Abstract
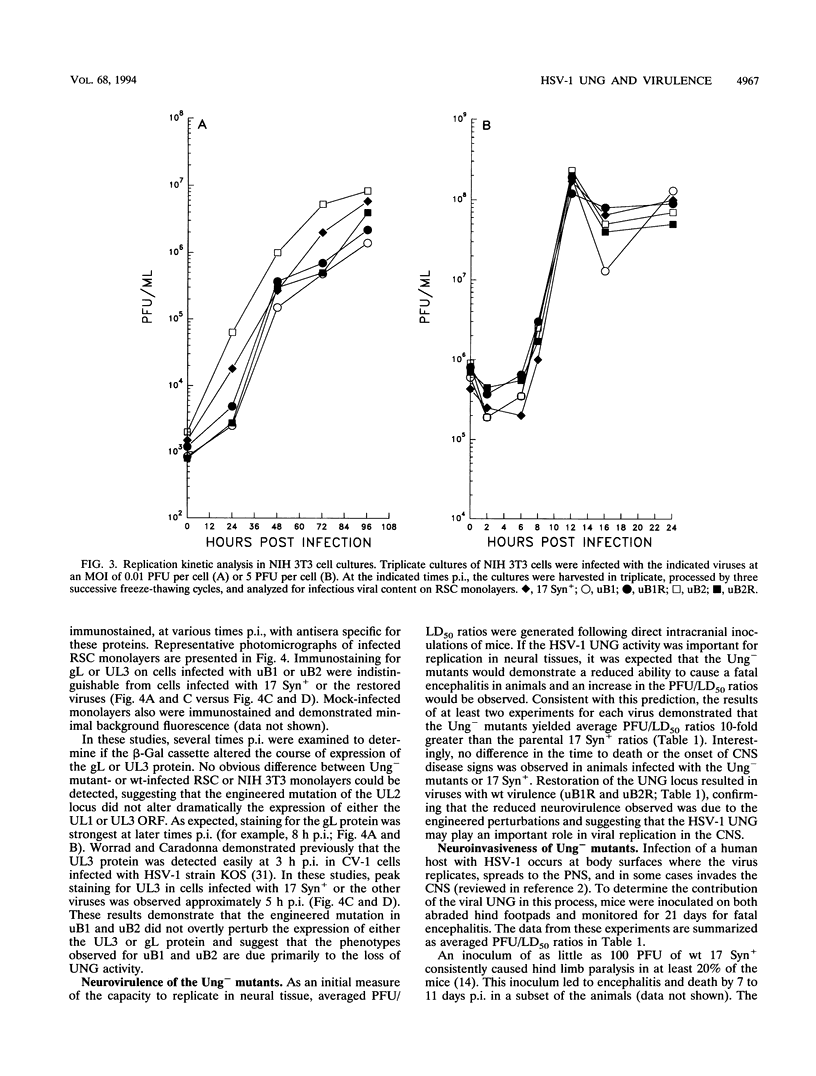
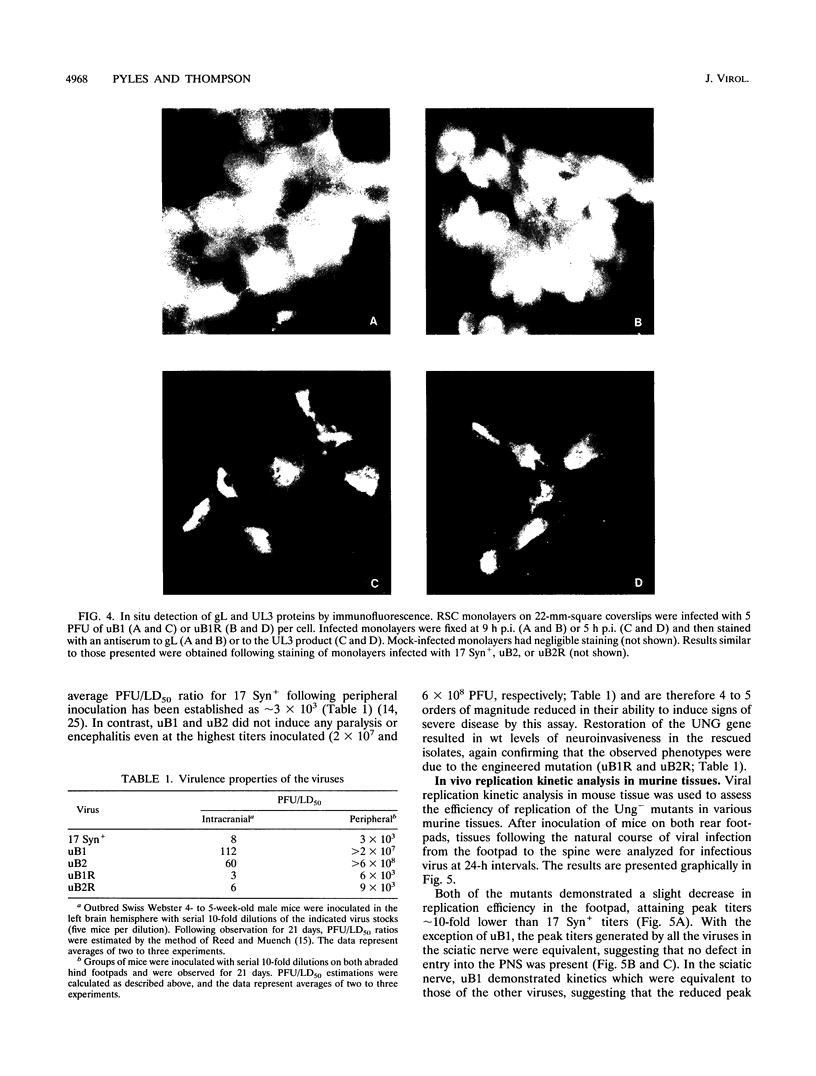
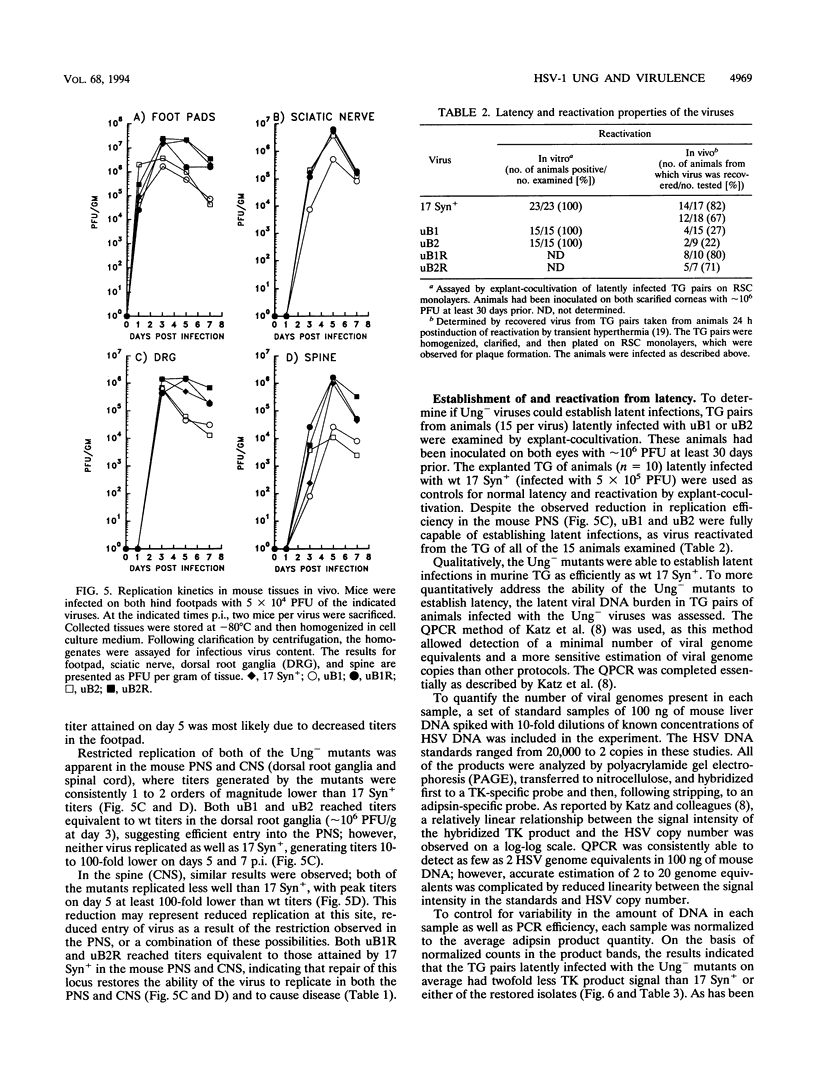
Herpes simplex virus (HSV) encodes a uracil DNA glycosylase (UNG; UL2), which has been shown to be dispensable for normal replication of HSV-1 in cultured cells (J. Mullaney, H.W. Moss, and D.J. McGeoch, J. Gen. Virol. 70:449-454, 1989). In adult neurons, UNG activity is undetectable (F. Focher, P. Mazzarello, A. Verri, U. Hubscher, and S. Spadari, Mutat. Res. 237:65-73, 1990), suggesting that the HSV-1 UNG may play an important role in viral replication in neurons acutely and/or following reactivation. To examine the contribution of the HSV-1 UNG in vivo, two independent strain 17 Syn+ Ung- mutants, designated uB1 and uB2, were examined in a mouse model of herpetic disease. Following direct intracranial inoculation, both mutants exhibited a 10-fold reduction in neurovirulence compared with the parental strain 17 Syn+. Inoculations by a peripheral route demonstrated that the Ung- mutants were at least 100,000-fold less neuroinvasive than 17 Syn+. Replication kinetics in vivo demonstrated that uB1 and uB2 replicated less well in both the mouse peripheral and central nervous systems. Latency was established by both of the mutants in 100% of the animals examined. Following transient hyperthermia, however, the frequency of reactivation of the mutants in vivo was dramatically reduced. Restoration of the UNG locus resulted in full neurovirulence, neuroinvasiveness, and the ability to reactivate in vivo. These findings suggest that the HSV-1 UNG plays an important role during acute viral replication in vivo and possibly in the reactivation process.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baines J. D., Roizman B. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J Virol. 1991 Feb;65(2):938–944. doi: 10.1128/jvi.65.2.938-944.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field H. J., Wildy P. The pathogenicity of thymidine kinase-deficient mutants of herpes simplex virus in mice. J Hyg (Lond) 1978 Oct;81(2):267–277. doi: 10.1017/s0022172400025109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focher F., Mazzarello P., Verri A., Hübscher U., Spadari S. Activity profiles of enzymes that control the uracil incorporation into DNA during neuronal development. Mutat Res. 1990 Mar;237(2):65–73. doi: 10.1016/0921-8734(90)90012-g. [DOI] [PubMed] [Google Scholar]
- Focher F., Verri A., Verzeletti S., Mazzarello P., Spadari S. Uracil in OriS of herpes simplex 1 alters its specific recognition by origin binding protein (OBP): does virus induced uracil-DNA glycosylase play a key role in viral reactivation and replication? Chromosoma. 1992;102(1 Suppl):S67–S71. doi: 10.1007/BF02451788. [DOI] [PubMed] [Google Scholar]
- Hutchinson L., Browne H., Wargent V., Davis-Poynter N., Primorac S., Goldsmith K., Minson A. C., Johnson D. C. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J Virol. 1992 Apr;66(4):2240–2250. doi: 10.1128/jvi.66.4.2240-2250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idowu A. D., Fraser-Smith E. B., Poffenberger K. L., Herman R. C. Deletion of the herpes simplex virus type 1 ribonucleotide reductase gene alters virulence and latency in vivo. Antiviral Res. 1992 Feb;17(2):145–156. doi: 10.1016/0166-3542(92)90048-a. [DOI] [PubMed] [Google Scholar]
- Katz J. P., Bodin E. T., Coen D. M. Quantitative polymerase chain reaction analysis of herpes simplex virus DNA in ganglia of mice infected with replication-incompetent mutants. J Virol. 1990 Sep;64(9):4288–4295. doi: 10.1128/jvi.64.9.4288-4295.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz B. A., Kohalmi S. E. Modulation of mutagenesis by deoxyribonucleotide levels. Annu Rev Genet. 1991;25:339–359. doi: 10.1146/annurev.ge.25.120191.002011. [DOI] [PubMed] [Google Scholar]
- Lindahl T. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog Nucleic Acid Res Mol Biol. 1979;22:135–192. doi: 10.1016/s0079-6603(08)60800-4. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Dalrymple M. A., Davison A. J., Dolan A., Frame M. C., McNab D., Perry L. J., Scott J. E., Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988 Jul;69(Pt 7):1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- Mullaney J., Moss H. W., McGeoch D. J. Gene UL2 of herpes simplex virus type 1 encodes a uracil-DNA glycosylase. J Gen Virol. 1989 Feb;70(Pt 2):449–454. doi: 10.1099/0022-1317-70-2-449. [DOI] [PubMed] [Google Scholar]
- Ostrander M., Cheng Y. C. Properties of herpes simplex virus type 1 and type 2 DNA polymerase. Biochim Biophys Acta. 1980 Sep 19;609(2):232–245. doi: 10.1016/0005-2787(80)90234-8. [DOI] [PubMed] [Google Scholar]
- Pyles R. B., Sawtell N. M., Thompson R. L. Herpes simplex virus type 1 dUTPase mutants are attenuated for neurovirulence, neuroinvasiveness, and reactivation from latency. J Virol. 1992 Nov;66(11):6706–6713. doi: 10.1128/jvi.66.11.6706-6713.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- Roop C., Hutchinson L., Johnson D. C. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J Virol. 1993 Apr;67(4):2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawtell N. M., Thompson R. L. Herpes simplex virus type 1 latency-associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. J Virol. 1992 Apr;66(4):2157–2169. doi: 10.1128/jvi.66.4.2157-2169.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawtell N. M., Thompson R. L. Rapid in vivo reactivation of herpes simplex virus in latently infected murine ganglionic neurons after transient hyperthermia. J Virol. 1992 Apr;66(4):2150–2156. doi: 10.1128/jvi.66.4.2150-2156.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J., Wagner E. K. Transcriptional analysis of the herpes simplex virus type 1 region containing the TRL/UL junction. Virology. 1993 Sep;196(1):220–231. doi: 10.1006/viro.1993.1470. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spector R., Boose B. Development and regional distribution of deoxyuridine 5'-triphosphatase in rabbit brain. J Neurochem. 1983 Oct;41(4):1192–1195. doi: 10.1111/j.1471-4159.1983.tb09073.x. [DOI] [PubMed] [Google Scholar]
- Thompson R. L., Devi-Rao G. V., Wagner E. K. DNA sequence and RNA transcription through a site of recombination in a non-neurovirulent herpes simplex virus intertypic recombinant. Virus Genes. 1988 Jun;1(3):275–286. doi: 10.1007/BF00572706. [DOI] [PubMed] [Google Scholar]
- Thompson R. L., Rogers S. K., Zerhusen M. A. Herpes simplex virus neurovirulence and productive infection of neural cells is associated with a function which maps between 0.82 and 0.832 map units on the HSV genome. Virology. 1989 Oct;172(2):435–450. doi: 10.1016/0042-6822(89)90186-4. [DOI] [PubMed] [Google Scholar]
- Thompson R. L., Wagner E. K. Partial rescue of herpes simplex virus neurovirulence with a 3.2 kb cloned DNA fragment. Virus Genes. 1988 Jun;1(3):261–273. doi: 10.1007/BF00572705. [DOI] [PubMed] [Google Scholar]
- Thompson R. L., Wagner E. K., Stevens J. G. Physical location of a herpes simplex virus type-1 gene function(s) specifically associated with a 10 million-fold increase in HSV neurovirulence. Virology. 1983 Nov;131(1):180–192. doi: 10.1016/0042-6822(83)90544-5. [DOI] [PubMed] [Google Scholar]
- Verri A., Mazzarello P., Biamonti G., Spadari S., Focher F. The specific binding of nuclear protein(s) to the cAMP responsive element (CRE) sequence (TGACGTCA) is reduced by the misincorporation of U and increased by the deamination of C. Nucleic Acids Res. 1990 Oct 11;18(19):5775–5780. doi: 10.1093/nar/18.19.5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verri A., Mazzarello P., Spadari S., Focher F. Uracil-DNA glycosylases preferentially excise mispaired uracil. Biochem J. 1992 Nov 1;287(Pt 3):1007–1010. doi: 10.1042/bj2871007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winters T. A., Williams M. V. Use of the PBS2 uracil-DNA glycosylase inhibitor to differentiate the uracil-DNA glycosylase activities encoded by herpes simplex virus types 1 and 2. J Virol Methods. 1990 Sep;29(3):233–242. doi: 10.1016/0166-0934(90)90051-g. [DOI] [PubMed] [Google Scholar]
- Worrad D. M., Caradonna S. Identification of the coding sequence for herpes simplex virus uracil-DNA glycosylase. J Virol. 1988 Dec;62(12):4774–4777. doi: 10.1128/jvi.62.12.4774-4777.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrad D. M., Caradonna S. The herpes simplex virus type 2 UL3 open reading frame encodes a nuclear localizing phosphoprotein. Virology. 1993 Aug;195(2):364–376. doi: 10.1006/viro.1993.1386. [DOI] [PubMed] [Google Scholar]
- Yamagami S., Mori K., Kawakita Y. Changes of thymidine kinase in the developing rat brain. J Neurochem. 1972 Feb;19(2):369–376. doi: 10.1111/j.1471-4159.1972.tb01346.x. [DOI] [PubMed] [Google Scholar]