Abstract
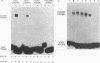
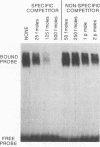
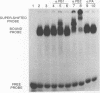
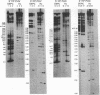
The enzymatic activity of recombinant influenza virus RNA polymerase is strictly dependent on the addition of a template RNA containing 5' and 3' viral sequences. Here we report the analysis of the binding specificity and physical characterization of the complex by using gel shift, modification interference, and density gradient techniques. The 13S complex binds specifically to short synthetic RNAs that mimic the partially double stranded panhandle structures found at the termini of both viral RNA and cRNA. The polymerase will also bind independently to the single-stranded 5' or 3' ends of viral RNA. It binds most strongly to specific sequences within the 5' end but is unable to bind these sequences in the context of a completely double stranded structure. Modification interference analysis identified the short sequence motifs at the 5' ends of the viral RNA and cRNA templates that are critical for binding.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BEERS R. F., Jr, SIZER I. W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem. 1952 Mar;195(1):133–140. [PubMed] [Google Scholar]
- Bartel D. P., Zapp M. L., Green M. R., Szostak J. W. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell. 1991 Nov 1;67(3):529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- Beaton A. R., Krug R. M. Transcription antitermination during influenza viral template RNA synthesis requires the nucleocapsid protein and the absence of a 5' capped end. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6282–6286. doi: 10.1073/pnas.83.17.6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd H. P., Tiley L. S., Cullen B. R. Specific binding of the human T-cell leukemia virus type I Rex protein to a short RNA sequence located within the Rex-response element. J Virol. 1992 Dec;66(12):7572–7575. doi: 10.1128/jvi.66.12.7572-7575.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouloy M., Morgan M. A., Shatkin A. J., Krug R. M. Cap and internal nucleotides of reovirus mRNA primers are incorporated into influenza viral complementary RNA during transcription in vitro. J Virol. 1979 Dec;32(3):895–904. doi: 10.1128/jvi.32.3.895-904.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey J., Cameron V., de Haseth P. L., Uhlenbeck O. C. Sequence-specific interaction of R17 coat protein with its ribonucleic acid binding site. Biochemistry. 1983 May 24;22(11):2601–2610. doi: 10.1021/bi00280a002. [DOI] [PubMed] [Google Scholar]
- Desselberger U., Racaniello V. R., Zazra J. J., Palese P. The 3' and 5'-terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene. 1980 Feb;8(3):315–328. doi: 10.1016/0378-1119(80)90007-4. [DOI] [PubMed] [Google Scholar]
- Detjen B. M., St Angelo C., Katze M. G., Krug R. M. The three influenza virus polymerase (P) proteins not associated with viral nucleocapsids in the infected cell are in the form of a complex. J Virol. 1987 Jan;61(1):16–22. doi: 10.1128/jvi.61.1.16-22.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiering S., Northrop J. P., Nolan G. P., Mattila P. S., Crabtree G. R., Herzenberg L. A. Single cell assay of a transcription factor reveals a threshold in transcription activated by signals emanating from the T-cell antigen receptor. Genes Dev. 1990 Oct;4(10):1823–1834. doi: 10.1101/gad.4.10.1823. [DOI] [PubMed] [Google Scholar]
- Fodor E., Seong B. L., Brownlee G. G. Photochemical cross-linking of influenza A polymerase to its virion RNA promoter defines a polymerase binding site at residues 9 to 12 of the promoter. J Gen Virol. 1993 Jul;74(Pt 7):1327–1333. doi: 10.1099/0022-1317-74-7-1327. [DOI] [PubMed] [Google Scholar]
- Fuerst T. R., Niles E. G., Studier F. W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen M., Chung T. D., Butcher J. A., Krystal M. Recombinant influenza virus polymerase: requirement of both 5' and 3' viral ends for endonuclease activity. J Virol. 1994 Mar;68(3):1509–1515. doi: 10.1128/jvi.68.3.1509-1515.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A. J., Abraham G., Skehel J. J., Smith J. C., Fellner P. Influenza virus messenger RNAs are incomplete transcripts of the genome RNAs. Nucleic Acids Res. 1977 Dec;4(12):4197–4209. doi: 10.1093/nar/4.12.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A. J., Lomniczi B., Bellamy A. R., Skehel J. J. Transcription of the influenza virus genome. Virology. 1977 Dec;83(2):337–355. doi: 10.1016/0042-6822(77)90179-9. [DOI] [PubMed] [Google Scholar]
- Hay A. J., Skehel J. J., McCauley J. Characterization of influenza virus RNA complete transcripts. Virology. 1982 Jan 30;116(2):517–522. doi: 10.1016/0042-6822(82)90144-1. [DOI] [PubMed] [Google Scholar]
- Honda A., Mukaigawa J., Yokoiyama A., Kato A., Ueda S., Nagata K., Krystal M., Nayak D. P., Ishihama A. Purification and molecular structure of RNA polymerase from influenza virus A/PR8. J Biochem. 1990 Apr;107(4):624–628. doi: 10.1093/oxfordjournals.jbchem.a123097. [DOI] [PubMed] [Google Scholar]
- Honda A., Uéda K., Nagata K., Ishihama A. RNA polymerase of influenza virus: role of NP in RNA chain elongation. J Biochem. 1988 Dec;104(6):1021–1026. doi: 10.1093/oxfordjournals.jbchem.a122569. [DOI] [PubMed] [Google Scholar]
- Hsu M. T., Parvin J. D., Gupta S., Krystal M., Palese P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc Natl Acad Sci U S A. 1987 Nov;84(22):8140–8144. doi: 10.1073/pnas.84.22.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T. S., Palese P., Krystal M. Determination of influenza virus proteins required for genome replication. J Virol. 1990 Nov;64(11):5669–5673. doi: 10.1128/jvi.64.11.5669-5673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura N., Nishida M., Nagata K., Ishihama A., Oda K., Nakada S. Transcription of a recombinant influenza virus RNA in cells that can express the influenza virus RNA polymerase and nucleoprotein genes. J Gen Virol. 1992 Jun;73(Pt 6):1321–1328. doi: 10.1099/0022-1317-73-6-1321. [DOI] [PubMed] [Google Scholar]
- Kjems J., Calnan B. J., Frankel A. D., Sharp P. A. Specific binding of a basic peptide from HIV-1 Rev. EMBO J. 1992 Mar;11(3):1119–1129. doi: 10.1002/j.1460-2075.1992.tb05152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Tuchiya K., Nagata K., Ishihama A. Reconstitution of influenza virus RNA polymerase from three subunits expressed using recombinant baculovirus system. Virus Res. 1992 Mar;22(3):235–245. doi: 10.1016/0168-1702(92)90055-e. [DOI] [PubMed] [Google Scholar]
- Li X., Palese P. Characterization of the polyadenylation signal of influenza virus RNA. J Virol. 1994 Feb;68(2):1245–1249. doi: 10.1128/jvi.68.2.1245-1249.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G. X., Luytjes W., Enami M., Palese P. The polyadenylation signal of influenza virus RNA involves a stretch of uridines followed by the RNA duplex of the panhandle structure. J Virol. 1991 Jun;65(6):2861–2867. doi: 10.1128/jvi.65.6.2861-2867.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín J., Albo C., Ortín J., Melero J. A., Portela A. In vitro reconstitution of active influenza virus ribonucleoprotein complexes using viral proteins purified from infected cells. J Gen Virol. 1992 Jul;73(Pt 7):1855–1859. doi: 10.1099/0022-1317-73-7-1855. [DOI] [PubMed] [Google Scholar]
- Nagata K., Takeuchi K., Ishihama A. In vitro synthesis of influenza viral RNA: biochemical complementation assay of factors required for influenza virus replication. J Biochem. 1989 Aug;106(2):205–208. doi: 10.1093/oxfordjournals.jbchem.a122833. [DOI] [PubMed] [Google Scholar]
- Ohlsson H., Edlund T. Sequence-specific interactions of nuclear factors with the insulin gene enhancer. Cell. 1986 Apr 11;45(1):35–44. doi: 10.1016/0092-8674(86)90535-0. [DOI] [PubMed] [Google Scholar]
- Parvin J. D., Palese P., Honda A., Ishihama A., Krystal M. Promoter analysis of influenza virus RNA polymerase. J Virol. 1989 Dec;63(12):5142–5152. doi: 10.1128/jvi.63.12.5142-5152.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotch S. J., Bouloy M., Krug R. M. Transfer of 5'-terminal cap of globin mRNA to influenza viral complementary RNA during transcription in vitro. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1618–1622. doi: 10.1073/pnas.76.4.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotch S. J., Bouloy M., Ulmanen I., Krug R. M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981 Mar;23(3):847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- Robertson J. S. 5' and 3' terminal nucleotide sequences of the RNA genome segments of influenza virus. Nucleic Acids Res. 1979 Aug 24;6(12):3745–3757. doi: 10.1093/nar/6.12.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J. S., Schubert M., Lazzarini R. A. Polyadenylation sites for influenza virus mRNA. J Virol. 1981 Apr;38(1):157–163. doi: 10.1128/jvi.38.1.157-163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong B. L., Brownlee G. G. A new method for reconstituting influenza polymerase and RNA in vitro: a study of the promoter elements for cRNA and vRNA synthesis in vitro and viral rescue in vivo. Virology. 1992 Jan;186(1):247–260. doi: 10.1016/0042-6822(92)90079-5. [DOI] [PubMed] [Google Scholar]
- Seong B. L., Brownlee G. G. Nucleotides 9 to 11 of the influenza A virion RNA promoter are crucial for activity in vitro. J Gen Virol. 1992 Dec;73(Pt 12):3115–3124. doi: 10.1099/0022-1317-73-12-3115. [DOI] [PubMed] [Google Scholar]
- Shapiro G. I., Krug R. M. Influenza virus RNA replication in vitro: synthesis of viral template RNAs and virion RNAs in the absence of an added primer. J Virol. 1988 Jul;62(7):2285–2290. doi: 10.1128/jvi.62.7.2285-2290.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. L., Levin J. Z., Palese P., Moss B. Synthesis and cellular location of the ten influenza polypeptides individually expressed by recombinant vaccinia viruses. Virology. 1987 Oct;160(2):336–345. doi: 10.1016/0042-6822(87)90004-3. [DOI] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Stern S., Wilson R. C., Noller H. F. Localization of the binding site for protein S4 on 16 S ribosomal RNA by chemical and enzymatic probing and primer extension. J Mol Biol. 1986 Nov 5;192(1):101–110. doi: 10.1016/0022-2836(86)90467-5. [DOI] [PubMed] [Google Scholar]
- Stoeckle M. Y., Shaw M. W., Choppin P. W. Segment-specific and common nucleotide sequences in the noncoding regions of influenza B virus genome RNAs. Proc Natl Acad Sci U S A. 1987 May;84(9):2703–2707. doi: 10.1073/pnas.84.9.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura A., Tobita K., Kilbourne E. D. Isolation and preliminary characterization of temperature-sensitive mutants of influenza virus. J Virol. 1972 Oct;10(4):639–647. doi: 10.1128/jvi.10.4.639-647.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiley L. S., Malim M. H., Tewary H. K., Stockley P. G., Cullen B. R. Identification of a high-affinity RNA-binding site for the human immunodeficiency virus type 1 Rev protein. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):758–762. doi: 10.1073/pnas.89.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H., Billeter M. A., Kahane S., Weissmann C., Hindley J., Porter A. Molecular basis for repressor activity of Q replicase. Nat New Biol. 1972 Jun 7;237(75):166–170. doi: 10.1038/newbio237166a0. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., Crothers D. M. RNA recognition by Tat-derived peptides: interaction in the major groove? Cell. 1991 Aug 9;66(3):577–588. doi: 10.1016/0092-8674(81)90020-9. [DOI] [PubMed] [Google Scholar]
- Weissmann C. The making of a phage. FEBS Lett. 1974 Mar 23;40(0):suppl–suppl:S18. doi: 10.1016/0014-5793(74)80684-8. [DOI] [PubMed] [Google Scholar]
- Wu H. N., Uhlenbeck O. C. Role of a bulged A residue in a specific RNA-protein interaction. Biochemistry. 1987 Dec 15;26(25):8221–8227. doi: 10.1021/bi00399a030. [DOI] [PubMed] [Google Scholar]
- Young R. J., Content J. 5'-terminus of influenza virus RNA. Nat New Biol. 1971 Mar 31;230(13):140–142. doi: 10.1038/newbio230140a0. [DOI] [PubMed] [Google Scholar]