Abstract
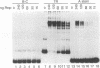
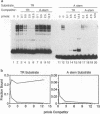
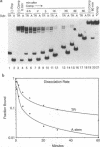
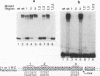
We have characterized a Rep binding sequence which is within the A stem region of the adeno-associated virus terminal repeat (TR) and compared its affinity with that of the complete hairpinned TR for pure Rep68. Both the A stem and the complete TR substrates produced a complex pattern of protein-DNA complexes in which at least six different bound species could be distinguished. Competition experiments suggested that the dissociation constant for the A stem sequence is approximately 125-fold higher than that for the complete TR. The competition experiments also suggested that the average number of Rep molecules per TR substrate molecule under conditions of saturating substrate is 3.7:1, while for the A stem substrate, the ratio is 10:1. In spite of the apparent difference in protein-to-DNA ratio in the complexes, no major difference was seen in the mobility or the pattern of the protein-DNA complexes with the two kinds of substrates, suggesting that the difference in protein-to-DNA ratio was due to the lower stability of the A stem complex rather than the actual number of Rep molecules per DNA molecule. At least some of the difference in stability of the two kinds of complexes was due to the fact that the dissociation rate of the A stem substrate from the protein-DNA complexes was approximately fourfold faster than that of the complete TR. The dissociation rate curves for both substrates, however, were complex, suggesting that substrate was being released from at least two different kinds of protein-DNA complexes at different rates. In addition, we have analyzed binding to several substitution mutants within the A stem of the TR. A five-base mutant near the terminal resolution site (trs site) had little effect on binding. Two other mutants produced seven- or five-base substitutions within the 25-bp sequence of the A stem that had been identified in the accompanying report (D. M. McCarty, D. J. Pereira, I. Zolotukhin, X. Zhou, J. H. Ryan, and N. Muzyczka, J. Virol. 68:4988-4997, 1994) as essential for binding. Each of these mutants eliminated some but not all of the repeating GAGC motifs in the 25-bp A stem region. Both of these mutants completely abolished binding to the A stem substrate but only partially reduced binding in the context of the complete hairpinned TR. Furthermore, neither mutant altered the pattern of Rep-DNA complexes produced.(ABSTRACT TRUNCATED AT 400 WORDS)
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ashktorab H., Srivastava A. Identification of nuclear proteins that specifically interact with adeno-associated virus type 2 inverted terminal repeat hairpin DNA. J Virol. 1989 Jul;63(7):3034–3039. doi: 10.1128/jvi.63.7.3034-3039.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaton A., Palumbo P., Berns K. I. Expression from the adeno-associated virus p5 and p19 promoters is negatively regulated in trans by the rep protein. J Virol. 1989 Oct;63(10):4450–4454. doi: 10.1128/jvi.63.10.4450-4454.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chejanovsky N., Carter B. J. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology. 1989 Nov;173(1):120–128. doi: 10.1016/0042-6822(89)90227-4. [DOI] [PubMed] [Google Scholar]
- Chiorini J. A., Weitzman M. D., Owens R. A., Urcelay E., Safer B., Kotin R. M. Biologically active Rep proteins of adeno-associated virus type 2 produced as fusion proteins in Escherichia coli. J Virol. 1994 Feb;68(2):797–804. doi: 10.1128/jvi.68.2.797-804.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauswirth W. W., Berns K. I. Adeno-associated virus DNA replication: nonunit-length molecules. Virology. 1979 Feb;93(1):57–68. doi: 10.1016/0042-6822(79)90275-7. [DOI] [PubMed] [Google Scholar]
- Hermonat P. L., Labow M. A., Wright R., Berns K. I., Muzyczka N. Genetics of adeno-associated virus: isolation and preliminary characterization of adeno-associated virus type 2 mutants. J Virol. 1984 Aug;51(2):329–339. doi: 10.1128/jvi.51.2.329-339.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im D. S., Muzyczka N. Factors that bind to adeno-associated virus terminal repeats. J Virol. 1989 Jul;63(7):3095–3104. doi: 10.1128/jvi.63.7.3095-3104.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im D. S., Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990 May 4;61(3):447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- Labow M. A., Hermonat P. L., Berns K. I. Positive and negative autoregulation of the adeno-associated virus type 2 genome. J Virol. 1986 Oct;60(1):251–258. doi: 10.1128/jvi.60.1.251-258.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty D. M., Christensen M., Muzyczka N. Sequences required for coordinate induction of adeno-associated virus p19 and p40 promoters by Rep protein. J Virol. 1991 Jun;65(6):2936–2945. doi: 10.1128/jvi.65.6.2936-2945.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty D. M., Ni T. H., Muzyczka N. Analysis of mutations in adeno-associated virus Rep protein in vivo and in vitro. J Virol. 1992 Jul;66(7):4050–4057. doi: 10.1128/jvi.66.7.4050-4057.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty D. M., Pereira D. J., Zolotukhin I., Zhou X., Ryan J. H., Muzyczka N. Identification of linear DNA sequences that specifically bind the adeno-associated virus Rep protein. J Virol. 1994 Aug;68(8):4988–4997. doi: 10.1128/jvi.68.8.4988-4997.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelson E., Trempe J. P., Carter B. J. Identification of the trans-acting Rep proteins of adeno-associated virus by antibodies to a synthetic oligopeptide. J Virol. 1986 Dec;60(3):823–832. doi: 10.1128/jvi.60.3.823-832.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T. H., Zhou X., McCarty D. M., Zolotukhin I., Muzyczka N. In vitro replication of adeno-associated virus DNA. J Virol. 1994 Feb;68(2):1128–1138. doi: 10.1128/jvi.68.2.1128-1138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens R. A., Weitzman M. D., Kyöstiö S. R., Carter B. J. Identification of a DNA-binding domain in the amino terminus of adeno-associated virus Rep proteins. J Virol. 1993 Feb;67(2):997–1005. doi: 10.1128/jvi.67.2.997-1005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder R. O., Im D. S., Muzyczka N. Evidence for covalent attachment of the adeno-associated virus (AAV) rep protein to the ends of the AAV genome. J Virol. 1990 Dec;64(12):6204–6213. doi: 10.1128/jvi.64.12.6204-6213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder R. O., Im D. S., Ni T., Xiao X., Samulski R. J., Muzyczka N. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J Virol. 1993 Oct;67(10):6096–6104. doi: 10.1128/jvi.67.10.6096-6104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder R. O., Samulski R. J., Muzyczka N. In vitro resolution of covalently joined AAV chromosome ends. Cell. 1990 Jan 12;60(1):105–113. doi: 10.1016/0092-8674(90)90720-y. [DOI] [PubMed] [Google Scholar]
- Srivastava A., Lusby E. W., Berns K. I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J Virol. 1983 Feb;45(2):555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus S. E., Sebring E. D., Rose J. A. Concatemers of alternating plus and minus strands are intermediates in adenovirus-associated virus DNA synthesis. Proc Natl Acad Sci U S A. 1976 Mar;73(3):742–746. doi: 10.1073/pnas.73.3.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tratschin J. D., Miller I. L., Carter B. J. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J Virol. 1984 Sep;51(3):611–619. doi: 10.1128/jvi.51.3.611-619.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tratschin J. D., Tal J., Carter B. J. Negative and positive regulation in trans of gene expression from adeno-associated virus vectors in mammalian cells by a viral rep gene product. Mol Cell Biol. 1986 Aug;6(8):2884–2894. doi: 10.1128/mcb.6.8.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Trempe J. P. Analysis of the terminal repeat binding abilities of mutant adeno-associated virus replication proteins. J Virol. 1993 Jul;67(7):4442–4447. doi: 10.1128/jvi.67.7.4442-4447.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]