Abstract
Background
One of the central issues in ecology is the question what allows sympatric occurrence of closely related species in the same general area? The non-biting midges Chironomus riparius and C. piger, interbreeding in the laboratory, have been shown to coexist frequently despite of their close relatedness, similar ecology and high morphological similarity.
Methodology/Principal Findings
In order to investigate factors shaping niche partitioning of these cryptic sister species, we explored the actual degree of reproductive isolation in the field. Congruent results from nuclear microsatellite and mitochondrial haplotype analyses indicated complete absence of interspecific gene-flow. Autocorrelation analysis showed a non-random spatial distribution of the two species. Though not dispersal limited at the scale of the study area, the sister species occurred less often than expected at the same site, indicating past or present competition. Correlation and multiple regression analyses suggested the repartition of the available habitat along water chemistry gradients (nitrite, conductivity, CaCO3), ultimately governed by differences in summer precipitation regime.
Conclusions
We show that these morphologically cryptic sister species partition their niches due to a certain degree of ecological distinctness and total reproductive isolation in the field. The coexistence of these species provides a suitable model system for the investigation of factors shaping the distribution of closely related, cryptic species.
Introduction
Competition for resources will generally be most severe among closely related species, because they tend to have, due to their shared phylogenetic history, the most similar demands [1], [2]. It is widely assumed that the sympatric coexistence of sibling or sister species requires some sort of resource partitioning under resource-limited conditions [3]–[5], but see [6]. This “limiting similarity” concept [7] may not hold under certain, narrowly defined circumstances [8], [9], but these instances are believed to be rather the exception from the rule [8] . Hence, testing for differences in the realised ecological niche will consequently be the first logical step in order to explain the coexistence of similar, closely related species. However, closely related species often tend to be morphologically similar for the same reason they are ecologically alike [10]. Therefore, proper species delimitation and unequivocal recognition in field studies are a necessary prerequisite, often requiring molecular methods [11] .
The dipteran midges Chironomus riparius Meigen 1804 (synonym C. thummi, respectively C. thummi thummi) and Chironomus piger Strenzke 1959 (synonym C. thummi piger) are sister taxa [12], [13]. Larvae of both species are widely distributed in small streams, ditches, ponds and puddles throughout the holarctic [14]. The life cycle of C. riparius and C. piger consists of four larval stages, a short pupal stage and the adult midge. Adults form large mating swarms. A few days after hatching, female midges usually produce a single egg mass containing several hundreds of eggs. The larvae hatch after a few days, and the whole life cycle may be completed within four weeks. Depending on the water temperature, both species are usually multivoltin, with a first generation emerging early in spring and the final generation swarming around late autumn [15]. The overwintering generation consists solely of later larval stages (L3, L4). The species are often dominating the local Chironomus community [16]. In the study region they are frequently found together at the same sites [14], making the species pair an interesting model for the investigation of mechanisms enabling the sympatric coexistence of sibling species.
As the two sister taxa are morphologically cryptic, safe species discrimination was only possible only by analysis of polytene chromosomal structure in the past [17]. Despite their morphological similarity, genome size differs by 30%, mainly due to repetitive DNA [18]. Not only their taxonomic status regarding species or subspecies rank is unclear, also the reports on their degree of reproductive isolation are inconsistent. Some degree of prezygotic isolation in the field is warranted by differential swarming behaviour [19]. While some studies indicate that C. riparius and C. piger readily form viable and fully fertile interspecific hybrids in the laboratory [20], others estimate fertile hybrids in the wild to be effectively absent, due to fertility reductions caused by hybrid dysgenesis syndromes [21]. The actual degree of hybridisation and reproductive isolation in the field, however, has not yet been explored.
In this study we aimed to investigate distributional patterns of both species in an area where both species co-occur, and to reveal ecological factors that may have shaped the observed distribution. To this end, we investigated genetic differentiation between the species using mitochondrial and nuclear markers and related their relative abundance with environmental parameters. In particular, we answered successfully the following questions:
What is the degree of reproductive isolation among C. piger and C. riparius in the field,
is there a non-random spatial pattern of distribution and co-occurrence, and
can we identify ecological parameters potentially structuring the species distribution?
Results
Species delimitation and identification
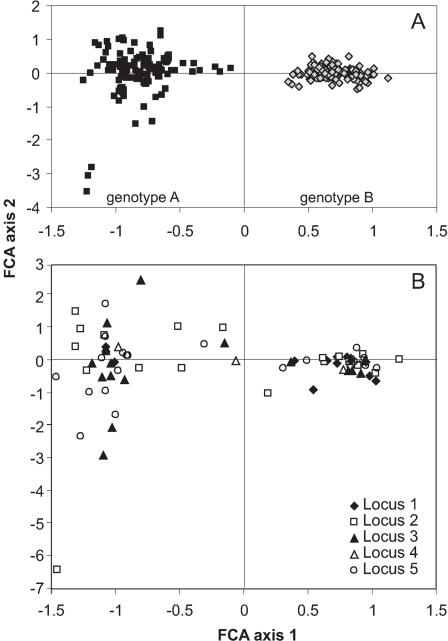
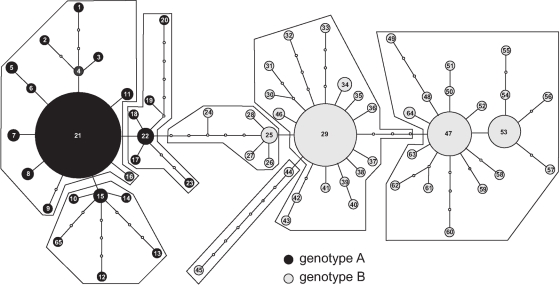
Two hundred sixty four individuals of C. riparius/piger were found at 34 sampling sites (Table 1). Microsatellite analysis detected a total of 76 alleles at the five loci (mean = 15.2, s.d. = 8.1). Factorial correspondence analysis on the microsatellite data revealed two distinct genotype clusters, termed A and B (Figure 1A). Their distinctness was due to both private alleles and frequency differences at all loci (Figure 1B). Identical results were obtained with other assignment methods like STRUCTURE (Pritchard et al., 1999) (results not shown). The statistical parsimony network revealed two major haplotype groups, linked by six mutational steps. Plotting the two nuclear genotypes on the haplotypes of the respective individuals revealed a complete congruence with these two haplogroups (Figure 2). Polytene chromosome preparations identified genotype A (black symbols) consistently as C. riparius and genotype B (grey symbols) as C. piger.
Table 1. Sampling sites, their geographical position , number of C. riparius/pigerindividuals found on 1 m2 and number of individuals used for genetic analysis.
| Site | Latitude | Longitude | Type of water body | Width (m) | Depth (m) | N C.riparius/C. piger | N µsats | N COI |
| ABR | 49.4777 | 8.3207 | ditch | 1.3 | 0.3 | 1/1 | 2 | 1 |
| BBB | 49.3677 | 9.0267 | creek | 1.0 | 0.2 | 7/5 | 12 | 11 |
| BBM | 49.5268 | 8.6142 | ditch | 1.0 | 0.2 | 5/7 | 12 | 12 |
| BGL | 49.5121 | 8.2977 | ditch | 1.0 | 0.2 | 2/10 | 12 | 12 |
| BMS | 49.4380 | 8.3392 | creek | 1.0 | 0.2 | -/8 | 5 | 8 |
| EBO | 49.2263 | 8.7006 | ditch | 1.5 | 0.3 | 1/- | - | 2 |
| FBD | 49.4363 | 8.3249 | creek | 1.5 | 0.3 | 1/- | 1 | 1 |
| FBL | 49.4246 | 8.3557 | creek | 1.0 | 0.1 | 8/- | 8 | 8 |
| GBH | 49.3144 | 8.5307 | ditch | 1.0 | 0.1 | 7/3 | 10 | 10 |
| GBR | 49.2932 | 8.4151 | creek | 1.5 | 0.3 | 4/- | 2 | 4 |
| HBD | 49.4499 | 8.6542 | ditch | 1.5 | 0.3 | 6/1 | 7 | 7 |
| HBS | 49.2730 | 8.3339 | creek | 0.6 | 0.2 | 8/- | 7 | 8 |
| KBH | 49.3296 | 8.4913 | ditch | 2.0 | 0.3 | 25/- | 25 | 24 |
| KBK | 49.2283 | 8.6405 | creek | 4.0 | 0.8 | 1/2 | 1 | 2 |
| KBT | 49.3313 | 8.5248 | creek | 4.0 | 0.6 | 1/2 | 2 | 1 |
| KLB | 49.2264 | 8.6429 | creek | 2.5 | 0.2 | -/1 | 1 | 1 |
| LBD | 49.4145 | 8.3165 | ditch | 1.0 | 0.1 | 2/10 | 9 | 11 |
| LGH | 49.5431 | 8.3168 | ditch | 2.5 | 0.3 | 5/17 | 22 | 11 |
| LGN | 49.3291 | 8.6814 | creek | 4.0 | 0.5 | 1/- | - | 1 |
| MBD | 49.4408 | 8.6674 | creek | 1.5 | 0.3 | 8/1 | 7 | 7 |
| MBF | 49.4710 | 8.2921 | ditch | 1.5 | 0.3 | 2/8 | 5 | 5 |
| NBL | 49.4996 | 8.2911 | ditch | 1.5 | 0.3 | -/7 | 9 | 6 |
| NBM | 49.4919 | 8.2894 | ditch | 1.5 | 0.2 | 1/5 | 5 | 4 |
| PBF | 49.4363 | 8.3249 | puddle | 0.4 | <0.1 | -/9 | 9 | 7 |
| PBR | 49.3789 | 8.3512 | puddle | 0.4 | <0.1 | -/10 | 10 | 6 |
| PFO | 49.2137 | 8.6936 | puddle | 0.5 | <0.1 | 1/6 | 7 | 5 |
| RBD | 49.4519 | 8.6422 | creek | 1.5 | 0.2 | 9/4 | 13 | 9 |
| SBB | 49.5579 | 8.3099 | creek | 3.0 | 0.4 | 2/4 | 6 | 5 |
| SBR | 49.2749 | 8.4628 | ditch | 2.5 | 0.3 | -/1 | 1 | - |
| SBS | 49.5813 | 8.6471 | creek | 0.5 | 0.1 | 21/- | 21 | 7 |
| TAS | 49.3025 | 8.7166 | pond | 10 | 1.5 | -/2 | - | 2 |
| TBN | 49.3207 | 8.7079 | pond | 6.0 | 1.3 | 1/- | - | 1 |
| WBH | 49.3145 | 8.3361 | creek | 4.0 | 0.4 | 2/1 | 3 | 1 |
| WGH | 49.3126 | 8.3947 | creek | 3.0 | 0.3 | 8/- | 8 | 3 |
| Σ 139/125 |
Figure 1. Factor score plot of A) individuals and B) microsatellite alleles on the first two axes of factorial correspondence analysis.
A) The two obvious groups of Chironomus individuals separated along axis 1 are termed genotype A (grey squares) and genotype B (black diamonds). B) Contribution of microsatellite alleles, coded after locus. Most microsatellite alleles are typical if not exclusive for one of the two genotypes, only few alleles occur in similar proportions in both clusters.
Figure 2. Statistical parsimony network of the mitochondrial COI haplotypes.
The size of the circles is proportional to the number of individuals with the respective haplotypes. Polygons mark the 3-step clades. The nuclear genotype of the respective individuals is indicated (A = grey, B = black).
Co-occurrence and population structure
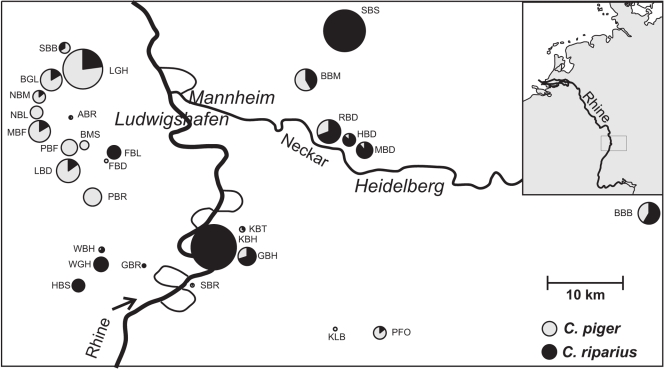
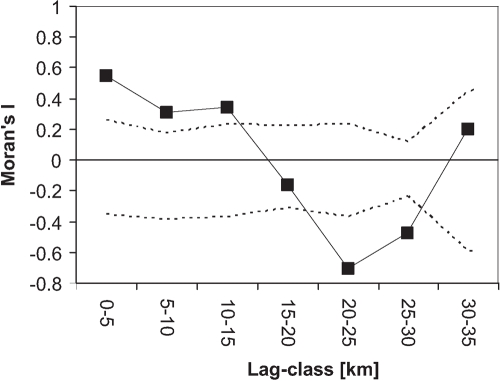
At about half of the sampling sites containing C. riparius or C. piger, both species co-occurred in varying proportions (Figure 3). However, individuals of the species occurred less often together than expected by chance (Fisher's exact test χ2 = 160, d.f. = 22, p<0.0001). The relative frequency of a species at a given site was not independent from their frequency at surrounding sites. We found a significant spatial autocorrelation of sampling sites up to 15 km apart (Figure 4). A significant, albeit very weak genetic population structure within both species was detected. For C. piger, a ΦST of 0.027 was calculated, while the estimate for C. riparius was 0.046 (Table 2), indicating a high amount of gene-flow among sampling sites among individuals of each species.
Figure 3. Map of the sampling points in the Rhine valley with pie charts of the relative frequency of Chironomus riparius (black) and C. piger (grey).
The size of the circles corresponds to the number of individuals sampled.
Figure 4. Spatial autocorrelation analysis of the relative C. riparius frequency for lag-classes of 5 km.
Dashed lines indicate the 95% confidence interval of observed Moran's I being significantly different from zero, derived from 999 permutations.
Table 2. Population structure estimated with AMOVA of microsatellite data for Chironomus piger and C. riparius.
| Source of variation | d.f. | SS | MS | Percent of total variance | ΦST | p |
| Among C. piger sampling sites | 10 | 38.673 | 3.867 | 3 | 0.027 | 0.040 |
| Within sampling sites | 85 | 265.460 | 3.123 | 97 | ||
| Total | 95 | 304.133 | 6.990 | |||
| Among C. riparius sampling sites | 8 | 44.717 | 5.590 | 5 | 0.046 | 0.010 |
| Within sampling sites | 91 | 334.972 | 3.681 | 95 | ||
| Total | 99 | 379.689 | 9.271 |
Significance of variance components was assessed with 9999 permutations.
Environmental correlates of relative species frequencies
Of the 38 variables tested, six were significantly correlated (p<0.05, q<0.10) with the relative C. riparius frequencies: conductivity (r = −0.624), Nitrite (−0.611), CaCO3 (−0.725), precipitation in the wettest month (+0.672), and precipitation in the warmest quarter (+0.582) (Table 3). Multiple regression retained only precipitation in the wettest month (July) and in the warmest quarter as significant (May–July, Table 4).
Table 3. Pearson correlation coefficients (r) of environmental variables with relative frequencies of C. riparius and the respective p and q values for multiple comparisons.
| variable | r | p | q |
| pH | 0.155 | 0.566 | 0.354 |
| Conductivity | *−0.624 | 0.010 | 0.048 |
| Velocity | 0.372 | 0.156 | 0.195 |
| Temperature | −0.246 | 0.359 | 0.346 |
| O2 | 0.418 | 0.108 | 0.178 |
| Nitrate | 0.169 | 0.531 | 0.354 |
| Nitrite | *−0.611 | 0.012 | 0.048 |
| Phosphate | 0.119 | 0.662 | 0.378 |
| Ammonium | −0.135 | 0.619 | 0.375 |
| CaCO3 | *−0.725 | 0.001 | 0.020 |
| Chloride | −0.274 | 0.304 | 0.320 |
| Organic content | 0.087 | 0.750 | 0.388 |
| >4mm | 0.071 | 0.795 | 0.388 |
| <4mm>2mm | 0.244 | 0.363 | 0.346 |
| <2mm>1mm | 0.292 | 0.273 | 0.304 |
| <1mm>630µm | 0.170 | 0.530 | 0.354 |
| <630µm>500µm | −0.127 | 0.638 | 0.375 |
| <500µm>250µm | −0.226 | 0.400 | 0.354 |
| <250µm | 0.007 | 0.978 | 0.436 |
| Annual Mean Temperature | 0.201 | 0.455 | 0.354 |
| Mean Monthly Temperature Range | 0.006 | 0.982 | 0.436 |
| Isothermality | −0.409 | 0.116 | 0.178 |
| Temperature Seasonality | 0.467 | 0.068 | 0.160 |
| Max Temp. of Warmest Month | 0.162 | 0.549 | 0.354 |
| Min Temperature of Coldest Month | −0.082 | 0.763 | 0.388 |
| Temp Annual Range | 0.291 | 0.274 | 0.304 |
| Mean Temp Wettest Quarter | 0.209 | 0.438 | 0.354 |
| Mean Temp Dryest Quarter | 0.054 | 0.844 | 0.402 |
| Mean Temp Warmest Quarter | 0.209 | 0.438 | 0.354 |
| Mean Temp Coldest Quarter | 0.075 | 0.784 | 0.388 |
| Annual Preciptiation | 0.446 | 0.083 | 0.166 |
| Prec Wettest Month | *0.672 | 0.004 | 0.040 |
| Prec Driest Month | 0.383 | 0.143 | 0.195 |
| Prec Seasonality | 0.164 | 0.544 | 0.354 |
| Mean Prec Wettest Quarter | *0.582 | 0.018 | 0.051 |
| Mean Prec Dryest Quarter | 0.374 | 0.153 | 0.195 |
| Mean Prec Warmest Quarter | *0.582 | 0.018 | 0.051 |
| Mean Prec Coldest Quarter | 0.193 | 0.474 | 0.354 |
Correlations with p<0.05 and q<0.1 are marked with asterisks.
Table 4. Multiple regression of environmental parameters significantly correlated with relative C. riparius frequencies (forward selection).
| variable | β | b | p |
| Prec Wettest Month (July) | 2.60 | 34.90 | 0.007 |
| Mean Prec Wettest Quarter (May–July) | 2.18 | 36.33 | 0.020 |
| CaCO3 | −0.69 | −0.60 | 0.102 |
| Conductivity | 0.47 | 0.89 | 0.225 |
| Nitrite | −0.20 | −1.78 | 0.309 |
The regression was significant (N = 18, F(5,12) = 6.7, p = 0.003, corr. r2 = 0.63).
Discussion
Chironomus riparius and C. piger behave as good species in the field
Microsatellite analysis showed the presence of two distinct genotype groups without intermediates, indicating complete reproductive isolation and the absence of putative hybrids (Figure 1A). Most alleles were specific to one of the cluster with only few alleles shared in similar proportions by the two taxa (Figure 1B). As only the lengths of PCR-fragments were scored, this partial overlap may be due to homoplasy or common ancestry. The results were so clear cut that the application of more sophisticated methods of hybrid detection (e.g. NewHybrids) was deemed unnecessary. The inference of reproductively isolated gene-pools is strengthened by the distinctness of the mitochondrial variation of the genotype groups (Figure 2), indicating long lasting isolation with absence of both current and past hybridisation [22]. Even though the generation of hybrids in the laboratory is possible to varying degrees [17], [21], both pre- and postzygotic isolation mechanisms [19], [21] seemed to have maintained complete reproductive isolation in the wild, despite the opportunity to interbred. Therefore, the two taxa conform to several species concepts, including the biological [10] at least in the area investigated and they should be consequently regarded as good species.
Spatial repartition of C. riparius and C. piger along ecological gradients
The unequivocal species assignment by molecular markers showed that larvae of C. riparius and C. piger occurred not only in the same general area, but in about half of the cases at the same site (Figure 3). Still, they were found less often syntopically than expected from their overall abundance. The virtual absence of population structure on the spatial scale of the study (Table 2) suggests that this is not due to dispersal restrictions or geographical obstacles. It indicates rather competitive interaction, either present or past [23]. C. piger was dominant mainly in the west of the area, while C. riparius occurred more frequently in the east (Figure 3), as mirrored in the significant spatial autocorrelation of relative species frequency (Figure 4). This pattern corresponds to the correlation of the species' relative frequencies with parameters measuring the average amount of precipitation during the summer months (Table 3). The latter are climatic parameters which generally tend to be spatially autocorrelated. Other parameters that covaried significantly with relative species abundances were water chemistry variables (conductivity, nitrite and CaCO3, Table 3). Multiple regression retained only the precipitation variables (Table 4). This suggests that differential desiccation resistance, as observed in other Chironomus species [24], could have caused the observed patterns. However, the absolute differences in precipitation (see Appendix) are probably too small for a differential desiccation risk of the water bodies in the area. Therefore, the climate constitutes probably merely the ultimate cause for the differential species distribution. The amount of rain during the warm summer months with their increased evaporation determines the concentrations of nitrite and other ions in the shallow puddles and ditches both species inhabit. It has been shown that both high salinity and high nitrite concentrations impart larval development in C. riparius/piger [25], [26]. Our results indicate that C. piger occurs in areas with less summer rain and tolerates higher nitrite concentrations and conductivity than C. riparius (Table 3). Therefore, the proximate cause for the observed correlation of the species frequencies with summer precipitation is more likely the gradient of water chemistry variables during the time of highest larval abundance [27].
Even though the inferred spatial repartition along ecological gradients is rather a hypothesis in need to be confirmed by subsequent experiments in the laboratory, it has become evident that C. riparius and C. piger are ecologically not completely equivalent. Although this has already been suspected before [14], our study is the first to demonstrate ecological partitioning among the species pair quantitatively in the field. Studies on the ecological differentiation of other Chironomus species have revealed a range of mechanisms that structure coexistence in sympatry. Dietary niche separation among two profundal species from the Chironomus plumosus-group has been suggested by stable isotope analysis [28]. The same species were found to differ in emergence time, suggesting also temporal niche separation [29]. Perhaps the most impressive example of interspecies competition avoidance is the spatial repartition of temporary rain water puddles by C. pulcher and C. imicola into shaded and sunny regions on a very small scale [30].
Despite the demonstrated spatial repartition along ecological gradients of C. riparius and C. piger, we found a substantial number of sites where both species co-occurred, indicating a substantial overlap in the realised ecological niche. Possible, not mutually exclusive explanations for this pattern include: i) a substantial stochasticity in the dispersal/colonisation of the sites. Even though the oviposition choice in another Chironomus species is influenced by nitrogenous compounds and conspecific larvae [31], a high degree of randomness regarding environmental conditions is generally assumed in the community assembly of chironomids [27]. Also which species arrived first at a yet unoccupied site may crucially influence the outcome of subsequent competition [32]. ii) Temporally fluctuating environmental conditions may also prevent complete competitive exclusion [33]. iii) Interaction with other species. Several other species of Chironomus are present at most of the investigated sites [16], as well as other mud dwelling taxa with similar requirements. iv) the abundance in the neighbourhood possibly also influences the local abundance of C. riparius and/or C. piger [33].
As this study documents, C. riparius and C. piger provide a promising model for the investigation of factors shaping the distribution of closely related, cryptic species. Currently ongoing experimental and ecological genomic studies on this emerging model system will help to gain a deeper understanding of the processes and factors that shape the realised niche of closely related species in sympatry. Understanding the internal factors and constraints shaping their distribution and coexistence will contribute to our mechanistical understanding of the processes shaping biodiversity in ecological communities.
Materials and Methods
Sampling
The sampling area lies in the middle of the upper Rhine valley in a rectangle of roughly 40 by 60 km between 49°09′–49°33′N and 8°10′–8°13′E. It comprises the Rhine valley plain, in the west limited by the mountains of the Pfälzer Wald and in the east by the rising hills of the Odenwald range. The area is hydrologically characterised by the presence of many drainage ditches, slowly flowing small streams, temporary puddles, the oxbows and the main stream of the river Rhine.
The sampling took place from mid September to November 2004, thus sampling the over-wintering generation of Chironomus larvae [34]. The sampling period was scheduled in autumn in order to avoid the large fluctuations in abundance among species throughout summer. Moreover, sampling the hibernating larvae assemblage that will foster next years first generation presents the result of competition processes during the growth season [35].
Sampling took place as described in [12]. Briefly, potential Chironomus habitats were considered opportunistically within the study region, but we mainly focused on typical Chironomus riparius/piger habitats (small streams, creeks, and ditches with fine, muddy sediment). An area of 1×1 m was sampled with a 30×40 cm net of 0.5 mm mesh size. Due to the small size and low depth of most water bodies, we did not consider different areas within a water body during sampling. All Chironomus larvae instar stage found (instar stage 3 and 4), as identified by the presence of ventral tubuli, were brought alive into the laboratory. For the present study, we chose all thirty-four sampling sites where C. riparius and/or C. piger which had been identified earlier using a COI barcoding approach [16].
DNA isolation and microsatellite analyses
Larvae were kept in the laboratory for at least 5 days without feeding, in order to remove potential PCR inhibiting substances from the gut [36]. Head and first body segments were removed for polytene chromosome analysis as described in [17]. Briefly, salivary glands were prepared from fresh larval tissue and fixed in 50% acetic acid. Chromosomes were stained in 2% orcein acetic acid for 15 min and fixed on glass slides for microscopical analysis. Remaining caudal tissue was homogenized in 700 µl standard CTAB buffer containing 0.1 mg/ml proteinase K. After digestion for at least 1 h at 62° C, chloroform/isoamyl alcohol 24:1 treatment was performed followed by 1 h precipitation at −20° C. DNA pellets were washed twice with ethanol 70% and resolved in 30 µl water.
Allelic variation was measured at five variable, unlinked microsatellite loci [37] for 255 individuals from 29 locations (Table 1). Microsatellite fragments were amplified as described in [37]. Amplified DNA fragments were diluted 1:25 prior to fragment length analysis (ALF sequencer, Pharmacia Biotech, Uppsala, Sweden) and alleles were scored using the ALFWIN 1.0 software (Pharmacia Biotech, Uppsala, Sweden).
Genetic structure, mitochondrial haplotype phylogeny and species identification
Factorial correspondence analysis (FCA) was applied on multilocus genotypes to explore the distribution of genetic variation graphically (GENETIX 4.04 software, [38]. Genetic population structure was assessed using the AMOVA approach [39] as implemented in the Excel add-in GenAlEx [40]. For this analysis only sampling sites with at least seven conspecific individuals were taken into account. C. riparius/piger COI haplotypes were identified from [16] (GenBank Accession numbers DQ910547-DQ910729). The phylogeny of the COI haplotypes was inferred using statistical parsimony (SP) [41]. The SP network was constructed with the program TCS v. 1.21 [42]. Nesting of clades followed the rules given in [43] and [44]. Inferred reproductively isolated entities were taxonomically identified using polytene chromosome preparations of a subset of individuals [17].
Co-occurrence
We used a Fisher's exact test (106 permutations) to investigate whether the co-occurrence of the identified taxa was random. Spatial patterns of the relative frequency of C. riparius and relevant environmental parameters at the sample sites with at least seven individuals found were inferred with spatial autocorrelation analysis. Seven mutually exclusive lag classes of 5000 m width were used to compute Moran's I spatial correlation coefficient for each class. Statistical significance of Moran's I was assessed with 999 Monte Carlo permutations. The Excel Add-in RookCase version 0.99 [45] was used for the calculations.
Physico-chemical and climatic characterisation of sampling sites
Thirty-eight ecological parameters were recorded in order to characterize abiotic habitat conditions at the respective sampling size. These parameters were chosen to cover a wide range of ecological conditions known to influence freshwater communities, and the distribution of chironomid species in particular. Recorded characteristics include physicochemical parameters [46], [47], sediment composition [48], climatic conditions [49], and structural habitat characteristics (e.g., size and depth of water body).
For the determination of sediment organic content, measured as loss on ignition, approximately 30 g of sediment sample were dried at 60°C for three days and weighed subsequently. Samples were then muffled at 550°C for 4 h, followed by determination of percentage weight loss.. For the identification of relative particle size composition of the samples, 150 g of homogenised sediment were washed through six sieves with decreasing mesh size and the content of each sieve was dried and weighted.
Conductivity, pH, water temperature and O2 saturation were measured with a WTW Multi 340i multimeter at each sampling site. Ammonium, nitrite and phosphate concentrations were calorimetrically determined using Aquamerk® quicktests. Chloride, CaCO3 and nitrate concentrations were measured with colour tests (Merkoquant®). The stream velocity was measured using an AMR ALMEMO® device.
Nineteen biologically meaningful climatic parameters were extracted for each sampling site from the BIOCLIM environmental layers with a spatial resolution of 0.5 min, implemented in the computer program DIVA-GIS version 4.2 [50]. Mean, median, standard deviation, minimum and maximum values for the recorded parameters are given in Appendix S1.
Statistical analysis
Means, standard deviations, median, minimum and maximum values for all 38 variables taken into account are given in the Appendix. All data with the exception of pH were either log10 (x+1; continuous variables) or arcsin (percentages) transformed to conform to the underlying assumptions of normality and heteroscedasticity in subsequent analyses. We calculated Pearson's correlation coefficients (r) between the relative C. riparius frequencies and all respective variables. Due to the multitude of comparisons, we calculated a q value for each test to estimate the minimum false discovery rate which is incurred when calling that test significant. Variables with p values<0.05 and q values<0.10 in correlation analysis were retained for a multiple regression (forward selection).
Supporting Information
Recorded environmental parameters.
(0.11 MB DOC)
Acknowledgments
We are grateful to Christoph Kley and Holger Geupel for their kind assistance in field and lab work.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research has been supported by the programme “Environmental Quality and its Security and Sustainment” (BW-Plus) with funding of the state Baden-Ẅurttemberg (project number BWR 22018). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Darwin C. London: John Murray; 1859. The origin of species by means of natural selection or the preservation of favoured races in the struggle for life. [PMC free article] [PubMed] [Google Scholar]
- 2.Wiens JJ, Graham CH. Niche conservatism: Integrating evolution, ecology, and conservation biology. Annual Review Of Ecology Evolution And Systematics. 2005;36:519–539. [Google Scholar]
- 3.McArthur R, Levins R. The limiting similarity, convergence and divergence of coexisting species. American Naturalist. 1967;101:377–385. [Google Scholar]
- 4.Meszena G, Gyllenberg M, Pasztor L, Metz JAJ. Competitive exclusion and limiting similarity: A unified theory. Theoretical Population Biology. 2006;69:68–87. doi: 10.1016/j.tpb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Amarasekare P. Competitive coexistence in spatially structured environments: a synthesis. Ecology Letters. 2003;6:1109–1122. [Google Scholar]
- 6.Hubbell SP. Princeton: Princeton University Press; 2001. The unified neutral theory of biodiversity and biogeography. [DOI] [PubMed] [Google Scholar]
- 7.Hutchinson GE. New York: Cold Spring Harbour Press; 1957. Concluding remarks. Population studies: Animal Ecology and Demography. pp. 415–427. [Google Scholar]
- 8.Chesson P. A need for niches? Trends in Ecology & Evolution. 1991;6:26–28. doi: 10.1016/0169-5347(91)90144-M. [DOI] [PubMed] [Google Scholar]
- 9.Zhang DY, Lin K, Hanski I. Coexistence of cryptic species. Ecology Letters. 2004;7:165–169. [Google Scholar]
- 10.Mayr E. New York: Dover Publications; 1942. Systematics and the Origin of Species. [Google Scholar]
- 11.Blaxter ML. The promise of a DNA taxonomy. Philosophical Transactions of the Royal Society of London - Series B: Biological Sciences. 2004;359:669–679. doi: 10.1098/rstb.2003.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunderina LI, Kiknadze II, Istomina AG, Gusev VD, Miroshnichenko LA. Divergence of the polytene chromosome banding sequences as a reflection of evolutionary rearrangements of genome linear structure. Russian Journal of Genetics. 2005;41:130–137. [PubMed] [Google Scholar]
- 13.Guryev V, Makarevitch I, Blinov A, Martin J. Phylogeny of the genus Chironomus (Diptera) inferred from DNA sequences of mitochondrial cytochrome b and cytochrome oxidase I. Molecular Phylogenetics & Evolution. 2001;19:9–21. doi: 10.1006/mpev.2001.0898. [DOI] [PubMed] [Google Scholar]
- 14.Strenzke K. Die systematische und ökologische Differnzierung der Gattung Chironomus. Annales Entomologici Fennici. 1957;26:111–139. [Google Scholar]
- 15.Armitage PD. Behaviour and ecology of adults. In: Armitage PD, Cranston PS, Pinder LCV, editors. The Chironomidae: The biology and ecology of non-biting midges. London: Chapman & Hall; 1995. pp. 194–224. [Google Scholar]
- 16.Pfenninger M, Steinke D, Nowak C, Kley C, Streit B. Utility of DNA-taxonomy and barcoding for the inference of larval community structure in morphologically cryptic Chironomus (Diptera) species. Molecular Ecology. 2007;16:1957–1968. doi: 10.1111/j.1365-294X.2006.03136.x. [DOI] [PubMed] [Google Scholar]
- 17.Keyl HG, Keyl I. Die cytologische Diagnostik der Chironomiden. I. Bestimmungstabelle für die Gattung Chironomus auf Grund der Speicheldrüsen-Chromosomen. Archiv für Hydrobiologie. 1959;58:43–57. [Google Scholar]
- 18.Ross R, Hankeln T, Schmidt ER. Complex evolution of tandem-repetitive DNA in the Chironomus thummi species group. Journal of Molecular Evolution. 1997;44:321–326. doi: 10.1007/pl00006149. [DOI] [PubMed] [Google Scholar]
- 19.Miehlbradt J. Reproductive isolation via optical swarming behaviour in sympatric Chironomus thummi and Chironomus piger. Behaviour. 1976;58:272. [Google Scholar]
- 20.Keyl HG, Strenzke K. Taxonomie und Cytologie von zwei Subspezies der Art Chironomus thummi. Zeitschrift für Naturforschung Part B. 1956;11:727–735. [Google Scholar]
- 21.Hägele K. Hybrid syndrome-induced postzygotic reproductive isolation: a second reproduction barrier in Chironomus thummi (Diptera, Chironomidae). Journal of Zoological Systematics and Evolutionary Research. 1999;37:161–164. [Google Scholar]
- 22.Arnold ML. Natural hybridization as an evolutionary process. Annu Rev Ecol Syst. 1992;23:237–261. [Google Scholar]
- 23.Connell JH. Diversity and the coevolution of competitors, or the ghost of competition past. Oikos. 1980;35:131–138. [Google Scholar]
- 24.Suemoto T, Kawai K, Imabayashi H. A comparison of desiccation tolerance among 12 species of chironomid larvae. Hydrobiologia. 2004;515:107–114. [Google Scholar]
- 25.Haas H, Strenzke K. Experimentelle Untersuchungen über den Einfluß der ionalen Zusammensetzung des Mediums auf die Entwicklung der Analpapillen von Chironomus thummi. Biologisches Zentralblatt. 1957;76:513–528. [Google Scholar]
- 26.Neumann D, Kramer M, Raschke I, Gräfe B. Detrimental effects of nitrite on the development of benthic Chironomus larvae, in relation to their settlement in muddy sediments. Archiv für Hydrobiologie. 2001;153:103–128. [Google Scholar]
- 27.Tokeshi M. Randomness and Aggregation-Analysis of Dispersion in an Epiphytic Chironomid Community (Vol 33, Pg 3, 1995). Freshwater Biology. 1995;34:567–578. [Google Scholar]
- 28.Kelly A, Jones RI, Grey J. Stable isotope analysis provides insights into dietary separation between Chironomus anthracinus and C. plumosus. Journal of the North American Benthological Society. 2004;23:287–296. [Google Scholar]
- 29.Johnson RK, Pejler B. Life histories and coexistence of the two profundal Chironomus species in Lake Erken, Sweden. Entomologica Scandinavica Suppl. 1987;29:233–238. [Google Scholar]
- 30.McLachlan A. Can two species of midge coexist in a single puddle of rain-water? Hydrobiologia. 1993;259:1–8. [Google Scholar]
- 31.Stevens MM, Warren GN, Braysher BD. Oviposition response of Chironomus tepperi to nitrogenious compounds and bioextracts in two-choice laboratory tests. Journal of Chemical Ecology. 2003;29:911–920. doi: 10.1023/a:1022983800421. [DOI] [PubMed] [Google Scholar]
- 32.De Meester L, Gómez A, Okamura B, Schwenk K. The monopolisation hypothesis and the dispersal gene-flow paradox in aquatic organisms. Acta Oecologia. 2002;23:121–135. [Google Scholar]
- 33.Chesson P. Mechanisms of maintenance of species diversity. Annual Review of Ecology and Systematics. 2000;31:343–366. [Google Scholar]
- 34.Tokeshi M. Life cycles and population dynamics. In: Armitage PD, Cranston PS, Pinder LCV, editors. The Chironomidae: The biology and ecology of non-biting midges. London: Chapman & Hall; 1995. pp. 225–268. [Google Scholar]
- 35.Tokeshi M. Species interactions and community structure. In: Armitage PD, Cranston PS, Pinder LCV, editors. The Chironomidae: The biology and ecology of non-biting midges. London: Chapman & Hall; 1995. pp. 297–335. [Google Scholar]
- 36.Carew ME, Pettigrove V, Hoffmann AA. Identifying chironomids (Diptera:Chironomidae) for Biological Monitoring with PCR-RFLP. Bulletin of Entemological Research. 2003;8:287–293. doi: 10.1079/ber2003268. [DOI] [PubMed] [Google Scholar]
- 37.Nowak C, Hankeln T, Schmidt ER, Schwenk K. Development and localization of microsatellite markes for the sibling species Chironomus riparius and Chironomus piger (Diptera: Chironomidae). Molecular Ecology Notes in press. 2006.
- 38.Belkhir K, Borsa P, Chikli L, Raufaste N, Bonhomme F. Montpellier: Laboratoire Génome, Université Montpellier; 1996. GENETIX 4.04, Logiciel Sous Windows TM Pour la Génétique des Populations. [Google Scholar]
- 39.Excoffier L, P.E. Smouse JMQ. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction sites. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peakall R, Smouse P. GenAlEx 6: Genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data.3. cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Molecular Ecology. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 43.Crandall KA. Multiple interspecies transmissions of human and simian T-cell leukemia/lymphoma virus type I sequences. Molecular Biology and Evolution. 1996;13:115–131. doi: 10.1093/oxfordjournals.molbev.a025550. [DOI] [PubMed] [Google Scholar]
- 44.Templeton AR. Nested clade analyses of phylogeographic data: testing hypotheses about gene flow and population history. Molecular Ecology. 1998;7:381–397. doi: 10.1046/j.1365-294x.1998.00308.x. [DOI] [PubMed] [Google Scholar]
- 45.Sawada M. ROOKCASE: An Excel 97/2000 Visual Basic (VB) Add-in for Exploring Global and Local Spatial Autocorrelation. Bulletin of the Ecological Society of America. 1999;80:231–234. [Google Scholar]
- 46.Hilsenhoff WL, Narff RP. Ecology of Chironomidae, Chaoboridae, and other benthos in fourteen Wisconsin Lakes. Annals of the Entomological Society of America. 1968;61:1173–1181. [Google Scholar]
- 47.Scharf BW. Experimentell-ökologische Untersuchungen an Chironomus thummi und Chironomus piger (Diptera, Chironomidae). Archiv für Hydrobiologie. 1973;72:225–244. [Google Scholar]
- 48.Vos JH, Teunissen M, Postma JF, van den Enden FP. Particle size effect on preference and growth of detritivorous Chironomus riparius larvae as influenced by food level. Archiv für Hydrobiologie. 2002;154:103–119. [Google Scholar]
- 49.Servia JM, Combo F, Gonzalez FA. Effects of short-term climatic variations on fluctuating asymmetry levels in Chironomus riparius larvae at a polluted site. Hydrobiologia. 2004;523:137–147. [Google Scholar]
- 50.Hijmans RJ, Guarino L, Cruz M, Rojas E. Computer tools for spatial analysis of plant genetic resources data: 1. DIVA-GIS. Plant Genetic Resources Newsletter. 2001;127:15–19. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Recorded environmental parameters.
(0.11 MB DOC)