Abstract
Background
The systematic study of the human genome indicates that the inter-individual variability is greater than expected and it is not only related to sequence polymorphisms but also to gene copy number variants (CNVs). Congenital Adrenal Hyperplasia due to 21-hydroxylase deficiency (21OHD) is the most common autosomal recessive disorder with a carrier frequency of 1∶25 to 1∶10. The gene that encodes 21-hydroxylase enzyme, CYP21A2, is considered to be one of the most polymorphic human genes. Copy number variations, such as deletions, which are severe mutations common in 21OHD patients, or gene duplications, which have been reported as rare events, have also been described. The correct characterization of 21OHD alleles is important for disease carrier detection and genetic counselling
Methodology and Findings
CYP21A2 genotyping by sequencing has been performed in a random sample of the Spanish population, where 144 individuals recruited from university students and employees of the hospital were studied. The frequency of CYP21A2 mutated alleles in our sample was 15.3% (77.3% were mild mutations, 9% were severe mutations and 13.6% were novel variants). Gene dosage assessment was also performed when CYP21A2 gene duplication was suspected. This analysis showed that 7% of individuals bore a chromosome with a duplicated CYP21A2 gene, where one of the copies was mutated.
Conclusions
As far as we know, the present study has shown the highest frequency of 21OHD carriers reported by a genotyping analysis. In addition, a high frequency of alleles with CYP21A2 duplications, which could be misinterpreted as 21OHD alleles, was found. Moreover, a high frequency of novel genetic variations with an unknown effect on 21-hydroxylase activity was also found. The high frequency of gene duplications, as well as novel variations, should be considered since they have an important involvement in carrier testing and genetic counseling.
Introduction
Congenital Adrenal Hyperplasia due to 21-hydroxylase deficiency (21OHD) (OMIM +201910) is the most common autosomal recessive disorder. This enzymatic deficiency impairs synthesis of cortisol and/or aldosterone from cholesterol in the adrenal cortex. The severe deficiency of steroid 21-hydroxylase enzyme (21OH; EC 1.14.99.10) results in the classic form of the disease, which is divided firstly, into the salt-wasting form, where neither cortisol nor aldosterone is synthesized; and secondly, into the simple virilizing form, where a residual activity of 21OH allows aldosterone synthesis. The moderate deficiency of 21OH results in the non classic form of the disease (NC21OHD), which is characterized by precocious pseudopuberty, acne, hirsutism and oligomenorrhea, although phenotypical expression is highly variable [1]–[3].
The incidence of the classic form of the disease is 1∶23000–1∶10000 depending on the populations studied [4]–[8] and the incidence of the NC21OHD is 1∶500–1∶100 in most populations. It can be more frequent in other populations like in the Ashkenazi Jews with a 1∶27 incidence [2], [9]. The carrier frequency of 21OHD, estimated on the basis of newborn screening, is between 0.011 and 0.020 for severe mutations and 0.08 to 0.18 for mild ones. On the other hand, genotyping analyses done in New Zealanders and Middle Europeans have shown higher frequencies for carriers of severe mutations (0.040–0.055) and lower for mild ones (0.02–0.04) [4], [10]. Most 21OHD patients are carriers of deletions of the gene encoding 21OH, CYP21A2, or of any of the 9 most frequent point mutations derived from the non-functional CYP21A1P pseudogene. The rest are due to rare genetic variants, descriptions of which are continuously increasing [11]–[20].
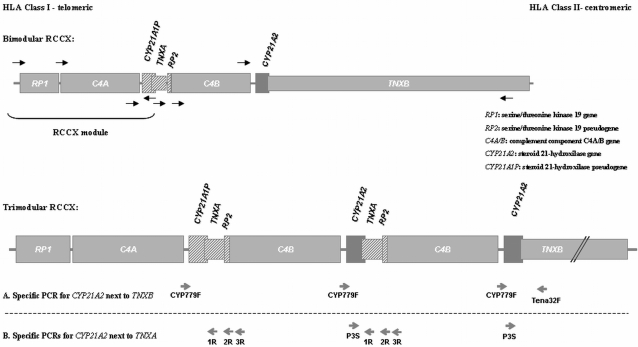
The CYP21A2 gene is 98% homologous to the CYP21A1P pseudogene in its coding sequence and 96% in introns. Both lie in the Major Histocompatibility Complex at chromosome 6p21.3, which is a complex organization of genes with variation in gene copy number and size [1], [21]. They constitute a genetic unit together with neighboring genes RP1, C4, TNXB, and their truncated pseudogenes RP2 and TNXA. This genetic unit is termed RCCX module (RP-C4-CYP21-TNX) and constitutes a highly variable stretch of DNA of approximately 30 kb [22] (Figure 1). Most chromosomes bear two of these modules, one with the CYP21A1P pseudogene and the other with the CYP21A2 gene. Monomodular and trimodular haplotypes have also been described [22]–[27].
Figure 1. Organization of RCCX module and Copy Number Variations at chromosome 6p21.
Black horizontal arrows denote gene orientation. A. Specific primers used for the amplification of CYP21A2 next to TNXB. B. Specific primers used for the amplification of CYP21A2 next to TNXA. Primer binding sites for each primer are indicated by grey horizontal arrows.
The high genetic variability at CYP21A2 locus makes the characterization of 21OHD alleles difficult, and, hence, complicates disease carrier detection and genetic counseling. In this paper, an in-depth study of the CYP21A2 gene in a group of 144 individuals is described. We found an unexpectedly high 21OHD carrier frequency, CYP21A2 gene duplications and CYP21A2 novel genetic variations. The results of the current work should be considered for 21OHD genetic diagnosis and counseling.
Results
Copy Number Variations: frequency of CYP21A2 gene duplications
The CYP21A2 gene dosage assessment by Real-Time PCR and by the specific amplification of each duplicated gene showed that 8 out of the 144 individuals were carriers of CYP21A2 gene duplication with one of the copies p.Gln318X mutated (UniProtKB ID P08686). In all the cases, the mutation was in linkage disequilibrium with two uncommon genetic variants, a G>A change in intron 2 at position -79 and a C>T change at nucleotide 13 in the 3′ UTR region (c.293-79G>A and c.*13C>T, respectively, GenBank Ref. ID NM000500.5). The PCRs for the amplification of the CYP21A2 duplicated genes confirmed the duplication. The specific sequencing of each gene showed the severe mutation on the CYP21A2 next to the TNXB gene, and the presence of a wild-type CYP21A2 next to the TNXA pseudogene.
Two other gene duplications were found by Real-Time PCR in two chromosomes bearing a chimeric CYP21A1P/CYP21A2. The duplications had been suspected by the presence of heterozygosity in the PCR products of a CYP21A2 fragment despite the fact that both individuals were carriers of p.Ile236Asn, p.Val237Glu, and p.Met239Lys mutations, which prevented the specific primer from its binding. The PCRs for the specific amplification of each CYP21A2 duplicated gene confirmed the duplication. The sequencing of each gene showed the chimeric CYP21A1P/CYP21A2 gene next to the TNXA pseudogene, and the presence of a wild-type CYP21A2 next to the TNXB gene.
The total number of duplications found in our sample results in a duplication allele frequency of 0.035, which meant a 7% frequency of carriers. Most of them bore the p.Gln318X mutation in one of the CYP21A2 genes.
Sequence Variations: frequency of putative disease-causing alleles
A total of 22 CYP21A2 mutated alleles were found (Table 1), which indicated that 16% of individuals were mutation carriers. No significant differences were found between males and females. Seventeen of these CYP21A2 genetic variants were known to maintain a residual 21-hydroxylase enzyme (21OH) activity, 2 completely impaired it, and 3 were novel variants with an unknown effect on the 21OH activity (Table 1).
Table 1. CYP21A2 genetic variations found in a random sample of 144 Spaniards.
| Type of Variation!! | Genetic variation† | Sequence Variation at protein level£ | 21OH& activity# | N° of alleles (allele frequency) |
| Mild | c.844G>T | p.Val281Leu | 20–50% | 16 (0.0555) |
| c.91C>T | p.Pro30Leu | 30–60% | 1 (0.0035) | |
| Severe | Conversion | * | 0% | 1 (0.0035) |
| c.874G>A | p.Gly291Ser | 0.8% | 1 (0.0035) | |
| Novel | c.1996C>A | p.Thr443Asn | unknown | 1 (0.0035) |
| c.553G>A | p.Asp184Asn | unknown | 1 (0.0035) | |
| c.69G>T | p.Trp22Cys | unknown | 1 (0.0035) | |
| Gene duplications | c.[Conversion;Wt] | p.[§;Wt] | 100% | 1 (0.0035) |
| c.[Conversion;Wt] | p.[‡;Wt] | 100% | 1 (0.0035) | |
| c.[955C>T;Wt] | p.[Gln318X;Wt] | 100% | 8 (0.0278) |
Single Nucleotide Polymorphisms and the insertion of CTG encoding Leu10 are not reported in this table.
GenBank Ref. ID NM_000500.5.
UniProtKB ID P08686.
21OH: 21-hydroxylase.
Data obtained from White et al. 2000.
Conversion: Chimeric CYP21A1P/CYP21A2.
p.[Pro30Leu;Gly110del8nt].
p.[Ile172Asn;Asp183Glu;Ile236Asn;Val237Glu;Met239Lys;Val281Leu;Phe306ins1T].
p.[Asp183Glu;Ile236Asn;Val237Glu;Met239Lys;Val281Leu;Phe306ins1T;Arg356Trp].
Mild mutations
The allele frequency for mild mutations was 0.059. The most frequent was p.Val281Leu, which was carried by 11% individuals.
Severe mutations
The allele frequency for severe mutations was 0.007. A chimeric CYP21A1P/CYP21A2 gene was carried by an individual and p.Gly291Ser was carried by another.
Novel genetic variations
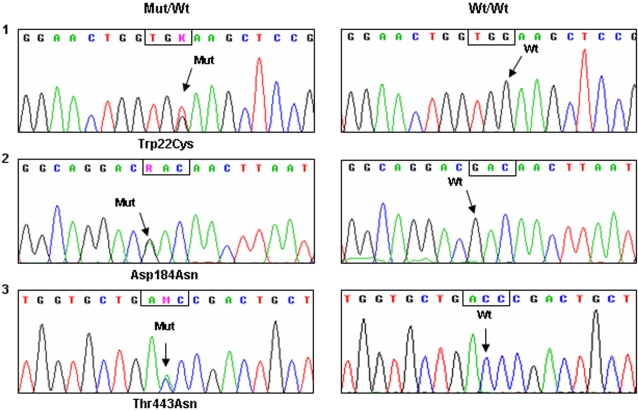
In our sample, 0.010 alleles carried novel genetic variants: p.Trp22Cys, p.Asp184Asn, and p.Thr443Asn. Each of these was found in different individuals (Figure 2). The effect of these mutations on the 21OH activity is unknown.
Figure 2. Electropherograms obtained for the three novel mutations in CYP21A2 gene and the corresponding wild-type alleles.
1. The base change from G to T at position 69 leads to the substitution of tryptophan 22 by cysteine; 2. The base change from G to A at position 553 replaces aspartic acid 184 by asparagine; 3. The base change from C to A at positition 1996 leads to the substitution of threonine 443 by asparagine. (GenBank Ref. ID NM_000500.5; UniProtKB ID P08686). Wt: wild-type; Mut: mutated.
CYP21A2 gene polymorphisms and haplotypes
In the 288 chromosomes analyzed, a total of 79 different genetic variants were found. They were distributed in the vicinity 5′ and 3′ UTR of the CYP21A2 gene as well as in introns and exons. Haplotype inference from data obtained for the 20 most polymorphic genetic variations enabled the identification of 75 different haplotypes. Most of them appeared only once, but others were recurrent. These latter were the haplotype associated to the p.Gln318X mutation, which is unique, or the haplotype associated to the p.Val281Leu mutation, which was present in 75% of these mutated alleles (data not shown).
Discussion
The estimated frequency of 21OHD allele carriers among different populations is 1.8% when inferred from the hormonal neonatal screening data [1], [28], which is designed to detect mostly the classic form of 21OHD. The frequency determined by CYP21A2 genotyping, which also detects NC21OHD, is from 4% to 10% [4], [10]. It has been reported that 14% of classic forms and 84% of non-classic forms are missed on the neonatal screening performed during the first days of life [29]. There are no 21OHD carrier frequency data for the Spanish population based on CYP21A2 genotyping. In this study, the complete analysis of the CYP21A2 gene has been performed in 144 random volunteers. The object of the study was firstly, to determine the frequency of 21OHD carriers and secondly, to characterize rare RCCX modules in alleles with duplicated CYP21A2. Although the sample size was small, our data concord with previously published studies (8); it may have been preferable to have determined the carrier frequency using a larger sample; this, however, was both time-consuming and expensive.
Copy number variations (CNVs) have been described for the RCCX locus: the presence of two RCCX modules is the standard and the presence of one, three or even four modules are rare arrangements. The trimodular organization accounts for only 14% of the chromosomes in population studies [23] and the majority carry two copies of the CYP21A1P pseudogene and one copy of the CYP21A2 gene. The trimodular haplotype has also been described with two copies of the CYP21A2 gene and one copy of the CYP21A1P pseudogene. This latter haplotype has been described, in some carriers of the p.Gln318X mutation and chimeric CYP21A1P/CYP21A2 genes, as a rare event [23]–[27]. Our series, however, have shown a high frequency of this arrangement, 7% of individuals. The specific amplification and sequencing of each duplicated gene localized the p.Gln318X mutation at the CYP21A2 gene next to TNXB gene and it showed a wild-type CYP21A2 at the 3′UTR of TNXA pseudogene. Thus, PCR approaches [30] based on the specific amplification of CYP21A2 like genes next to TNXB would fail in the detection of the wild-type gene and would misdiagnose these duplicated alleles as pathologic, which they are certainly not.
The duplicated CYP21A2 alleles found bear one inactivated gene due to the severe mutations and a wild-type gene which abolishes the pathological effect of the mutation. Furthermore, CYP21A2 duplications have recently been reported as a risk factor for de novo mutations in the offspring [13]. Hence, an awareness of the presence of these types of gene duplications is important if a misdiagnosis is to be avoided. A possible misdiagnosis could be the erroneous identification of a wild type allele as pathological. This is critical for the molecular diagnosis of 21OHD, as well as for prenatal testing and for genetic counseling.
We found that 15.3% of individuals from our sample were carriers of CYP21A2 mutations including novel variants. As far as we know, this is the highest carrier frequency described so far.
The percentage of individuals who were carriers of NC21OHD was 12%. The application of Hardy-Weinberg equilibrium to our frequency data, estimated the incidence of the mild form of the disease in 1∶225. This value is a more precise estimation than the incidence estimated by clinical diagnosis because the latter may underdiagnose patients with mild phenotypes [2]. The frequency of NC21OHD carrier found by genotyping in unselected Middle Europeans and New Zealanders was lower than ours [4], [10]. This may reflect ethnic differences between the populations studied. Although it should be noted that we sequenced the complete CYP21A2 gene whereas the other genotyping based studies only analyzed the 9–11 most common mutations.
In this investigation, as the Middle Europeans and New Zealanders studies, the p.Val281Leu mutation was the most common mutation among NC21OHD alleles (94% in our sample). This was consistent with the reported mutational spectrum of non-classic patients [2], [18].
The frequency of carriers of severe mutations found (1.4%) corresponded with the incidence of the classic form observed in ours, as well as other populations. It is also similar to data obtained from genotyping in New Zealanders [10], [21]. Nevertheless, it is much lower than the frequency described for Middle European population (5.5%) [4]. This may reflect a different incidence of classic 21OHD when European populations were compared [6]–[8]. The frequency of CYP21A2 deletions and chimeric CYP21A1P/CYP21A2 genes found was lower than the 15–20% described by other authors [31], [32]. However, it corresponded with our previous work [18] and it is similar to the frequency observed in classic 21OHD patients from Italy, France or England [33]–[35].
Three novel genetic variants were found, (p.Trp22Cys, p.Asp184Asn, and p.Thr443Asn) which account for approximately 1% of chromosomes. Two of them, p.Trp22Cys and p.Thr443Asn, are predicted to damage 21-hydroxylase activity (PolyPhen tool (http://genetics.bwh.harvard.edu/pph/)). The high frequency of novel genetic variants shows the importance of sequencing the whole CYP21A2 gene before it can be considered as wild-type. In some cases, the effect of novel genetic variants may be deduced from the phenotype of the patients. Nevertheless, in vitro functional studies are needed to ascertain them and, if possible, they should be made before considering a variation as a pathological mutation or as a functional polymorphism.
The CYP21A2 analysis in our Spanish random population has confirmed that this gene is one of the most polymorphic human ones, as it has already been described by Cargill and co-workers [36]. We found up to 79 different genetic variations at this locus. Twenty of these genetic variations showed a minor allele frequency higher than 0.01. These polymorphic variations rendered up to 73 different haplotypes (data not shown), most of them occurred only once.
In conclusion, as far as we know, the present study has shown the highest frequency of 21OHD carriers reported by a genotyping analysis. In addition, CYP21A2 gene duplications with one of the copies mutated have been found to be a common event. Moreover, a high frequency of novel genetic variants with an unknown effect on 21OH activity was found and represents a source of uncertainty in 21OHD genetic diagnosis. The number of novel variants, as well as gene duplications found here, could be as high in other populations and should be considered when the 21OHD genetic diagnosis and genetic counseling is carried out.
Materials and Methods
DNA samples
The CYP21A2 gene was studied in a random sample of 144 individuals (95 females, 49 males) recruited from the university students and employees of the hospital. Information about their parents' and grandparents' geographic origins was provided. All these individuals gave informed consent for the analysis, and the procedures were conducted according to the principles expressed in the Declaration of Helsinki.
Genetic analysis
DNA was extracted from peripheral blood leucocytes by standard procedures.
Sequencing
The CYP21A2 gene and the closest 5′ and 3′ UTR were sequenced after being amplified in two overlapping fragments by the polymerase chain reaction method as previously described [18].
CYP21A2 copy number assessment:
For the CYP21A2 gene dosage assessment, 100 ng of DNA were used. The analytical procedure was based on a Real-Time PCR method, using specific primers for the amplification of the CYP21A2 gene, and TaqMan® (Applied Biosystems) probes as previously described [37].
Specific amplification of each duplicated CYP21A2 gene
Different DNAs previously characterized by southern blot and the Real-Time PCR method were used as controls for the following PCRs
A.- For the amplification of the CYP21A2 at the 3′ UTR of TNXB, the strategy described by Lee HH and co-workers was followed [30]. This is based on the amplification of an 8515 bp fragment by the use of a reverse primer (Tena32F), which anneals specifically to exon 32 of TNXB. The forward primer (CYP779F) is common for the 5′ UTR of CYP21A2 and CYP21A1P. The samples with a single copy of CYP21A2 on a chromosome, as well as those samples with two copies of CYP21A2 on the same chromosome, have to be positive for this PCR.
B.- For the amplification of the CYP21A2 gene at the 3′ UTR of TNXA, we designed three different PCRs using a forward primer (P3S) that anneals specifically to exon 3 of CYP21A2 and: B1, a reverse primer (1R), which anneals to TNXA producing a fragment of 4611 bp. B2, with reverse primer (2R), which specifically anneals to RP2 and forward primer (P3S) resulting in a 7042 bp fragment and B3, with a reverse primer (3R), which anneals to C4 producing a 7475 bp fragment. (Figure 1) The samples with a single copy CYP21A2 on a chromosome have to be negative for these PCRs, while samples with two CYP21A2 copies on a chromosome have to be positive for these PCRs. (Table S1. PCR conditions under request).
The duplicated CYP21A2 alleles found were subjected to these PCRs and the products obtained were sequenced in order to know whether the p.Gln318X mutated copy and the chimeric CYP21A1P/CYP21A2 genes were next to TNXB or to TNXA.
Haplotype construction
The Bayesian statistical method implemented in the program PHASE v2.1.1 was used for the haplotype construction. Only DNA variations, pathologic or not, with a minor allele frequency higher than 0.01 were considered. This selection criterion resulted in 20 genetic variations distributed all over the CYP21A2 gene, as well as the closest 5′ and 3′ UTR. Eighteen of these variations were biallelic markers, and two were triallelic markers: rs6451 and rs6467. It was also included the polymorphic insertion of an extra leucine in exon 1, and the deletion of a G at position -106 of intron 2 (rs41315224).
Supporting Information
Sequences of primers.
(0.03 MB DOC)
Acknowledgments
We would like to thank all volunteers for their contribution to the present work. We thank Peter Rees for the English editing. This work is part of S.P. PhD thesis program at Santiago de Compostela University.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors have no support or funding to report.
References
- 1.Speiser PW, White PC. Congenital adrenal hyperplasia. N Engl J Med. 2003;349:776–788. doi: 10.1056/NEJMra021561. [DOI] [PubMed] [Google Scholar]
- 2.New MI. Extensive clinical experience: nonclassical 21-hydroxylase deficiency. J Clin Endocrinol Metab. 2006;91:4205–4214. doi: 10.1210/jc.2006-1645. [DOI] [PubMed] [Google Scholar]
- 3.Krone N, Dhir V, Ivison HE, Arlt W. Congenital adrenal hyperplasia and P450 oxidoreductase deficiency. Clin Endocrinol (Oxf) 2007;66:162–172. doi: 10.1111/j.1365-2265.2006.02740.x. [DOI] [PubMed] [Google Scholar]
- 4.Baumgartner-Parzer SM, Nowotny P, Heinze G, Waldhausl W, Vierhapper H. Carrier frequency of congenital adrenal hyperplasia (21-hydroxylase deficiency) in a middle European population. J Clin Endocrinol Metab. 2005;90:775–778. doi: 10.1210/jc.2004-1728. [DOI] [PubMed] [Google Scholar]
- 5.Brosnan PG, Brosnan CA, Kemp SF, Domek DB, Jelley DH, et al. Effect of newborn screening for congenital adrenal hyperplasia. Arch Pediatr Adolesc Med. 1999;153:1272–1278. doi: 10.1001/archpedi.153.12.1272. [DOI] [PubMed] [Google Scholar]
- 6.Steigert M, Schoenle EJ, Biason-Lauber A, Torresani T. High reliability of neonatal screening for congenital adrenal hyperplasia in Switzerland. J Clin Endocrinol Metab. 2002;87:4106–4110. doi: 10.1210/jc.2002-012093. [DOI] [PubMed] [Google Scholar]
- 7.Thil'en A, Nordenstrom A, Hagenfeldt L, von Dobeln U, Guthenberg C, et al. Benefits of neonatal screening for congenital adrenal hyperplasia (21-hydroxylase deficiency) in Sweden. Pediatrics. 1998;101:E11. doi: 10.1542/peds.101.4.e11. [DOI] [PubMed] [Google Scholar]
- 8.van der Kamp HJ, Wit JM. Neonatal screening for congenital adrenal hyperplasia. Eur J Endocrinol. 2004;151(Suppl 3):U71–75. doi: 10.1530/eje.0.151u071. [DOI] [PubMed] [Google Scholar]
- 9.Zerah M, Ueshiba H, Wood E, Speiser PW, Crawford C, et al. Prevalence of nonclassical steroid 21-hydroxylase deficiency based on a morning salivary 17-hydroxyprogesterone screening test: a small sample study. J Clin Endocrinol Metab. 1990;70:1662–1667. doi: 10.1210/jcem-70-6-1662. [DOI] [PubMed] [Google Scholar]
- 10.Fitness J, Dixit N, Webster D, Torresani T, Pergolizzi R, et al. Genotyping of CYP21, linked chromosome 6p markers, and a sex-specific gene in neonatal screening for congenital adrenal hyperplasia. J Clin Endocrinol Metab. 1999;84:960–966. doi: 10.1210/jcem.84.3.5550. [DOI] [PubMed] [Google Scholar]
- 11.Baradaran-Heravi A, Vakili R, Robins T, Carlsson J, Ghaemi N, et al. Three novel CYP21A2 mutations and their protein modelling in patients with classical 21-hydroxylase deficiency from northeastern Iran. Clin Endocrinol (Oxf) 2007;67:335–341. doi: 10.1111/j.1365-2265.2007.02886.x. [DOI] [PubMed] [Google Scholar]
- 12.Barbaro M, Baldazzi L, Balsamo A, Lajic S, Robins T, et al. Functional studies of two novel and two rare mutations in the 21-hydroxylase gene. J Mol Med. 2006;84:521–528. doi: 10.1007/s00109-006-0043-7. [DOI] [PubMed] [Google Scholar]
- 13.Baumgartner-Parzer SM, Fischer G, Vierhapper H. Predisposition for de novo gene aberrations in the offspring of mothers with a duplicated CYP21A2 gene. J Clin Endocrinol Metab. 2007;92:1164–1167. doi: 10.1210/jc.2006-2189. [DOI] [PubMed] [Google Scholar]
- 14.Friaes A, Rego AT, Aragues JM, Moura LF, Mirante A, et al. CYP21A2 mutations in Portuguese patients with congenital adrenal hyperplasia: identification of two novel mutations and characterization of four different partial gene conversions. Mol Genet Metab. 2006;88:58–65. doi: 10.1016/j.ymgme.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Grischuk Y, Rubtsov P, Riepe FG, Grotzinger J, Beljelarskaia S, et al. Four novel missense mutations in the CYP21A2 gene detected in Russian patients suffering from the classical form of congenital adrenal hyperplasia: identification, functional characterization, and structural analysis. J Clin Endocrinol Metab. 2006;91:4976–4980. doi: 10.1210/jc.2006-0777. [DOI] [PubMed] [Google Scholar]
- 16.Krone N, Braun A, Roscher AA, Schwarz HP. A novel frameshift mutation (141delT) in exon 1 of the 21-hydroxylase gene (CYP21) in a patient with the salt wasting form of congenital adrenal hyperplasia. Mutation in brief no. 255. Online. Hum Mutat. 1999;14:90–91. doi: 10.1002/(sici)1098-1004(1999)14:1<90::aid-humu20>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 17.Krone N, Riepe FG, Partsch CJ, Vorhoff W, Bramswig J, et al. Three novel point mutations of the CYP21 gene detected in classical forms of congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Exp Clin Endocrinol Diabetes. 2006;114:111–117. doi: 10.1055/s-2005-872841. [DOI] [PubMed] [Google Scholar]
- 18.Loidi L, Quinteiro C, Parajes S, Barreiro J, Leston DG, et al. High variability in CYP21A2 mutated alleles in Spanish 21-hydroxylase deficiency patients, six novel mutations and a founder effect. Clin Endocrinol (Oxf) 2006;64:330–336. doi: 10.1111/j.1365-2265.2006.02465.x. [DOI] [PubMed] [Google Scholar]
- 19.Robins T, Bellanne-Chantelot C, Barbaro M, Cabrol S, Wedell A, et al. Characterization of novel missense mutations in CYP21 causing congenital adrenal hyperplasia. J Mol Med. 2007;85:247–255. doi: 10.1007/s00109-006-0121-x. [DOI] [PubMed] [Google Scholar]
- 20.Stikkelbroeck NM, Hoefsloot LH, de Wijs IJ, Otten BJ, Hermus AR, et al. CYP21 gene mutation analysis in 198 patients with 21-hydroxylase deficiency in The Netherlands: six novel mutations and a specific cluster of four mutations. J Clin Endocrinol Metab. 2003;88:3852–3859. doi: 10.1210/jc.2002-021681. [DOI] [PubMed] [Google Scholar]
- 21.White PC, Speiser PW. Congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Endocr Rev. 2000;21:245–291. doi: 10.1210/edrv.21.3.0398. [DOI] [PubMed] [Google Scholar]
- 22.Yang Z, Mendoza AR, Welch TR, Zipf WB, Yu CY. Modular variations of the human major histocompatibility complex class III genes for serine/threonine kinase RP, complement component C4, steroid 21-hydroxylase CYP21, and tenascin TNX (the RCCX module). A mechanism for gene deletions and disease associations. J Biol Chem. 1999;274:12147–12156. doi: 10.1074/jbc.274.17.12147. [DOI] [PubMed] [Google Scholar]
- 23.Blanchong CA, Zhou B, Rupert KL, Chung EK, Jones KN, et al. Deficiencies of human complement component C4A and C4B and heterozygosity in length variants of RP-C4-CYP21-TNX (RCCX) modules in caucasians. The load of RCCX genetic diversity on major histocompatibility complex-associated disease. J Exp Med. 2000;191:2183–2196. doi: 10.1084/jem.191.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koppens PF, Hoogenboezem T, Degenhart HJ. Duplication of the CYP21A2 gene complicates mutation analysis of steroid 21-hydroxylase deficiency: characteristics of three unusual haplotypes. Hum Genet. 2002;111:405–410. doi: 10.1007/s00439-002-0810-7. [DOI] [PubMed] [Google Scholar]
- 25.Sinnott PJ, Costigan C, Dyer PA, Harris R, Strachan T. Extended MHC haplotypes and CYP21/C4 gene organisation in Irish 21-hydroxylase deficiency families. Hum Genet. 1991;87:361–366. doi: 10.1007/BF00200920. [DOI] [PubMed] [Google Scholar]
- 26.Wedell A, Stengler B, Luthman H. Characterization of mutations on the rare duplicated C4/CYP21 haplotype in steroid 21-hydroxylase deficiency. Hum Genet. 1994;94:50–54. doi: 10.1007/BF02272841. [DOI] [PubMed] [Google Scholar]
- 27.Haglund-Stengler B, Martin Ritzen E, Gustafsson J, Luthman H. Haplotypes of the steroid 21-hydroxylase gene region encoding mild steroid 21-hydroxylase deficiency. Proc Natl Acad Sci U S A. 1991;88:8352–8356. doi: 10.1073/pnas.88.19.8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rey Liste MT, García Caeiro AL. Cribado neonatal de la hiperplasia suprarrenal congénita. Aplicabilidad en Galicia. Axencia de Avaliación de Tecnoloxías Sanitarias de Galicia avalia-t. 2004 [Google Scholar]
- 29.Therrell BL, Jr, Berenbaum SA, Manter-Kapanke V, Simmank J, Korman K, et al. Results of screening 1.9 million Texas newborns for 21-hydroxylase-deficient congenital adrenal hyperplasia. Pediatrics. 1998;101:583–590. doi: 10.1542/peds.101.4.583. [DOI] [PubMed] [Google Scholar]
- 30.Lee HH, Lee YJ, Lin CY. PCR-based detection of the CYP21 deletion and TNXA/TNXB hybrid in the RCCX module. Genomics. 2004;83:944–950. doi: 10.1016/j.ygeno.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 31.Krone N, Braun A, Roscher AA, Knorr D, Schwarz HP. Predicting phenotype in steroid 21-hydroxylase deficiency? Comprehensive genotyping in 155 unrelated, well defined patients from southern Germany. J Clin Endocrinol Metab. 2000;85:1059–1065. doi: 10.1210/jcem.85.3.6441. [DOI] [PubMed] [Google Scholar]
- 32.Wedell A, Thilen A, Ritzen EM, Stengler B, Luthman H. Mutational spectrum of the steroid 21-hydroxylase gene in Sweden: implications for genetic diagnosis and association with disease manifestation. J Clin Endocrinol Metab. 1994;78:1145–1152. doi: 10.1210/jcem.78.5.8175971. [DOI] [PubMed] [Google Scholar]
- 33.Rumsby G, Avey CJ, Conway GS, Honour JW. Genotype-phenotype analysis in late onset 21-hydroxylase deficiency in comparison to the classical forms. Clin Endocrinol (Oxf) 1998;48:707–711. doi: 10.1046/j.1365-2265.1998.00402.x. [DOI] [PubMed] [Google Scholar]
- 34.Balsamo A, Cacciari E, Baldazzi L, Tartaglia L, Cassio A, et al. CYP21 analysis and phenotype/genotype relationship in the screened population of the Italian Emilia-Romagna region. Clin Endocrinol (Oxf) 2000;53:117–125. doi: 10.1046/j.1365-2265.2000.01048.x. [DOI] [PubMed] [Google Scholar]
- 35.Deneux C, Tardy V, Dib A, Mornet E, Billaud L, et al. Phenotype-genotype correlation in 56 women with nonclassical congenital adrenal hyperplasia due to 21-hydroxylase deficiency. J Clin Endocrinol Metab. 2001;86:207–213. doi: 10.1210/jcem.86.1.7131. [DOI] [PubMed] [Google Scholar]
- 36.Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 37.Parajes S, Quinterio C, Dominguez F, Loidi L. A simple and robust quantitative PCR assay to determine CYP21A2 gene dose in the diagnosis of 21-hydroxylase deficiency. Clin Chem. 2007;53:1577–1584. doi: 10.1373/clinchem.2007.087361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of primers.
(0.03 MB DOC)