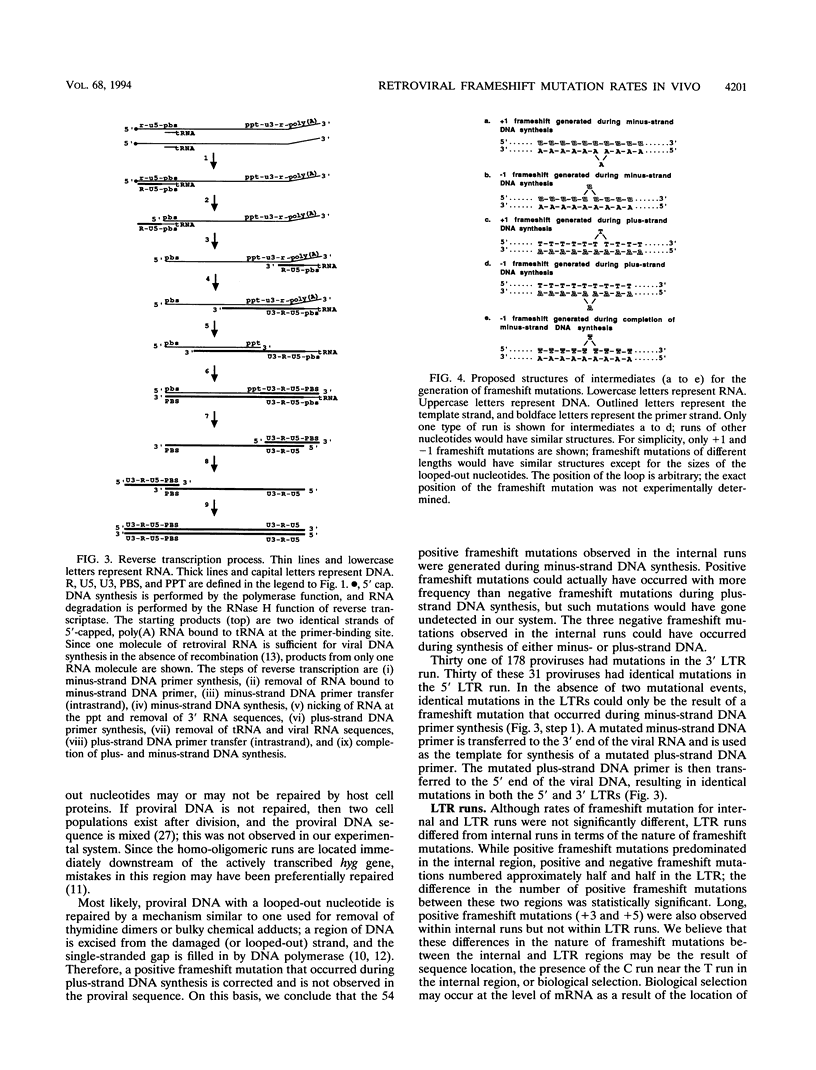
Homo-oligomeric runs were inserted into a spleen necrosis virus-based retrovirus vector to determine the nature and rate of mutations within runs of 10 to 12 identical nucleotides during a single replication cycle. Clones of helper cells containing integrated copies of retroviral vectors were used to produce virus for infection of target (nonhelper) cells. Proviral sequences from target cell clones were compared with proviral sequences from helper cell clones to study mutations that occurred during a single cycle of replication. In addition to the internal region spanning the homo-oligomeric inserts, a naturally occurring run of 10 T's in the long terminal repeat (LTR) also was sequenced. Rates of mutation ranged from < 0.01 to 0.38 frameshift mutations per run per cycle for different nucleotide runs. Frameshift mutations ranged from deletions of 2 bases to additions of 5 bases; the most common mutations were +1 and -1. Frameshift mutation rates did not increase as the run length increased from 10 to 12 bases. Rates of frameshift mutation for runs of T's and A's were significantly higher than rates for runs of C's and G's, and rates for runs of pyrimidines were significantly higher than those for runs of purines. Interestingly, the vast majority of frameshift mutations in the internal region (95%) were positive, suggesting that the primer strand tends to slip backward on the template in this region. LTR runs had a significantly lower number of positive frameshift mutations than the internal runs. By analyzing the types of frameshift mutations within runs and by comparing the patterns of frameshift mutations in the 5' and 3' LTRs of individual proviruses, we conclude that the majority of mutations observed in our system occurred during minus-strand DNA synthesis of reverse transcription.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Battula N., Loeb L. A. On the fidelity of DNA replication. Lack of exodeoxyribonuclease activity and error-correcting function in avian myeloblastosis virus DNA polymerase. J Biol Chem. 1976 Feb 25;251(4):982–986. [PubMed] [Google Scholar]
- Bebenek K., Abbotts J., Roberts J. D., Wilson S. H., Kunkel T. A. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989 Oct 5;264(28):16948–16956. [PubMed] [Google Scholar]
- Bebenek K., Joyce C. M., Fitzgerald M. P., Kunkel T. A. The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J Biol Chem. 1990 Aug 15;265(23):13878–13887. [PubMed] [Google Scholar]
- Boyer J. C., Bebenek K., Kunkel T. A. Unequal human immunodeficiency virus type 1 reverse transcriptase error rates with RNA and DNA templates. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):6919–6923. doi: 10.1073/pnas.89.15.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupples C. G., Cabrera M., Cruz C., Miller J. H. A set of lacZ mutations in Escherichia coli that allow rapid detection of specific frameshift mutations. Genetics. 1990 Jun;125(2):275–280. doi: 10.1093/genetics/125.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty J. P., Temin H. M. Determination of the rate of base-pair substitution and insertion mutations in retrovirus replication. J Virol. 1988 Aug;62(8):2817–2822. doi: 10.1128/jvi.62.8.2817-2822.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty J. P., Temin H. M. High mutation rate of a spleen necrosis virus-based retrovirus vector. Mol Cell Biol. 1986 Dec;6(12):4387–4395. doi: 10.1128/mcb.6.12.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty J. P., Wisniewski R., Yang S. L., Rhode B. W., Temin H. M. New retrovirus helper cells with almost no nucleotide sequence homology to retrovirus vectors. J Virol. 1989 Jul;63(7):3209–3212. doi: 10.1128/jvi.63.7.3209-3212.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresco J. R., Alberts B. M. THE ACCOMMODATION OF NONCOMPLEMENTARY BASES IN HELICAL POLYRIBONUCLEOTIDES AND DEOXYRIBONUCLEIC ACIDS. Proc Natl Acad Sci U S A. 1960 Mar;46(3):311–321. doi: 10.1073/pnas.46.3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson J., Munn M., Rupp W. D., Kahn R., Wood R. D. Localization of DNA repair synthesis by human cell extracts to a short region at the site of a lesion. J Biol Chem. 1989 Dec 25;264(36):21788–21792. [PubMed] [Google Scholar]
- Hoeijmakers J. H. Nucleotide excision repair. II: From yeast to mammals. Trends Genet. 1993 Jun;9(6):211–217. doi: 10.1016/0168-9525(93)90121-w. [DOI] [PubMed] [Google Scholar]
- Huang J. C., Svoboda D. L., Reardon J. T., Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5' and the 6th phosphodiester bond 3' to the photodimer. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. S., Allan R. W., Temin H. M. One retroviral RNA is sufficient for synthesis of viral DNA. J Virol. 1994 Jan;68(1):207–216. doi: 10.1128/jvi.68.1.207-216.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai S., Nishizawa M. New procedure for DNA transfection with polycation and dimethyl sulfoxide. Mol Cell Biol. 1984 Jun;4(6):1172–1174. doi: 10.1128/mcb.4.6.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Alexander P. S. The base substitution fidelity of eucaryotic DNA polymerases. Mispairing frequencies, site preferences, insertion preferences, and base substitution by dislocation. J Biol Chem. 1986 Jan 5;261(1):160–166. [PubMed] [Google Scholar]
- Kunkel T. A., Hamatake R. K., Motto-Fox J., Fitzgerald M. P., Sugino A. Fidelity of DNA polymerase I and the DNA polymerase I-DNA primase complex from Saccharomyces cerevisiae. Mol Cell Biol. 1989 Oct;9(10):4447–4458. doi: 10.1128/mcb.9.10.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Misalignment-mediated DNA synthesis errors. Biochemistry. 1990 Sep 4;29(35):8003–8011. doi: 10.1021/bi00487a001. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. The mutational specificity of DNA polymerase-beta during in vitro DNA synthesis. Production of frameshift, base substitution, and deletion mutations. J Biol Chem. 1985 May 10;260(9):5787–5796. [PubMed] [Google Scholar]
- Kunkel T. A. The mutational specificity of DNA polymerases-alpha and -gamma during in vitro DNA synthesis. J Biol Chem. 1985 Oct 15;260(23):12866–12874. [PubMed] [Google Scholar]
- Leis J., Baltimore D., Bishop J. M., Coffin J., Fleissner E., Goff S. P., Oroszlan S., Robinson H., Skalka A. M., Temin H. M. Standardized and simplified nomenclature for proteins common to all retroviruses. J Virol. 1988 May;62(5):1808–1809. doi: 10.1128/jvi.62.5.1808-1809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansky L. M., Temin H. M. Lower mutation rate of bovine leukemia virus relative to that of spleen necrosis virus. J Virol. 1994 Jan;68(1):494–499. doi: 10.1128/jvi.68.1.494-499.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani A., Temin H. M. Purification and properties of spleen necrosis virus DNA polymerase. J Virol. 1975 Oct;16(4):797–806. doi: 10.1128/jvi.16.4.797-806.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., Bell J. B., Kunkel T. A., Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3'----5' exonuclease activity. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak V. K., Temin H. M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6024–6028. doi: 10.1073/pnas.87.16.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak V. K., Temin H. M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs J. L., McAllister R. M., Lennette E. H. Immunofluorescent studies of RD-114 virus replication in cell culture. J Gen Virol. 1974 Oct;25(1):21–29. doi: 10.1099/0022-1317-25-1-21. [DOI] [PubMed] [Google Scholar]
- Roberts J. D., Bebenek K., Kunkel T. A. The accuracy of reverse transcriptase from HIV-1. Science. 1988 Nov 25;242(4882):1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Owen J. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics. 1985 Apr;109(4):633–659. doi: 10.1093/genetics/109.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin H. M. Studies on carcinogenesis by avian sarcoma viruses. 8. Glycolysis and cell multiplication. Int J Cancer. 1968 Mar 15;3(2):273–282. doi: 10.1002/ijc.2910030213. [DOI] [PubMed] [Google Scholar]
- Temin H. M. The DNA provirus hypothesis. Science. 1976 Jun 11;192(4244):1075–1080. doi: 10.1126/science.58444. [DOI] [PubMed] [Google Scholar]
- Varela-Echavarría A., Garvey N., Preston B. D., Dougherty J. P. Comparison of Moloney murine leukemia virus mutation rate with the fidelity of its reverse transcriptase in vitro. J Biol Chem. 1992 Dec 5;267(34):24681–24688. [PubMed] [Google Scholar]
- Watanabe S., Temin H. M. Construction of a helper cell line for avian reticuloendotheliosis virus cloning vectors. Mol Cell Biol. 1983 Dec;3(12):2241–2249. doi: 10.1128/mcb.3.12.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]



