Abstract
CD14 functions as a key pattern recognition receptor for a diverse array of gram-negative and gram-positive cell-wall components in the host innate immune response by binding to pathogen-associated molecular patterns (PAMPs) at partially overlapping binding site(s). To determine the potential contribution of CD14 residues in this pattern recognition, we have examined using solution NMR spectroscopy the binding of three different endotoxin ligands, Lipopolysaccharide, lipoteichoic acid and a PGN-derived compound, muramyl dipeptide to a 15N isotopically labeled 152-residue N-terminal fragment of sCD14 expressed in Pichia pastoris. Mapping of NMR spectral changes upon addition of ligands revealed that the pattern of residues affected by binding of each ligand is partially similar and partially different. This first direct structural observation of the ability of specific residue combinations of CD14 to differentially affect endotoxin binding may help explain the broad specificity of CD14 in ligand recognition and provide a structural basis for pattern recognition. Another interesting finding from the observed spectral changes is that the mode of binding may be dynamically modulated and could provide a mechanism for binding endotoxins with structural diversity through a common binding site.
Keywords: NMR, human CD14 structure, CD14-endotoxin complex, endotoxin, pattern recognition, pichia pastoris expression, isotopic labeling, cell-based assay, lipopolysaccharide, ReLPS, LTA, BODIPY
Clinical manifestation of sepsis typically involves a complex interaction between the host immune system and major bacterial cell wall constituents known as endotoxins. The cluster differentiation antigen, CD14, has been shown to be a key high-affinity cellular receptor for these bacterial endotoxins and plays an important role in endotoxin-induced activation of innate immune cells [1]. In humans, CD14 is constitutively expressed on cell surfaces of monocytes/macrophages as a 55 kDa membrane-bound protein and also exists in a soluble form (sCD14), in serum and bodily fluids in concentrations of 2-6 μg/ml [2, 3]. Recognition and binding of different microbial endotoxins to both mCD14 and sCD14 initiates a signaling cascade mediated by Toll-like receptors (TLRs) that promotes the synthesis and secretion of multiple host-derived inflammatory mediators [4], overactivation of which is ultimately responsible for the deleterious effects of sepsis. Understanding mechanism of endotoxin recognition is therefore critical in development of effective therapeutic strategies targeted against CD14 for prevention of sepsis.
To date, several studies have confirmed interactions of CD14 with a variety of microbial ligands such as lipopolysaccaride (LPS) from gram-negative bacteria, lipoteichoic acid (LTA) and peptidoglycan (PGN) from gram-positive bacteria, lipoproteins, lipoarabinomannan, polymannuronic acids and even with synthetic molecules [1, 5-9]. The ability of CD14 to interact with such structurally diverse endotoxins has led to the hypothesis that CD14 functions as a pattern recognition receptor by recognizing and binding to different pathogen-associated molecular patterns [7]. It is believed that this pattern recognition may be important for high selectivity of CD14-mediated cellular activation and distinctiveness of the signaling pathway in response to specific ligands [10]. However, the mode of recognition of these structurally distinct patterns by CD14 is not clear, since structural details for CD14 in complex with any of its ligands is not available. Deletion mutagenesis experiments and epitope mapping studies with blocking antibodies and proteases have previously identified broad regions on CD14 that are crucial for binding of various microbial ligands (residues 39-44, 57-64, 81-100) and which possess overlapping sites of interaction for these ligands [11-15]. More recently, a high-resolution X-ray crystal structure for mouse sCD14 by itself has been determined [16], in which a potential endotoxin binding pocket can be seen at the N-terminus, with the binding regions identified previously from the mutagenesis experiments clustered around this pocket. However, apart from these general interpretations, no direct structural evidence for differential binding to specific residues of CD14 is available, which is critical in order to confirm the role of CD14 as a pattern recognition receptor and provide a structural basis for endotoxin recognition and binding by CD14.
In order to determine the potential contribution of CD14 residues to endotoxin pattern recognition, we have analyzed the binding of various endotoxin ligands to CD14 at a molecular level using solution NMR spectroscopy. NMR spectral changes were mapped out for binding interactions between residues of a fully functional 152-residue N-terminal domain of CD14 expressed and isotopically labeled in Pichia pastoris, and the cell-wall components from Gram-negative and Gram-positive bacteria, that differ with respect to both structure and ability to stimulate innate host responses to CD14. Comparison of the pattern of residues in CD14 affected by binding of each ligand reveals differential changes in NMR spectra of CD14 for each ligand. However, the pattern of residues affected by binding of these ligands is partially similar and partially different, providing for the first time, direct structural evidence for endotoxin-specific recognition by CD14 and supporting the pattern recognition hypothesis. Another interesting finding from the observed spectral changes is that the mode of binding may be dynamically modulated and may provide a mechanism for pattern recognition.
MATERIALS AND METHODS
Expression and purification of isotopically labeled recombinant sCD14 in Pichia pastoris
The gene sequence encoding the N-terminal endotoxin binding domain of human sCD14 (sCD14-EBD), consisting of 152 residues after the signal peptide sequence (20-171), was synthesized with codon-optimization for yeast (BIO S&T) and cloned into the pPICZαA vector using the manufacturer’s protocol (Invitrogen). The sCD14-EBD pPICZαA construct was transformed into wild-type X-33 Pichia pastoris cells, plated on zeocin-containing (200 μg/mL) agar plates and incubated for 3-4 days until colonies appeared. Individual colonies were screened for selection of best CD14-expressing clones using small-scale cultures. Expression levels were determined by SDS-PAGE analysis of secreted protein in the medium.
For large-scale expression in shake flasks, 1 liter of BMD medium (Invitrogen) was inoculated with the best expressing colony and grown up to an OD600 of 12 (∼24 hrs). The culture was then spun down and the cells resuspended in 250 mL of BMMY medium (Invitrogen). The yeast extract and peptone were filtered through acrodisc mustang E filters (Pall) to reduce endotoxin levels in the media. Isotopic labeling was accomplished by substituting the primary nitrogen source ammonium sulfate with 15N labeled ammonium chloride. Protein expression was carried out for 4 days with addition of methanol everyday at 0.5% of the culture volume. At the end of expression period, cells were spun down and purification of sCD14-EBD initiated by loading the supernatant onto a Talon cobalt-affinity column resin (Clontech). The column was washed with 20 column volumes of PBS buffer (pH 7.4) containing 20 mM imidazole, and the bound CD14 eluted with PBS containing 200 mM imidazole. The protein was further purified by size exclusion chromatography using a HiPrep S-100 column (GE Healthcare) equilibrated with PBS. Pure endotoxin-free fractions of sCD14-EBD were pooled, concentrated and used for characterization by SDS-PAGE and Western blot analysis. Samples of sCD14-EBD (1 μg) were loaded on Novex 18% tris-glycine gels (Invitrogen) in duplicate and run following the manufacturer’s recommendations. Thereafter, half of the gel was Coomassie-blue stained and de-stained overnight while the other half of the gel was transferred onto a PVDF membrane (Pierce) for Western blot analysis. The membrane was incubated with the monoclonal antibody for CD14, MEM-18 and developed in metal enhanced DAB (Pierce) as per manufacturer’s directions.
Circular dichroism spectra in the far-UV region (190-250 nm) were collected for 10 μM samples of sCD14-EBD with and without endotoxins in PBS buffer on an AVIV Model 202 spectrometer at 25 °C. Molar ellipticity values were calculated and plotted against wavelength for displaying the circular dichroism spectrum.
Biological activity assays on recombinant sCD14
For the activity assay, THP1-Blue cells (Invivogen) stably transfected with a NF-κB-inducible reporter secreted embryonic alkaline phosphatase (SEAP) plasmid system were cultured in supplemented RPMI 1640 medium at 37 °C in a 5% CO2 environment. Cells were spun down and resuspended at 1 × 105 cells/mL in fresh growth media. 100 μl of cell suspension was added to each well of a 96 well plate along with sCD14-EBD or rhCD14, rhLBP and individual ligands. The ligands or proteins alone served as positive controls with endotoxin free water and growth media as negative controls. After incubation for 20-24 hours at 37 °C under 5% CO2, 10 μl of cell supernatant was added to 200 μl of Quanti-Blue reagent (Invivogen) to assay for SEAP, further incubated at 37 °C for 18-20 hours, and subsequently read at 630 nm using a Biotek Synergy2 plate reader.
NMR studies of sCD14-endotoxin interactions
CD14 protein samples for the NMR experiments were prepared in PBS buffer with typical concentrations of ∼ 0.2 mM. Stock solutions of endotoxin ligand compounds, ReLPS (List Biological Laboratories), LTA (Invivogen), and MDP (Sigma-Aldrich) ligands were all prepared as stock solutions at 5 mg/mL concentration in D2O to ensure minimal dilution of protein upon addition of the ligand. The solutions were extensively sonicated to ensure complete solubility of the ligands. NMR samples of protein:ligand were prepared in a molar ratio of 1:1, except for MDP, where a ratio of 1:10 was used. rhLBP was also present in all samples at a concentration of 5 μM. Protein:ligand samples were incubated for 30 min at 37 °C to ensure complete binding of ligand [17]. Sensitivity-enhanced 1H-15N HSQC spectra were recorded for each sample with and without the ligand. Data was collected with 1024 complex points in the direct dimension and 128 complex points in the indirect dimension. All NMR experiments were carried out on a Varian Unity Inova 600 MHz spectrometer at 17 °C equipped with a coldprobe. NMR data was processed and analyzed with Felix software package (Molecular Simulations).
Results and Discussion
Expression and isotopic labeling of sCD14 in Pichia pastoris
Expression and purification of recombinant sCD14 from Pichia pastoris has been reported previously by other groups [18, 19]. However, isotopic labeling of sCD14 in an endotoxin-free form for NMR characterization has not been demonstrated in these studies. Herein, we were able to accomplish 15N isotopic labeling in Pichia pastoris in an inexpensive manner with 15N ammonium chloride as the nitrogen source, which allowed us to obtain milligram quantities of uniformly 15N labeled samples of a fully functional fragment of sCD14 corresponding to the first 152 residues at the N-terminus of the molecule that also includes the region for endotoxin binding [20]. Utilization of the Pichia pastoris system with an α-factor signal sequence (Fig. 1) allowed the protein to be secreted into the expression medium, considerably easing purification of the protein. A two-step purification scheme employed for sCD14-EBD resulted in a single band for sCD14-EBD corresponding to a MW of ∼32 kDa as deduced from both SDS-PAGE gel and Western blot analysis (Fig. 1), indicating that the protein is expressed as a single isoform and in an endotoxin-free form, which is quite valuable for high-resolution NMR structural studies. The observed molecular weight is higher than the expected molecular weight of ∼18 kDa, which may be due to the presence of glycosylation at two potential N-glycosylation sites on sCD14-EBD, corresponding to Asn18 and Asn132 [21]. Deglycosylation studies with an enzyme PNGase F that deglycosylates N-glycosylated sites resulted in disappearance of the 32 kDa band and appearance of a single band at ∼20 kDa (data not shown), supporting the presence of glycosylation.
FIG. 1.
(A) Schematic for cloning of sCD14-EBD sequence (residues 20-171) in pPICZα A vector along with a His6-tag for expression in Pichia pastoris. Recombinant sCD14-EBD is secreted into the extracellular medium by the yeast α-factor secretion signal via cleavage at the Kex2 protease site. (B) Analysis of sCD14-EBD expression in Pichia pastoris by SDS-PAGE and Western blot of recombinant sCD14-EBD.
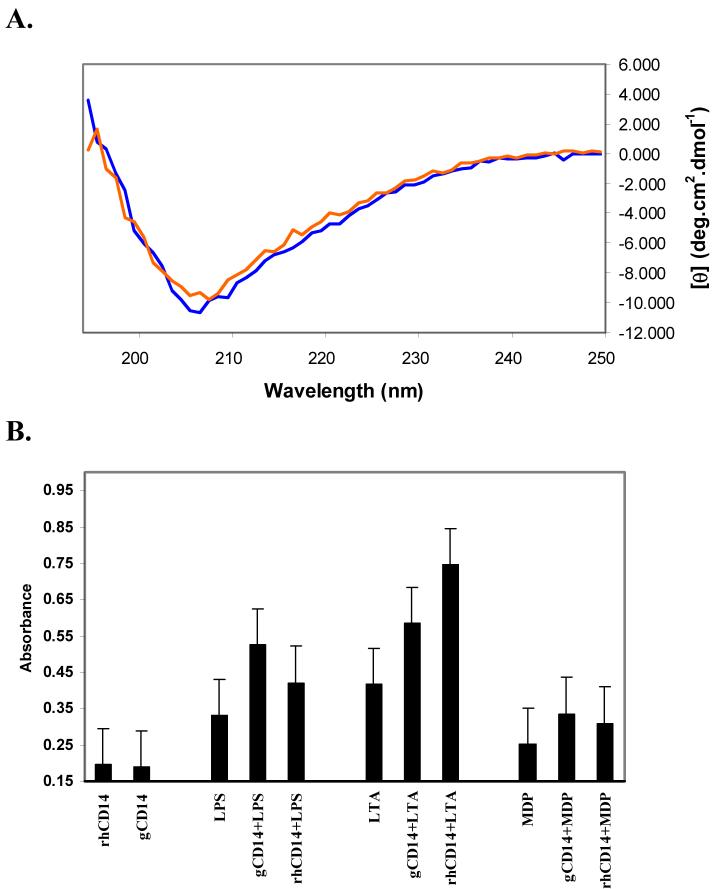
In order to investigate if the purified protein was correctly folded, Circular Dichroism (CD) spectroscopy was used to study the solution secondary structure of CD14-EBD. Fig. 2 shows the far-UV CD spectra of purified sCD14-EBD. As can be seen from the CD spectrum, CD14-EBD contains a mixture of α-helical and β-sheet type structures. This is confirmed by a recent X-ray structure of mouse sCD14 which depicts a α/β fold for the protein [16]. The spectrum of sCD14-EBD changes minimally upon addition of the endotoxin ligand ReLPS, suggesting there are no global conformational changes in the protein upon ligand binding [22]. Similar results were obtained upon binding of other ligands such as LTA and MDP (data not shown).
FIG. 2.
(A) Far-UV circular dichroism spectra of sCD14-EBD (blue line) and sCD14-EBD -LPS complex (red line). Both spectra are average of five scans. (B) Biological activity analysis of sCD14-EBD with endotoxin ligands. Responses of THP1-blue cells mediated by full-length wild-type sCD14 (rhCD14, 400 ng/mL) and purified sCD14-EBD (gCD14, 200 ng/mL) upon stimulation with three endotoxins, LPS (1 μg/mL), LTA (1 μg/mL) and MDP (200 ng/mL). After 24 h incubation, endotoxin stimulation was assessed by measuring the levels of NF-κB inducible reporter alkaline phosphatase (SEAP) using a SEAP detection medium with absorbance values at 630 nm. Control experiments in form of responses to rhCD14 and gCD14 alone are shown as well.
Determination of biological activity of sCD14-EBD
To test the biological activity of sCD14-EBD, the ability of various endotoxin ligands such as LPS, LTA and MDP to induce NF-κB cytokine production was examined within a cell-based assay and whether this effect was enhanced via interaction of the ligands with sCD14-EBD. This assay relies on detection of a NF-κB-inducible reporter, SEAP, upon activation of an established cell line such as THP1-Blue by endotoxins in presence of CD14 (Fig. 2). Compared to activation of THP1-Blue by the protein or ligands alone, addition of LPS to sCD14-EBD and LBP produced almost a three-fold increase in production of NF-κB while LTA showed almost a five-fold increase in line with the observation that LPS and LTA are highly potent activators in mammals. On the other hand, PGN-derived ligand, MDP, showed only a small activation compared to protein or ligand alone. It has been reported previously that sCD14 shows low levels of cellular responsiveness to MDP and that too only at saturating concentrations [8]. Based on similar levels of NF-κB response observed for all ligands in presence of sCD14-EBD and rhCD14, it can also be inferred that they both possess similar abilities to induce NF-κB release by human monocytes [24]. Results from these experiments therefore indicate that our recombinant soluble CD14-EBD can bind and induce cellular effects in response to different endotoxins similar to wild-type protein, confirming its biological activity.
Changes in NMR spectra of CD14 upon endotoxin binding provides structural evidence for pattern recognition
In order to gain a structural perspective regarding the mechanism by which CD14 is able to recognize and differentially bind a diverse array of endotoxin patterns using a common binding site(s), we have investigated the interaction of endotoxins, ReLPS, LTA and MDP using 1H-15N HSQC NMR spectroscopy. These three ligands represent the most common and potent activators of inflammatory response in mammals. While ReLPS and LTA are known agonists, MDP, a PGN-derived ligand, displays similar agonist properties as PGN since it is the minimal structure required for adjuvant activity of PGN and has been known to competitively inhibit soluble PGN binding to CD14 [8,11]. The binding studies described here constitute the first molecular level characterization of structural interactions between a protein receptor and endotoxins in the innate immune system.
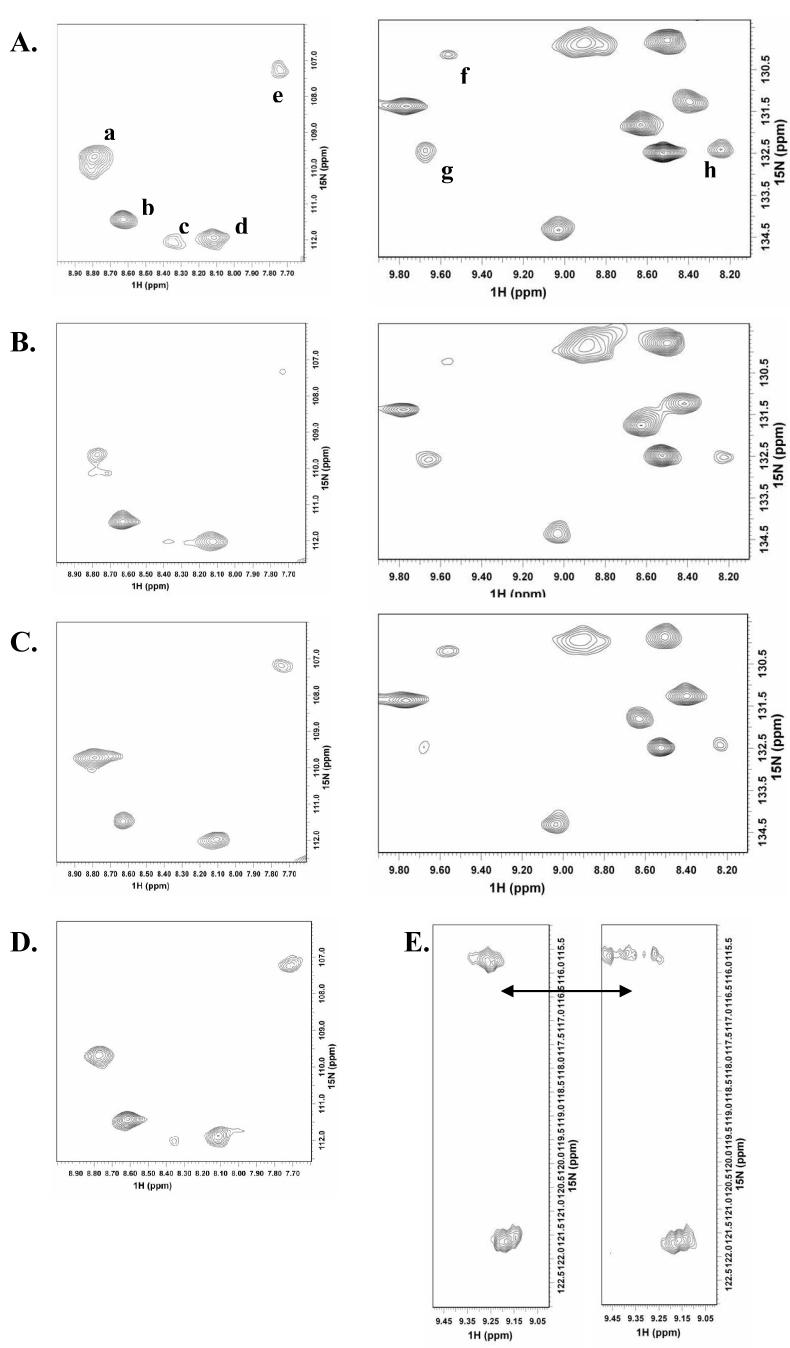
Analysis of NMR spectra with and without the endotoxin ligands reveals no drastic overall change in the protein HSQC spectra, which is in line with the circular dichroism analysis where no major global structural changes are observed. However, several of the 1H and 15N backbone amide peaks show specific changes in their resonances with the addition of each ligand, possibly from local structural changes. Both line-broadening and chemical shift changes are observed in the spectra with ReLPS and LTA, but in the case of MDP, only small chemical shift perturbations can be seen. A comparison of selected regions in the HSQC spectra that exhibit largest changes upon ligand addition is shown in Fig. 3 (A-D). The most striking characteristic observed in these regions is the pattern of CD14 resonances affected by each ligand. It is evident from the spectra that all three ligands affect similar resonances in the CD14 spectra. However, the identity of resonances affected by individual ligands differs. For example, ReLPS and LTA both affect peaks “c” and “h” in the spectra, with all three ligands affecting peak “c”, however peaks “e” and “f” are affected only by ReLPS, while LTA affects only peaks “b” and “g” in the regions shown. Our observation here of the ability of specific residues to differentially affect endotoxin binding therefore demonstrates previously unobserved residue level binding characteristics for CD14 and provides a structural basis for pattern recognition. It is possible that each endotoxin may affect directly or indirectly a different set of residues depending on the characteristic molecular pattern it possesses giving broad ligand specificity to CD14, while several of the structural features may be shared between the binding endotoxins to maintain spatial and functional similarity in the interaction site. Since assignments for CD14 are not available right now, it is not possible to locate the exact regions of ligand binding on the sequence of CD14. However, it is likely that the regions shown are likely the binding sites of the ligands, as chemical shift perturbations in case of MDP and broadening in case of ReLPS and LTA are much smaller in other spectral regions of CD14, although spectral changes due to remote conformational changes in the protein upon ligand binding cannot be ruled out at this stage. Regardless of whether the changes are a result of direct binding or caused indirectly, it is clear that recognition of endotoxin ligands depends on specific contributions of select CD14 residues that may have implications for pattern recognition.
FIG. 3.
Comparison of changes upon endotoxin ligand addition for a representative set of resonances in the 600 MHz 15N,1H HSQC spectra of 15N labeled sCD14-EBD. (A) sCD14-EBD only, (B) sCD14-EBD in 1:1 complex with ReLPS, (C) sCD14-EBD in 1:1 complex with LTA and (D) sCD14-EBD in 1:10 complex with MDP. (E) Splitting of a representative resonance in sCD14-EBD (left sub-spectrum) into multiple peaks upon complexation with ReLPS (right sub-spectrum), indicating change in dynamics to slower exchange on the NMR time scale.
Another significant feature observed in the NMR spectra of CD14, particularly with bound ReLPS and LTA is that, in a number of cases, the resonances affected by binding are split into multiple peaks (Fig. 3E). We interpret this as resulting from a change in dynamics of flexible regions of CD14 from fast to slow exchange on the NMR chemical shift time scale, signifying occupancy of various conformational substates selected for binding from within a dynamic ensemble of interconverting population. Interestingly, regions previously identified in endotoxin binding map to a highly flexible hydrophilic rim and also the bottom of a large hydrophobic pocket based on the crystal structure of mouse sCD14 [16]. While the hydrophobic pocket is fairly rigid, the residues on the hydrophilic rim exhibit multiple conformations in addition to high average temperature factors. The electron density for this region is also quite poorly defined in the crystal structure. Based on these observations, we surmise that the flexibility of the hydrophilic rim would allow it to interact with structurally diverse sugar moieties and concurrently, the rigid but large nature of the hydrophobic pocket would allow it to accommodate ligands such as LPS and LTA with structural variations in the hydrophobic portion. We therefore propose that while structural characteristics of the binding site incorporating specific residue combinations can help impart CD14 broad specificity in ligand recognition, this dynamic modulation of CD14 may also play an important role in providing a mechanism for binding endotoxins with structural diversity through a common binding site. The NMR observations on structural and dynamic changes presented here certainly lend credence to this possibility.
Acknowledgement
This work was supported by a National Institutes of Health grant R01-AI060818 to Nitin Jain.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding-protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- [2].Lee JD, Kravchenko V, Kirkland TN, Han J, Mackman N, Moriarty A, Leturcq D, Tobias PS, Ulevitch RJ. Glycosyl-phosphatidylinositol-anchored or integral membrane forms of CD14 mediate identical cellular-responses to endotoxin. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:9930–9934. doi: 10.1073/pnas.90.21.9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Frey EA, Miller DS, Jahr TG, Sundan A, Bazil V, Espevik T, Finlay BB, Wright SD. Soluble CD14 participates in the response of cells to lipopolysaccharide. Journal of Experimental Medicine. 1992;176:1665–1671. doi: 10.1084/jem.176.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, Schroeder L, Aderem A. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between Toll-like receptors. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dziarski R, Ulmer AJ, Gupta D. Karger; Basel: 2000. Interactions of CD14 with components of gram-positive bacteria, CD14 in the Inflammatory Response; pp. 83–107. [DOI] [PubMed] [Google Scholar]
- [6].Cleveland MG, Gorham JD, Murphy TL, Tuomanen E, Murphy KM. Lipoteichoic acid preparations of grain-positive bacteria induce interleukin-12 through a CD14-dependent pathway. Infection and Immunity. 1996;64:1906–1912. doi: 10.1128/iai.64.6.1906-1912.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pugin J, Heumann D, Tomasz A, Kravchenko VV, Akamatsu Y, Nishijima M, Glauser MP, Tobias PS, Ulevitch RJ. CD14 is a pattern recognition receptor. Immunity. 1994;1:509–516. doi: 10.1016/1074-7613(94)90093-0. [DOI] [PubMed] [Google Scholar]
- [8].Dziarski R, Tapping RI, Tobias PS. Binding of bacterial peptidoglycan to CD14. Journal of Biological Chemistry. 1998;273:8680–8690. doi: 10.1074/jbc.273.15.8680. [DOI] [PubMed] [Google Scholar]
- [9].Weidemann B, Brade H, Rietschel ET, Dziarski R, Bazil V, Kusumoto S, Flad HD, Ulmer AJ. Soluble peptidoglycan-induced monokine production can be blocked by anti-CD14 monoclonal-antibodies and by Lipid A partial structures. Infection and Immunity. 1994;62:4709–4715. doi: 10.1128/iai.62.11.4709-4715.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Delude RL, Savedra R, Zhao HL, Thieringer R, Yamamoto S, Fenton MJ, Golenbock DT. CD14 enhances cellular-responses to endotoxin without imparting ligand-specific recognition. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:9288–9292. doi: 10.1073/pnas.92.20.9288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dziarski R, Viriyakosol S, Kirkland TN, Gupta D. Soluble CD14 enhances membrane CD14-mediated responses to peptidoglycan: Structural requirements differ from those for responses to lipopolysaccharide. Infection and Immunity. 2000;68:5254–5260. doi: 10.1128/iai.68.9.5254-5260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Juan TSC, Hailman E, Kelley MJ, Busse LA, Davy E, Empig CJ, Narhi LO, Wright SD, Lichenstein HS. Identification of a lipopolysaccharide-binding domain in CD14 between amino-acid 57 and amino-acid 64. Journal of Biological Chemistry. 1995;270:5219–5224. doi: 10.1074/jbc.270.10.5219. [DOI] [PubMed] [Google Scholar]
- [13].Stelter F, Bernheiden M, Menzel R, Jack RS, Witt S, Fan XL, Pfister M, Schutt C. Mutation of amino acids 39-44 of human CD14 abrogates binding of lipopolysaccharide and Escherichia coli. European Journal of Biochemistry. 1997;243:100–109. doi: 10.1111/j.1432-1033.1997.00100.x. [DOI] [PubMed] [Google Scholar]
- [14].Voss S, Welte S, Fotin-Mleczek M, Fischer R, Ulmer AJ, Jung G, Wiesmuller KH, Brock R. A CD14 domain with lipopolysaccharide-binding and -neutralizing activity. Chembiochem. 2006;7:275–286. doi: 10.1002/cbic.200500257. [DOI] [PubMed] [Google Scholar]
- [15].Cunningham MD, Shapiro RA, Seachord C, Ratcliffe K, Cassiano L, Darveau RP. CD14 employs hydrophilic regions to “capture” lipopolysaccharides. Journal of Immunology. 2000;164:3255–3263. doi: 10.4049/jimmunol.164.6.3255. [DOI] [PubMed] [Google Scholar]
- [16].Kim JI, Lee CJ, Jin MS, Lee CH, Paik SG, Lee H, Lee JO. Crystal structure of CD14 and its implications for lipopolysaccharide signaling. Journal of Biological Chemistry. 2005;280:11347–11351. doi: 10.1074/jbc.M414607200. [DOI] [PubMed] [Google Scholar]
- [17].Yu B, Wright SD. Catalytic properties of lipopolysaccharide (LPS) binding protein - Transfer of LPS to soluble CD14. Journal of Biological Chemistry. 1996;271:4100–4105. doi: 10.1074/jbc.271.8.4100. [DOI] [PubMed] [Google Scholar]
- [18].Majerle A, Kidric J, Jerala R. Expression and refolding of functional fragments of the human lipopolysaccharide receptor CD14 in Escherichia coli and Pichia pastoris. Protein Expression and Purification. 1999;17:96–104. doi: 10.1006/prep.1999.1109. [DOI] [PubMed] [Google Scholar]
- [19].Nomura S, Inamori K, Muta T, Yamazaki S, Sunakawa Y, Iwanaga S, Takeshige K. Purification and characterization of human soluble CD14 expressed in Pichia pastoris. Protein Expression and Purification. 2003;28:310–320. doi: 10.1016/s1046-5928(02)00705-2. [DOI] [PubMed] [Google Scholar]
- [20].Juan TSC, Kelley MJ, Johnson DA, Busse LA, Hailman E, Wright SD, Lichenstein HS. Soluble CD14 truncated at amino-acid 152 binds lipopolysaccharide (LPS) and enables cellular-response to LPS. Journal of Biological Chemistry. 1995;270:1382–1387. doi: 10.1074/jbc.270.3.1382. [DOI] [PubMed] [Google Scholar]
- [21].Stelter F, Pfister M, Bernheiden M, Jack RS, Bufler P, Engelmann H, Schutt C. The myeloid differentiation antigen CD14 is N- and O-glycosylated - Contribution of N-linked glycosylation to different soluble CD14 isoforms. European Journal of Biochemistry. 1996;236:457–464. doi: 10.1111/j.1432-1033.1996.00457.x. [DOI] [PubMed] [Google Scholar]
- [22].McGinley MD, Narhi LO, Kelley MJ, Davy E, Robinson J, Rohde MF, Wright SD, Lichenstein HS. CD14 - Physical properties and identification of an exposed site that is protected by lipopolysaccharide. Journal of Biological Chemistry. 1995;270:5213–5218. doi: 10.1074/jbc.270.10.5213. [DOI] [PubMed] [Google Scholar]
- [23].Tobias PS, Soldau K, Gegner JA, Mintz D, Ulevitch RJ. Lipopolysaccharide binding protein-mediated complexation of lipopolysaccharide with soluble CD14. Journal of Biological Chemistry. 1995;270:10482–10488. doi: 10.1074/jbc.270.18.10482. [DOI] [PubMed] [Google Scholar]
- [24].Kirkland TN, Finley F, Leturcq D, Moriarty A, Lee JD, Ulevitch RJ, Tobias PS. Analysis of lipopolysaccharide binding by CD14. Journal of Biological Chemistry. 1993;268:24818–24823. [PubMed] [Google Scholar]