Abstract
The ab initio structures of 2,7,9-tricarboxypyrroloquinoline quinone (PQQ), semiquinone (PQQH), and dihydroquinone (PQQH2) have been determined and compared with ab initio structures of the (PQQ)Ca2+, (PQQH)Ca2+, and (PQQH2)Ca2+ complexes as well as the x-ray structure of (PQQ)Ca2+ bound at the active site of the methanol dehydrogenase (MDH) of methyltropic bacteria. Plausible mechanisms for the MDH oxidation of methanol involving the (PQQ)Ca2+ complex are explored via ab initio computations and discussed. Considering the reaction of methanol with PQQ in the absence of Ca2+, nucleophilic addition of methanol to the PQQ C-5 carbonyl followed by a retro-ene elimination is deemed unlikely due to large energy barrier. A much more favorable disposition of the methanol C-5 adduct to provide formaldehyde involves proton ionization of the intermediate followed by elimination of methoxide concerted with hydride transfer to the oxygen of the C-4 carbonyl. Much the same transition state is reached if one searches for the transition state beginning with Asp-303–CO2−general-base removal of the methanol proton of the (PQQ)Ca2+O(H)CH3 complex concerted with hydride transfer to the oxygen at C-4. For such a mechanism the role of the Ca2+ moiety would be to (i) contribute to the formation of the ES complex (ii) provide a modest decrease in the pKa of methanol substrate,; and (iii) polarize the oxygen at C-5.
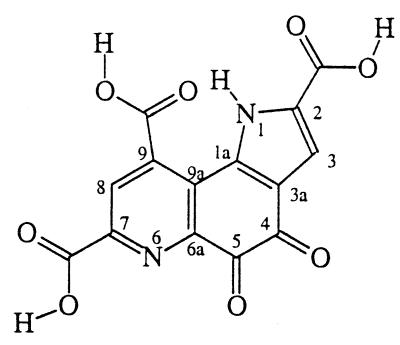
We have had an extended interest in the chemistry of 2,7,9-tricarboxypyrroloquinoline quinone (pyrroloquinoline quinone, PQQ, and the older name methoxatin) and the mechanism of amine oxidation by PQQ (Structure s1) (1–5).
Methanol dehydrogenase (MDH; EC 1.1.99.8) is a quinoprotein that requires Ca2+ as well as PQQ cofactor (6–8) for activity in the oxidation of methanol to formaldehyde (9–15). Three-dimensional structures of MDH from methylotropic bacteria have been solved independently by two groups (16–19). These crystal structures provide detailed information regarding the interaction between PQQ and MDH (13–19). According to these crystal structures, Ca2+ binds to PQQ through N-6, C-5 carbonyl oxygen, and C-7 carboxylate. Protein Glu and Asn side chains occupy the remaining Ca2+ coordination sites. The protein structure must be responsible for ligation of Ca2+ to the N-6, C-5 carbonyl oxygen, and C-7 carboxylate functions of PQQ. Thus, in water the ligating agent α-picolinate (pyridine-2-carboxylate) has little affinity for Ca2+, whereas transition metals are ligated by the pyridine nitrogen and carboxylate oxygen. The role of Ca2+ has been assumed to be structural, although there have been persistent speculations (13–15) that it may also play a catalytic role. Indeed, a recent study by Itoh et al. (20) has undoubtedly demonstrated a catalytic role for Ca2+ in a model system. Their study revealed that the trimethyl ester of PQQ (PQQ–TME) is capable of oxidizing methanol to formaldehyde in anhydrous acetonitrile when treated with Ca(ClO4)2 in the presence of an organic base (20). In these experiments the outer sphere ClO4− ligand and aprotic solvent should enhance the complexing ability of the Ca2+ catalyst. On the other hand the carboxyl groups of PQQ–TME should have little affinity for Ca2+.
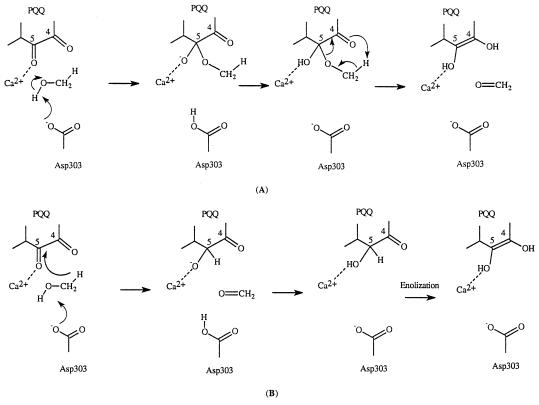
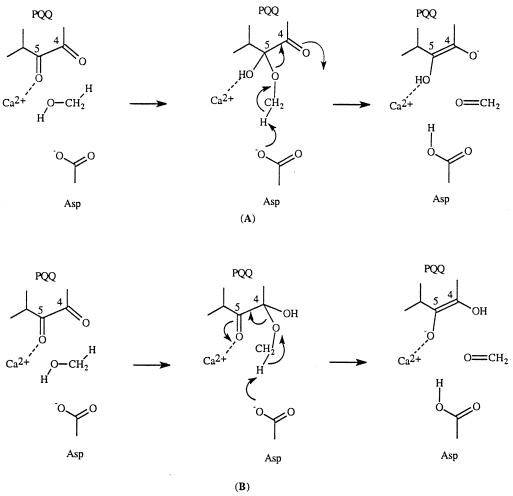
A number of possible mechanisms have been proposed for MDH over the years (13–15). Several of these proposed mechanisms have been proved to be invalid when the x-ray crystal structures became available. Two frequently mentioned mechanisms are shown in Scheme S2. The first mechanism involves a general base (Asp-303–CO2−) catalyzed addition of methanol to the PQQ carbonyl group at C5 followed by a retro-ene reaction and the second mechanism involves general base (Asp-303–CO2−) catalyzed proton abstraction concerted with hydride transfer. The retro-ene reaction in the first mechanism is not expected to be facile. Thus, retro-ene reactions are normally extremely slow and are generally carried out at high temperature (21, 22).
Scheme 2.
In this study we provide a theoretical approach to evaluate the role of Ca2+ in the catalytic mechanism of MDH using an active site model based on the coordinates of the PQQ/Ca2+/MDH structure. In the course of the study we investigated the structure of PQQ in its different oxidation states and how its structure and conformation change in the presence of Ca2+.
THEORETICAL PROCEDURE
All ab initio and semiempirical molecular orbital calculations were carried out using the Guassian 94 program (23). For the ab initio calculations, geometry optimizations were performed at the Hartree–Fock (HF) level with two different basis sets [3-21G(d) and 6-31G(d)]. For radicals, the unrestricted HF (24) method was employed. Semiempirical molecular orbital calculations were done using the PM3 (25) Hamiltonian; these calculations were carried out to provide reasonable initial geometries for the ab initio molecular orbital calculations. The fully reduced PQQ is designated as PQQH2 (dihydroquinone PQQ) and the semiquinone PQQ is called PQQH.
The reaction of PQQ with methanol was examined prior to studies of the effect of Ca2+ on this reaction. In following calculations, only PQQ, methoxide, and Ca2+ were included, and the protein ligands around Ca2+ were not included. As a result, the calculated energetics are only approximate. A larger and more accurate active site model would have to include the protein ligands and some of the polar groups in the active site, which is presently not treatable by high level ab initio molecular orbital theory. However, the essential chemistry should be valid even with the small active site model.
RESULTS AND DISCUSSION
Conformation of PQQ.
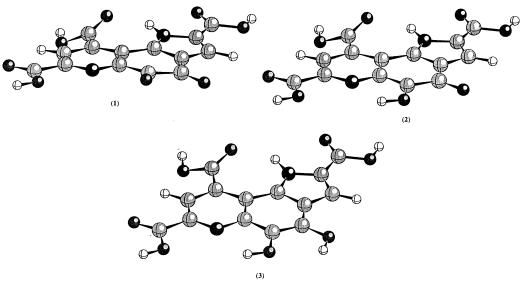
PQQ is very susceptible to nucleophilic attack and it readily forms adducts with acetone, alcohols, and other compounds. The first crystallographic study was actually on PQQ–acetone adduct, not on PQQ itself (6). Later on, it was possible to get crystals of Na+ and K+ salts of PQQ (26, 27). Crystallographic studies with the salts have shown that the tricylic ring of PQQ is planar; the three carboxylate groups can be either coplanar with the ring or twisted out of the plane. In these crystals, PQQ forms extended hydrogen bonding and metal ion coordination networks, and each layer of PQQs stacks nicely on top of each other. As a result, it is not clear whether the observed planarity of the tricyclic ring is an inherent property of PQQ itself or a manifestation of crystal packing forces. Further, there is scarce information concerning the structure of PQQH radicals and the reduced PQQ. To address this issue, we examined the structures of PQQ in different oxidation states using ab initio molecular orbital theory at the HF/6-31G(d) level. The optimized geometries and the calculated electronic energies are given in Fig. 1 and Table 1, respectively.
Figure 1.
The optimized geometries for 1–3 at HF/6-31G(d) level of theory.
Table 1.
Some geometrical parameters for PQQ, PQQH, and PQQH2 at the HF/6-31G(d) level of theory
| Bond | PQQ | PQQH | PQQH2 |
|---|---|---|---|
| N1-C2 | 1.3645 | 1.3629 | 1.3595 |
| N1-C1a | 1.3402 | 1.3618 | 1.3570 |
| C2-C3 | 1.3611 | 1.3754 | 1.3597 |
| C3-C3a | 1.4096 | 1.4075 | 1.4160 |
| C3a-C1a | 1.3875 | 1.4061 | 1.3971 |
| C3a-C4 | 1.4545 | 1.4510 | 1.4199 |
| C4-C5 | 1.5338 | 1.4409 | 1.3463 |
| C4-O | 1.1888 | 1.2316 | 1.3417 |
| C5-O | 1.1813 | 1.3290 | 1.3572 |
| C5-C6a | 1.5191 | 1.4238 | 1.4317 |
| C6a-C9a | 1.4127 | 1.4517 | 1.4251 |
| C9a-C1a | 1.4787 | 1.4392 | 1.4374 |
| C6a-N6 | 1.3118 | 1.3442 | 1.3362 |
| N6-C7 | 1.3110 | 1.3226 | 1.2964 |
| C7-C8 | 1.3811 | 1.4011 | 1.3974 |
| C8-C9 | 1.3865 | 1.4008 | 1.3723 |
| C9-C9a | 1.4079 | 1.4283 | 1.4236 |
Note that energy is given in a.u. (where 1 a.u. = 627.5 kcal/mol): PQQ, −1241.50639; PQQH, −1242.11445; and PQQH2, −1242.68725.
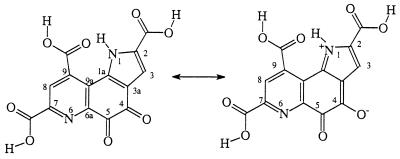
As revealed by Fig. 1, the free PQQ is not completely planar. There is small distortion in the tricyclic ring. The C-4 and C-5 carbonyl oxygens of PQQ are slightly twisted out of plane to alleviate repulsive interactions. There is a good hydrogen bond between the N1-H and the carbonyl oxygen at C-9. Two of the three carboxyl groups, C-2 and C-7 carboxyl groups, are coplanar with the rings, but the C-9 carboxyl group is twisted out of the pyridine ring by about 28°. Because these carboxyl groups are connected to the ring system through C—C single bonds, it would not be energetically very costly for these carboxyl groups to move out of the tricyclic ring plane. Therefore, the carboxyl groups can be either coplanar or twisted out of plane depending on the environment. In the crystal structure of MDH (16, 17) and the crystal structures of PQQ–Na+ and PQQ–K+ salts (26, 27), the C-2 and C-7 carboxyls are indeed coplanar with the tricyclic ring and the C-9 carboxyl is twisted out of plane. However, in a crystal structure of PQQ(H2O) (monohydrate of PQQ) (27), the C-9 carboxyl group appears to be coplanar with the tricyclic ring system. As indicated by the calculated geometrical parameters, the two quinone carbonyl groups are not well conjugated with either the pyrrole or the pyridine ring while the adjacency of the pyrrole ring decreases the electron density of the quinone. As a result, these two quinone carbonyls are very reactive. Based on the geometrical parameters, the C-5 carbonyl is expected to be more active than the C-4 one, in agreement with experimental observation (13–15, 28, 29) that C-5 adducts are favored at neutral pH. However, owing to the resonance contribution as shown in Scheme S3, the C-4 carbonyl oxygen is more basic than the C-5 carbonyl oxygen; therefore, under acidic condition, formation of the C-4 adduct would be preferred as observed experimentally (29). We also calculated the energy difference between the two PQQ–methanol adducts (C-5 and C-4 adducts) with the C-5 adduct being favored by only 0.9 kcal/mol at the HF/6-31G(d) level of theory. Semiempirical molecular orbital calculations give a larger energy difference with the C-5 adduct being favored (29). The radical anion of PQQ is also planar.
Scheme 3.
Semiquinone Radical (PQQH).
As shown in Fig. 1, the unrestricted HF/6-31G(d) optimized structure of neutral semiquinone radical is planar. According to our calculation, the spin density is distributed over the entire tricyclic ring. Although no direct structural information is available for PQQH, early ESR studies indicated that the spin density is distributed over the tricyclic ring, implying planarity (4, 30). Replacement of the 9-carboxyl group by a hydrogen did not alter the spin density significantly (4), which seems to suggest that the spin density on the 9-carboxyl group in PQQH is very small. However, in a recently solved x-ray crystal structure of MDH by Ghosh and coworkers (18, 19), they reported that PQQH is present in the active site and the C-4 carbonyl oxygen of PQQH appeared to be bent out of plane of the tricyclic ring. This finding is surprising because one would expect stronger repulsion between the quinone carbonyls in PQQ than in PQQH. In isolated PQQ, the C-4 carbonyl oxygen atom is only slightly out of the ring plane as shown by our calculations. In the x-ray crystallographic study of MDH from another source, it was found that PQQ in the active site is planar (16, 17). Our calculation gave essentially a planar semiquinone radical even in the presence of the Ca2+ (see below for further discussion). Presently, it remains an unanswered question as to why in the x-ray crystal structure by Ghosh and coworkers (18, 19), the C-4 carbonyl oxygen is out of plane. PQQ is a reactive compound and it has been shown that MDH can be easily contaminated during purification (31). Whether the unusual structure is caused by contamination or other factors remains to be seen.
PQQH2.
PQQH2 is an aromatic compound and the tricyclic ring is expected to be planar. Indeed, in the HF/6-31G(d) optimized structure of PQQH2, the tricyclic ring is planar (see Fig. 1). Carboxyl groups at C-2 and C-7 are coplanar with the ring, but the C-9 carboxyl is twisted out of plane by 22°. The most significant change in bond length involves the C4-C5 bond, which is a single bond in PQQ, but a double bond in PQQH2.
Effect of Ca2+.
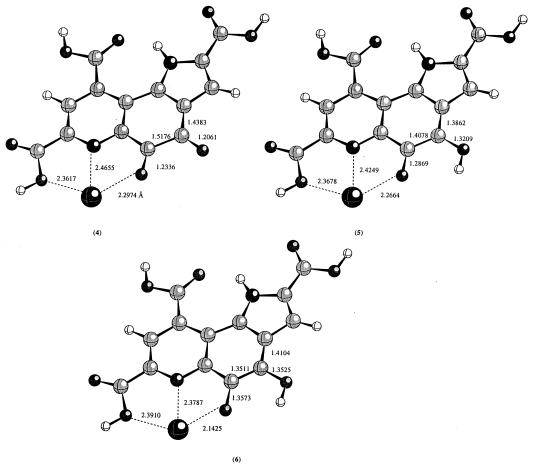
The metal ion binding site in PQQ and its derivatives is defined by C-5 carbonyl oxygen, pyridine nitrogen, and the C-7 carboxyl oxygen. The requirement for C-7 carboxyl was confirmed by the inability of 7-decarboxyl PQQ to bind metal ion (3). To examine the geometrical perturbation caused by the presence of Ca2+, we optimized the metal ion bound PQQ, PQQH, and PQQH2 using HF/3-21G(d) level of theory. The calculated structures are shown in Fig. 2. PQQ, PQQH, and PQQH2 are all planar. Deprotonation of C-7 carboxyl does not change the conformation of PQQ, PQQH, and PQQH2. The binding of Ca2+ introduces some changes to the structures. As seen from Fig. 2, the distances between Ca2+ and its ligands change as the oxidation state of PQQ changes. In PQQ, the Ca2+–O5, Ca2+–N, and Ca2+–O7a are 2.297 Å, 2.466 Å, and 2.362 Å; the corresponding values are 2.266 Å, 2.425 Å, and 2.368 Å in PQQH. In the reduced PQQH2 form, these distances are 2.332 Å, 2.379 Å, and 2.391 Å, respectively. Removal of the proton from the C-7 carboxyl group does not change the conformations (data not shown).
Figure 2.
The optimized geometries in the presence of Ca2+ ion at the HF/3-21G(d) level of theory.
Reaction of PQQ with Methanol.
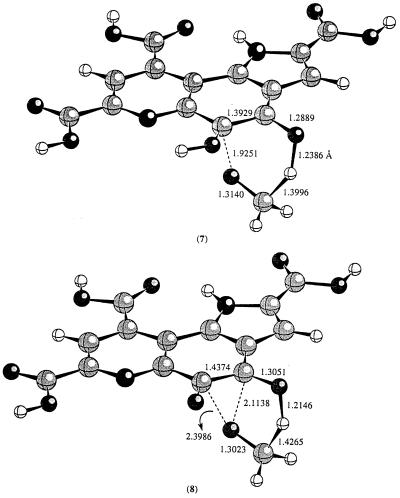
Because PQQ forms adducts easily with a number of compounds such as acetone and alcohols, it has been generally believed that the first step in the reaction of PQQ with methanol is the formation of a PQQ–methanol adduct. The decomposition of this adduct would give rise to PQQH2 and formaldehyde via a retro-ene reaction. First, we examined this pathway. The transition state for this retro-ene reaction (7) was located at the HF/3-21G(d) level of theory (Fig. 3). Because it is known that the inclusion of electron correlation effect is important for correct estimation of the reaction barriers for retro-ene and ene reactions (21), we also carried out energy calculations using a hybrid density functional theory [B3LYP/3-21G(d)]. The estimated barrier at B3LYP/3-21G(d) is about 33.0 kcal/mol. The structure of the transition state for the retro-ene reaction looks normal. The breaking C—O distance is 1.9251 Å and the forming O—H distance is 1.2386 Å (7, Fig. 3).
Figure 3.
The calculated transition state structures for the retro-ene reaction at HF/3-21G(d) level.
In the laboratory, retro-ene reactions are normally very slow and only occur at high temperature (21, 22). It is unclear how methanol dehydrogenase could employ such a mechanism. However, it was demonstrated many years ago that the product of alkoxide addition to a carbonyl function could undergo a rapid pericyclic reaction (32, 33). Evans and Golob (32) showed that rate accelerations of 1010–1017 can be easily accomplished by simple deprotonation of alcohol adducts of carbonyl functions. Furthermore, they also demonstrated that the acceleration in rate depends on the counter ion present and maximum acceleration is achieved when the counter ion is removed. It should be pointed out that, owning to the presence of Ca2+ in the active site of MDH, we do not expect to see such great rate enhancement as these should such a mechanism be in effect. Can such a mechanism be operative in MDH catalyzed reaction? To test this possibility, we examined the reaction of the deprotonated PQQ–methanol adduct. Fig. 3 displays the calculated transition state for such a process. The transition state (8) is very different from the transition state (7) for the decomposition of the neutral PQQ–methanol adduct. In 8, the C5—O distance is much longer (2.3986 Å vs. 1.9251 Å); the transition state looks much like a transition state that one would expect for a direct hydride transfer from methoxide to the C-4 carbonyl oxygen of PQQ. The calculated barrier for this reaction is about 10.5 kcal/mol. Indeed, the alkoxide substituent could provide an enormous driving force for the breakdown of PQQ–methoxide adduct. The caveat to this proposal is how to generate methoxide in the active site of MDH given that the only base available is the weakly basic carboxylate of Asp-303.
Catalytic Mechanism of MDH.
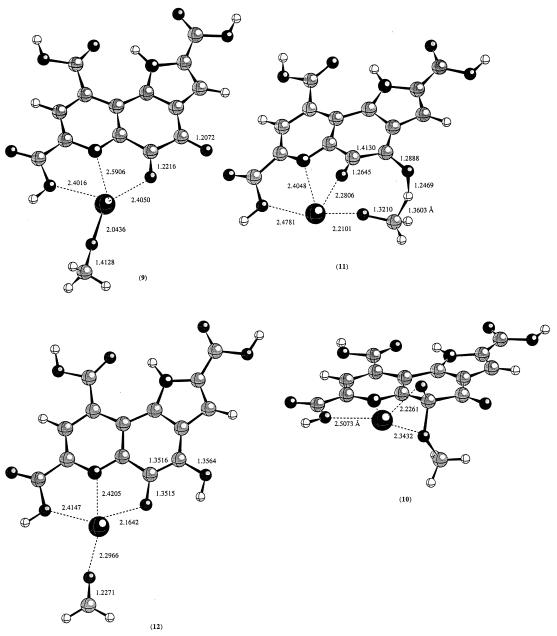
A reasonable mechanism would involve a ground state with the CH3OH substrate ligated to the Ca2+ center of the Ca2+/PQQ/MDH complex. Ligation of H2O to Ca2+ results in a decrease in its pKa from 15.5 to 12.7. A similar modest decrease in pKa of CH3OH would be expected in the CH3(H)O–Ca2+/PQQ/MDH complex. This should assist in lowering the rate constant by several orders of magnitude for the Asp-303–CO2− general base removal of the methanol proton in a reaction concerted with hydride transfer (Scheme S4). In such a reaction the carboxylate to proton bond formation should be late to provide the driving force for hydride transfer. The late transition state for the proton transfer is akin to starting with a ground state of structure CH3O−/Ca2+/PQQ/MDH. The latter complex could exist either as a Ca2+ bound methoxide (9) or a C-5 covalent adduct (10). According to the present calculations, 9 is more favored by 9.6 kcal/mol. Starting with either 9 or 10, the transition state connecting the reactant to the product was searched. Surprisingly, both converged to the same transition state. The transition state structure is shown in Fig. 4. In the transition state (11), Ca2+ coordinates to both C-5 carbonyl oxygen and the methoxide oxygen. The barrier from 9 to 11 is about 28.1 kcal/mol at B3LYP/3-21G(d) level of theory; the barrier from 10 to 11 is about 18.5 kcal/mol at the same level of theory. Metal ion-assisted hydride transfer is known for several enzymatic reactions such as horse liver alcohol dehydrogenase where a Zn2+ ion is involved (34). The influence of Zn2+ ion in lowering the pKa of ligated alcohol is, of course, more pronounced than that seen with Ca2+ ion.
Scheme 4.
Figure 4.
The calculated transition state structure for the hydride transfer reaction from a Ca2+ bound methoxide to the C-4 carbonyl oxygen at the HF/3-21G(d) level of theory.
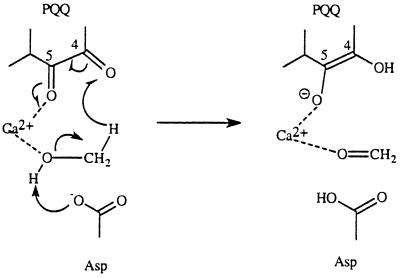
The proposed formation of alcohol–metal ion complex is consistent with experimental observation of formation of a C-5 propanal adduct of PQQ when MDH is treated with cyclopropanol (35). General-base deprotonation of the hydroxyl group of the initially formed cyclopropanol-Ca2+ complex by an active site Asp, results in an apparently concerted ring opening forming the C-5 propanal adduct.
One may argue that the initially formed Ca2+–methanol complex could convert to a covalent PQQ–methoxide adduct via a general-base proton abstraction by active site Asp-303 concerted with nucleophilic attack at C-5 carbonyl carbon. However, the formation of such a covalent PQQ–methoxide adduct is probably counterproductive because, as shown from our calculations, both the direct hydride transfer and the decomposition of the covalent PQQ–methoxide adduct seem to go through the same transition state (11) to form formaldehyde. Therefore, we prefer the direct hydride transfer process.
Another possible mechanism worthy of comment is shown in Scheme S5. A similar mechanism is operative in amine oxidation (1, 2, 5). According to this mechanism, a C-5 covalent PQQ–methanol adduct is formed first during the catalytic reaction (pathway A in Scheme S5). Deprotonation of a methyl hydrogen by Asp-303 initiates an E2-type of elimination process. This possibility has not been ruled out. However, the pKa values of the hydrogens of the methoxy group are considerably higher than the pKa of Asp in the active site. In the case of amine oxidations, the hydrogen involved is an allylic hydrogen and its pKa is much lower; the resulting anion can also be stabilized by resonance. A kinetic study using isotopically labeled substrate (e.g., 18O-labeled methanol) might provide useful information regarding the possible involvement of this mechanism. However, even if this possibility exists, the C-4 adduct should be preferred to the C-5 adduct because the Ca2+ can assist the breakdown of the C-4 adduct (pathway B in Scheme S5).
Scheme 5.
CONCLUSIONS
The conformation of PQQ and the role of Ca2+ in the catalytic mechanism of quinoprotein methanol dehydrogenase were examined using a quantum mechanical method. The tricyclic ring system of PQQ remains essentially planar (or very close to being planar) regardless of its oxidation state. The binding of Ca2+ does not change this picture. The carboxyl groups at C-2 and C-7 positions are coplanar to the tricyclic ring; however the carboxyl group at C-9 position can be either coplanar or twisted out of the plane of the tricyclic ring. This is in agreement with experimental observations that the orientation of the carboxyl groups depends on the environment. The computed planar structure for PQQH contradicts a recently reported crystal structure of MDH in which one of the carbonyl oxygens of PQQH is out of the plane of the ring. Presently, it is unclear what is the cause of this discrepancy.
We also investigated the possible mechanistic pathways employed by methanol dehydrogenase and the role of Ca2+ in the catalytic mechanism. Based on our theoretical study, a molecular level mechanism is proposed (Scheme S4). In this mechanism the quinoprotein methanol dehydrogenase catalyzed oxidation of methanol consists of the following steps: (i) association of methanol to active site Ca2+; (ii) deprotonating Ca2+-bound methanol by an active site Asp in concert with; (iii) hydride transfer from Ca2+-bound substrate to the carbonyl at C-4 position and formation of a Ca2+ bound formaldehyde; and (iv) releasing of formaldehyde from the active site of the enzyme. The catalytic role of PQQ complexed Ca2+ is 3-fold: (i) modest reduction of the pKa of CH3-OH, (ii) polarizing the carbonyl group at the C-5 position of the PQQ moiety, and (iii) placing the reaction components in position to react. The proposed role of Ca2+ is also in agreement with a recent study on strontium (Sr2+) substituted MDH (36). Similar mechanisms could also be operative in other quinoproteins such as other alcohol and glucose dehydrogenases.
Acknowledgments
We thank Dale Edmondson for suggestions. This work is supported by the Petroleum Research Fund of the American Chemical Society. We also thank the National Center for Supercomputing Applications (Champaign, IL) for allocation of supercomputing resources.
ABBREVIATIONS
- PQQ
2,7,9-tricarboxypyrroloquinoline quinone
- PQQH
semiquinone
- PQQH2
dihydroquinone
- MDH
methanol dehydrogenase
- HF
Hartree–Fock
References
- 1.Eckert T S, Bruice T C. J Am Chem Soc. 1983;105:4431–4441. [Google Scholar]
- 2.Sleath P R, Noar J B, Eberlein G A, Bruice T C. J Am Chem Soc. 1985;107:3328–3338. [Google Scholar]
- 3.Noar J B, Rodriguez E J, Bruice T C. J Am Chem Soc. 1985;107:7198–7199. [Google Scholar]
- 4.Rodriguez E J, Bruice T C, Edmondson D E. J Am Chem Soc. 1987;109:532–537. [Google Scholar]
- 5.Rodriguez E J, Bruice T C. J Am Chem Soc. 1989;111:7947–7956. [Google Scholar]
- 6.Salisbury S A, Forrest H S, Cruse W B T, Kennard O. Nature (London) 1979;280:843–844. doi: 10.1038/280843a0. [DOI] [PubMed] [Google Scholar]
- 7.Anthony C, Zatman L J. Biochem J. 1967;104:960–969. doi: 10.1042/bj1040960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duine J A, Frank J, Verwiel P E J. Eur J Biochem. 1981;118:395–399. doi: 10.1111/j.1432-1033.1981.tb06415.x. [DOI] [PubMed] [Google Scholar]
- 9.Anthony C, Zatman C J. Biochem J. 1964;92:614–621. doi: 10.1042/bj0920614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anthony C. In: Principles and Applications of Quinoproteins. Davidson V L, editor. New York: Dekker; 1993. pp. 17–45. [Google Scholar]
- 11.Duine J A, Frank J, van Zeeland J K. FEBS Lett. 1979;108:443–446. doi: 10.1016/0014-5793(79)80584-0. [DOI] [PubMed] [Google Scholar]
- 12.Duine J A, Frank J, Verwiel P E J. Eur J Biochem. 1980;108:187–192. doi: 10.1111/j.1432-1033.1980.tb04711.x. [DOI] [PubMed] [Google Scholar]
- 13.Anthony C, Ghosh M, Blake C C F. Biochem J. 1994;304:665–674. doi: 10.1042/bj3040665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anthony C. Biochem J. 1996;320:697–711. doi: 10.1042/bj3200697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anthony C, Ghosh M. Curr Sci. 1997;72:716–727. [Google Scholar]
- 16.White S, Boyd G, Mathews F S, Xia Z-X, Dai W-W, Zhang Y-F, Davidson V L. Biochemistry. 1993;32:12955–12958. doi: 10.1021/bi00211a002. [DOI] [PubMed] [Google Scholar]
- 17.Xia Z-X, Dai W-W, Zhang Y-F, White S A, Boyd G D, Mathews F S. J Mol Biol. 1996;259:480–501. doi: 10.1006/jmbi.1996.0334. [DOI] [PubMed] [Google Scholar]
- 18.Blake C C F, Ghosh M, Harlos K, Avezoux A, Anthony C. Nat Struct Biol. 1994;1:102–105. doi: 10.1038/nsb0294-102. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh M, Anthony C, Harlos K, Goodwin M G, Blake C. Structure. 1995;3:177–187. doi: 10.1016/s0969-2126(01)00148-4. [DOI] [PubMed] [Google Scholar]
- 20.Itoh S, Kawakami H, Fukuzumi S. J Am Chem Soc. 1997;119:439–440. [Google Scholar]
- 21.Loncharich R J, Houk K N. J Am Chem Soc. 1987;109:6947–6952. [Google Scholar]
- 22.Snider B B. Acc Chem Res. 1980;13:426–432. [Google Scholar]
- 23.Frisch M J, Trucks G W, Schlegel H B, Gill P M W, Johnson B G, et al. Gaussian 94. Pittsburgh: Gaussian; 1995. , Revision B.2. [Google Scholar]
- 24.Pople J A, Nesbet R K. J Chem Phys. 1954;22:571–574. [Google Scholar]
- 25.Stewart J J P. J Comput Chem. 1989;10:209–220. [Google Scholar]
- 26.Ushida T, Doi M, Tomita K, Hayashi H, Inoue M, Urakami T. J Am Chem Soc. 1989;111:6822–6828. [Google Scholar]
- 27.van Koningsveld H, Jongejan J A, Duine J A. In: PQQ and Quinoproteins. Jongejan J A, Duine J A, editors. Dordrecht, The Netherlands: Kluwer; 1989. pp. 243–251. [Google Scholar]
- 28.Dekker R H, Duine J A, Frank J, Verwiel P, Westerling J. Eur J Biochem. 1982;125:69–73. doi: 10.1111/j.1432-1033.1982.tb06652.x. [DOI] [PubMed] [Google Scholar]
- 29.Itoh S, Ogino M, Fukui Y, Murao H, Komatsu M, Ohshiro Y, Inoue T, Kai Y, Kasai N. J Am Chem Soc. 1993;115:9960–9967. [Google Scholar]
- 30.Duine J A, Frank J, De Beer R. Arch Biochem Biophys. 1984;233:708–711. doi: 10.1016/0003-9861(84)90497-1. [DOI] [PubMed] [Google Scholar]
- 31.Ghosh R, Quayle J R. Biochem J. 1981;199:245–250. doi: 10.1042/bj1990245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans D A, Golob A M. J Am Chem Soc. 1975;97:4765–4766. [Google Scholar]
- 33.Bunnage M E, Nicolaou K C. Chem Eur J. 1997;3:187–192. doi: 10.1002/chem.19970030204. [DOI] [PubMed] [Google Scholar]
- 34.Lippard S J, Berg J M. Principles of Bioinorganic Chemistry. Mill Valley, CA: University Science Books; 1994. [Google Scholar]
- 35.Frank J, Dijkstra M, Duine J A. Eur J Biochem. 1988;174:331–338. doi: 10.1111/j.1432-1033.1988.tb14102.x. [DOI] [PubMed] [Google Scholar]
- 36.Harris T K, Davidson V L. Biochem J. 1994;300:175–182. doi: 10.1042/bj3000175. [DOI] [PMC free article] [PubMed] [Google Scholar]