Abstract
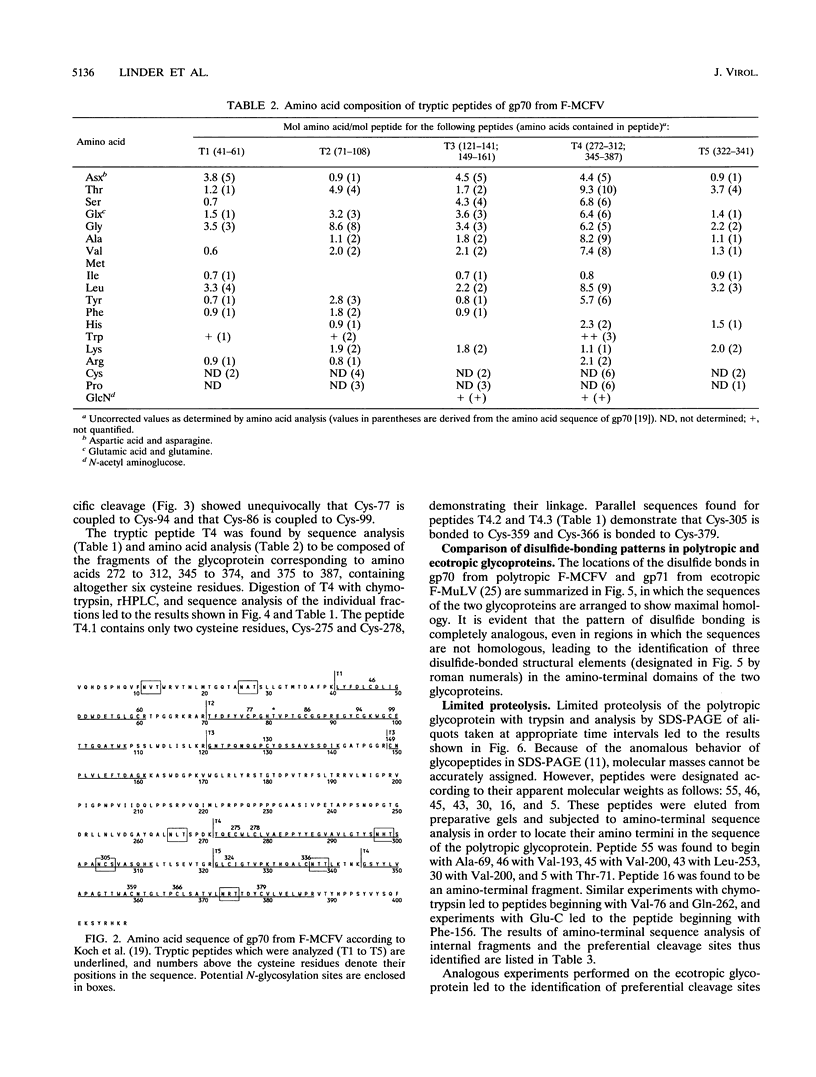
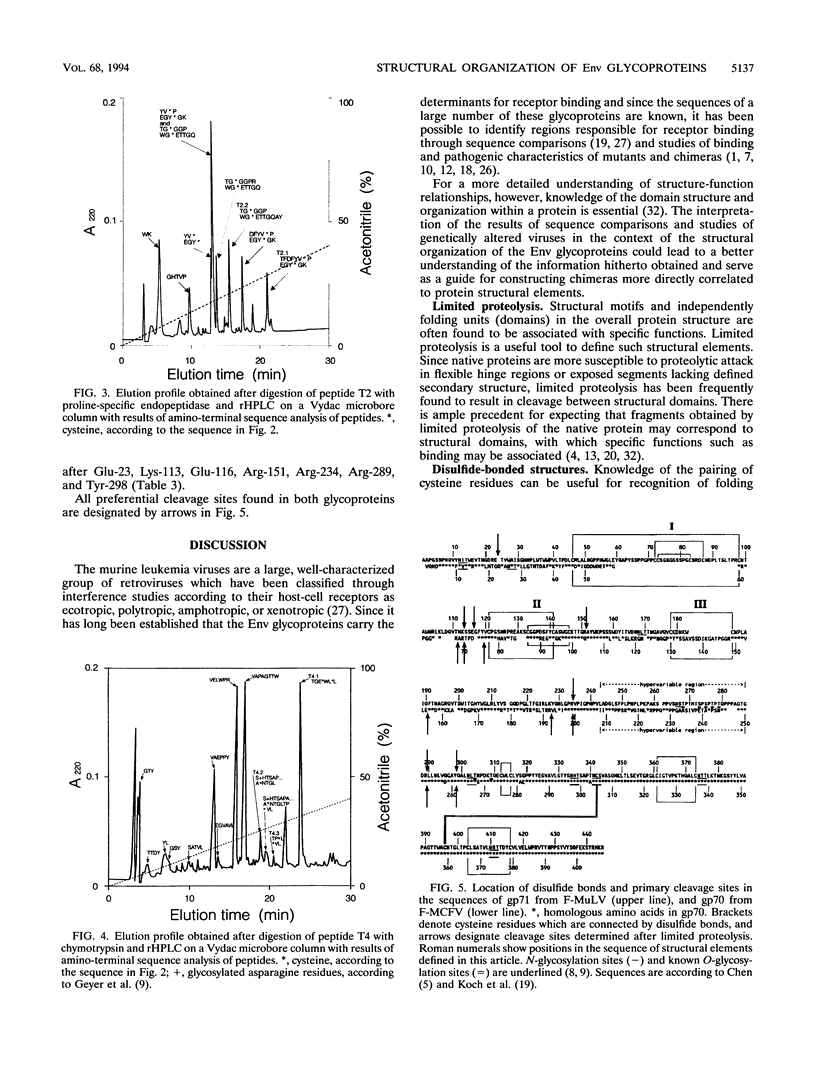
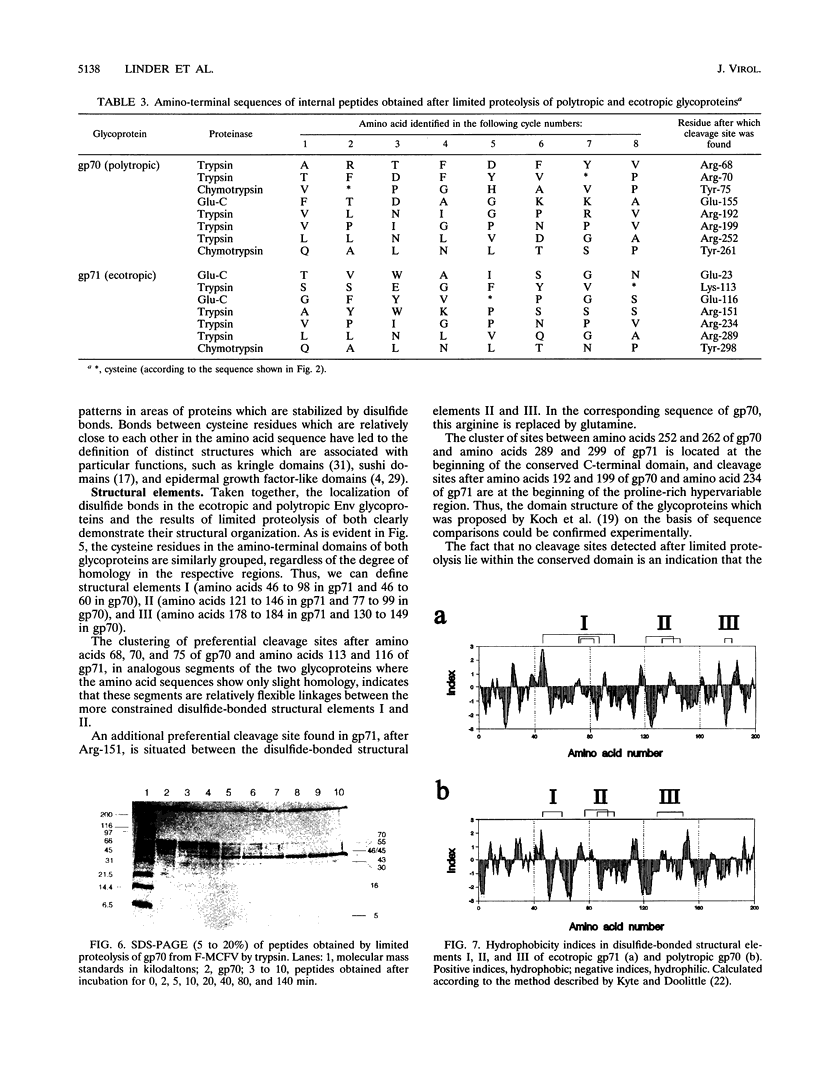
The disulfide-bonding pattern of glycoprotein 70 (gp70), the surface glycoprotein (SU) encoded by the envelope gene of polytropic Friend milk cell focus-inducing virus, was elucidated and compared with that of glycoprotein 71 (gp71), the corresponding glycoprotein of the ecotropic Friend murine leukemia virus, which had previously been determined (M. Linder, D. Linder, J. Hahnen, H.-H. Schott, and Stirm, Eur. J. Biochem. 203:65-73, 1992). In the carboxy-terminal constant domain, in which these glycoproteins have about 97% sequence homology, the location of the four disulfide bonds was found to be analogous. In the amino-terminal differential domain, with about 37% sequence homology, 8 of the 12 cysteine residues of the ecotropic SU are conserved in the polytropic SU. In this domain, a similar clustering of disulfide bonds was detected, which led to the identification of three distinct disulfide-bonded regions in both glycoproteins. However, because of deletions and sequence deviations, the glycoproteins must have significantly different three-dimensional structures in these regions. Since the receptor-binding functions of both glycoproteins have been attributed to their amino-terminal domains and since each binds to a different receptor, these disulfide-bonded structures are likely candidates for receptor-binding functions. Limited proteolysis of both glycoproteins with various endoproteinases led to the identification of preferential proteolytic sites between disulfide-bonded regions, at the beginning of the hypervariable proline-rich region, and between differential and constant domains, further confirming the structural organization of the folded glycoproteins.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Battini J. L., Heard J. M., Danos O. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992 Mar;66(3):1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan J. F. A novel family of growth factor receptors: a common binding domain in the growth hormone, prolactin, erythropoietin and IL-6 receptors, and the p75 IL-2 receptor beta-chain. Biochem Biophys Res Commun. 1989 Oct 31;164(2):788–795. doi: 10.1016/0006-291x(89)91528-3. [DOI] [PubMed] [Google Scholar]
- Bean M. F., Carr S. A. Characterization of disulfide bond position in proteins and sequence analysis of cystine-bridged peptides by tandem mass spectrometry. Anal Biochem. 1992 Mar;201(2):216–226. doi: 10.1016/0003-2697(92)90331-z. [DOI] [PubMed] [Google Scholar]
- Bogusky M. J., Dobson C. M., Smith R. A. Reversible independent unfolding of the domains of urokinase monitored by 1H NMR. Biochemistry. 1989 Aug 8;28(16):6728–6735. doi: 10.1021/bi00442a028. [DOI] [PubMed] [Google Scholar]
- Chen R. Complete amino acid sequence and glycosylation sites of glycoprotein gp71A of Friend murine leukemia virus. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5788–5792. doi: 10.1073/pnas.79.19.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emini E. A., Hughes J. V., Perlow D. S., Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol. 1985 Sep;55(3):836–839. doi: 10.1128/jvi.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felkner R. H., Roth M. J. Mutational analysis of the N-linked glycosylation sites of the SU envelope protein of Moloney murine leukemia virus. J Virol. 1992 Jul;66(7):4258–4264. doi: 10.1128/jvi.66.7.4258-4264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer R., Dabrowski J., Dabrowski U., Linder D., Schlüter M., Schott H. H., Stirm S. Oligosaccharides at individual glycosylation sites in glycoprotein 71 of Friend murine leukemia virus. Eur J Biochem. 1990 Jan 12;187(1):95–110. doi: 10.1111/j.1432-1033.1990.tb15281.x. [DOI] [PubMed] [Google Scholar]
- Gray K. D., Roth M. J. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J Virol. 1993 Jun;67(6):3489–3496. doi: 10.1128/jvi.67.6.3489-3496.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard J. M., Danos O. An amino-terminal fragment of the Friend murine leukemia virus envelope glycoprotein binds the ecotropic receptor. J Virol. 1991 Aug;65(8):4026–4032. doi: 10.1128/jvi.65.8.4026-4032.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard M. J., Klee C. B. Functional domain structure of calcineurin A: mapping by limited proteolysis. Biochemistry. 1989 Feb 21;28(4):1868–1874. doi: 10.1021/bi00430a066. [DOI] [PubMed] [Google Scholar]
- Jentoft N. Why are proteins O-glycosylated? Trends Biochem Sci. 1990 Aug;15(8):291–294. doi: 10.1016/0968-0004(90)90014-3. [DOI] [PubMed] [Google Scholar]
- Kabat D. Molecular biology of Friend viral erythroleukemia. Curr Top Microbiol Immunol. 1989;148:1–42. doi: 10.1007/978-3-642-74700-7_1. [DOI] [PubMed] [Google Scholar]
- Kato H., Enjyoji K. Amino acid sequence and location of the disulfide bonds in bovine beta 2 glycoprotein I: the presence of five Sushi domains. Biochemistry. 1991 Dec 17;30(50):11687–11694. doi: 10.1021/bi00114a012. [DOI] [PubMed] [Google Scholar]
- Kayman S. C., Kopelman R., Projan S., Kinney D. M., Pinter A. Mutational analysis of N-linked glycosylation sites of Friend murine leukemia virus envelope protein. J Virol. 1991 Oct;65(10):5323–5332. doi: 10.1128/jvi.65.10.5323-5332.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W., Zimmermann W., Oliff A., Friedrich R. Molecular analysis of the envelope gene and long terminal repeat of Friend mink cell focus-inducing virus: implications for the functions of these sequences. J Virol. 1984 Mar;49(3):828–840. doi: 10.1128/jvi.49.3.828-840.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo Y., Ogura N., Nakagawa H. Limited proteolysis of the nitrate reductase from spinach leaves. J Biol Chem. 1988 Dec 25;263(36):19684–19689. [PubMed] [Google Scholar]
- Kurth J., Stoffel W. A facile method for the isolation and preparation of proteins and peptides for sequence analysis in the picomolar range. Biol Chem Hoppe Seyler. 1990 Aug;371(8):675–685. [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Linder D., Stirm S., Schneider J., Hunsmann G., Smythers G., Oroszlan S. Glycoproteins of friend murine leukemia virus: separation and NH2-terminal amino acid sequences of gp69 and gp71. J Virol. 1982 Apr;42(1):352–355. doi: 10.1128/jvi.42.1.352-355.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder M., Linder D., Hahnen J., Schott H. H., Stirm S. Localization of the intrachain disulfide bonds of the envelope glycoprotein 71 from Friend murine leukemia virus. Eur J Biochem. 1992 Jan 15;203(1-2):65–73. doi: 10.1111/j.1432-1033.1992.tb19828.x. [DOI] [PubMed] [Google Scholar]
- Matano T., Odawara T., Ohshima M., Yoshikura H., Iwamoto A. trans-dominant interference with virus infection at two different stages by a mutant envelope protein of Friend murine leukemia virus. J Virol. 1993 Apr;67(4):2026–2033. doi: 10.1128/jvi.67.4.2026-2033.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott D., Friedrich R., Rein A. Sequence analysis of amphotropic and 10A1 murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J Virol. 1990 Feb;64(2):757–766. doi: 10.1128/jvi.64.2.757-766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage C. R., Jr, Hash J. H., Cohen S. Epidermal growth factor. Location of disulfide bonds. J Biol Chem. 1973 Nov 25;248(22):7669–7672. [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Wilhelm O. G., Jaskunas S. R., Vlahos C. J., Bang N. U. Functional properties of the recombinant kringle-2 domain of tissue plasminogen activator produced in Escherichia coli. J Biol Chem. 1990 Aug 25;265(24):14606–14611. [PubMed] [Google Scholar]
- Wilson J. E. The use of monoclonal antibodies and limited proteolysis in elucidation of structure-function relationships in proteins. Methods Biochem Anal. 1991;35:207–250. doi: 10.1002/9780470110560.ch4. [DOI] [PubMed] [Google Scholar]




