Abstract
Immunoglobulin (Ig) GM and KM allotypes—genetic markers of γ and κ chains, respectively—are associated with the outcome of hepatitis C virus (HCV) infection, but the underlying mechanisms are not well understood. We hypothesized that GM and KM allotypes could contribute to the outcome of HCV infection by influencing the levels of IgG antibodies to the HCV glycoproteins E1E2. We serologically allotyped 100 African Americans with persistent HCV infection for GM and KM markers and measured anti-E1E2 antibodies. Subjects with the GM 1,17 5,13 phenotype had significantly higher levels of anti-E1E2 antibodies than the subjects who lacked this phenotype (p = 0.008). Likewise, subjects with the KM 1-carrying phenotypes had higher levels of anti-E1E2 antibodies than the subjects who lacked these phenotypes (p = 0.041). Median titers were fourfold higher in persons expressing both GM 1,17 5,13 and KM 1-carrying phenotypes compared to those who lacked these phenotypes (p = 0.011). Interactive effects of these GM-KM phenotypes were previously found to be highly significantly associated with spontaneous HCV clearance. Results presented here show that Ig allotypes contribute to the inter-individual differences in humoral immunity to the HCV epitopes, a finding that may provide a mechanistic explanation for their involvement in the outcome of HCV infection.
Keywords: Immunoglobulin, GM and KM allotypes, IgG antibodies, Proteins E1 and E2, Hepatitis C virus, Humoral immunity
INTRODUCTION
Hepatitis C virus is a common infection and a major health problem worldwide, and there is as yet no vaccine licensed to protect against infection. Understanding of the immunological mechanisms underlying the resolution of acute HCV infection would provide valuable insights in devising novel therapeutic strategies, including the designing of a vaccine, to combat this infection. Our previous studies have shown that GM and KM allotypes are involved in the immunobiology of hepatitis C [1–5]. In particular, we have reported that certain combinations of GM and KM phenotypes are associated with HCV clearance and persistence [1]. The immunogenetic mechanisms responsible for this association are not completely understood.
Immune responses to some viruses are associated with particular GM and KM phenotypes (6–8). Antibody responses to most viral epitopes appear to be restricted to the IgG subclasses—IgG1 and IgG3—that harbor the majority of the GM determinants [9–13]. A recent study has established that amino acid sequence polymorphisms in the constant (C) region of the Ig molecule affect the secondary structure of the antigen-binding site in the variable (V) region [14]. Amino acid substitutions associated with various GM allotypic determinants cause structural changes in the C region, which could impose structural constraints (conformation) on the V region, resulting in changes in antibody specificity. Thus, isotype restriction of antiviral antibodies may be a reflection of structural constraints imposed by C-region allotypes on the antibody V regions engaged in antigen binding.
These observations, together with the increasing evidence that antibodies contribute to viral clearance [15], led us to hypothesize that GM and KM allotypes influence the levels of antibodies to HCV epitopes. Participation of the glycoproteins E1 and E2 in HCV cell entry [16], the importance of E2 epitopes in HCV neutralization [17], and the involvement of anti-E2 IgG antibodies in antibody dependent cellular cytotoxicity (ADCC) against HCV-infected cells [18], provide a good rationale for finding putative mechanisms for the inter-individual differences in antibody responses to these envelope proteins.
MATERIALS AND METHODS
Subjects
Between 1988 and 1989, a cohort was recruited in Baltimore, Maryland of persons who had injected illicit drugs in the preceding 10 years, were greater than 17 years of age, and free of manifestations of AIDS [19]. Within this cohort, a subset of 1667 individuals was identified as the HCV subcohort because they had antibodies to HCV and at least one follow-up visit. The HCV subcohort was further characterized as to whether HCV infection was ongoing or had cleared [20]. An additional 419, 246 and 50 participants were recruited into the cohort in 1994, 1998 and 2000, respectively. For the present study, 100 subjects with persistent HCV infection were selected. The median duration of infection was 20.02 years, range 7.59 to 22.51. All subjects were HIV negative and African American. Most subjects were male (63%). The median age was 41 years, range 30 to 60. Our cohort was > 90% HCV genotype/serotype 1.
Serologic Testing
HCV antibody testing was done using a second or third generation Ortho HCV EIA (Ortho Diagnostic Systems, Raritan, NJ). In antibody positive sera, HCV RNA was assessed in two samples, separated by a minimum of six months. HCV RNA testing was done using a branched DNA (bDNA) assay (Quantiplex HCV RNA 2.0 assay, Chiron Corporation, Emeryville, CA) and/or HCV COBAS AMPLICOR (COBAS AMPLICOR HCV, Roche Diagnostics, Branchburg, NJ). When HCV RNA was classified as ‘undetectable’ in sera from the two visits, this was always by the more sensitive HCV COBAS AMPLICOR system. In addition, antibody status was confirmed by a recombinant immunoblot assay (RIBA 3.0) (Chiron Corporation, Emeryville, CA). Subjects with two positive bDNA assays were included in this study. HIV-1 testing was done by EIA and positive specimens were confirmed by Western blot. All assays were performed according to manufacturers’ specifications. All samples used for testing had been stored at −80°C after processing, and sera used for RNA testing had not been previously used for other assays.
GM and KM Allotyping
Serum samples were typed for G1M (1/a, 2/x, 3/f, 17/z), G2M (23/n), G3M (5/b1, 6/c3, 13/b3, 21/g), and KM 1 and 3 allotypes by a standard hemagglutination-inhibition method [21,22]. In brief, a mixture containing human blood group ORh+ erythrocytes coated with anti-Rh antibodies of known GM/KM allotypes, the test sera, and monospecific anti-allotype antibodies were incubated in a microtiter plate. Test sera containing IgG of particular allotype inhibited hemagglutination by the anti-allotype antibody, whereas negative sera did not. Linkage disequilibrium in the GM system is almost absolute and the determinants are transmitted as a group called haplotypes [23–25]. The notation follows the international system for human gene nomenclature, in which haplotypes and phenotypes are written by grouping together the markers that belong to each IgG subclass, by the numerical order of the marker and of the subclass; markers belonging to different subclasses are separated by a space, while allotypes within a subclass are separated by commas.
Anti-E1E2 IgG Antibody Detection
Antibody responses to E1 and E2 envelope glycoproteins were determined by using HCV genotype 1a antigens in an ELISA, as described elsewhere [9]. Specifically, 293T cells were transfected with an HCV H77 E1E2 expression construct and cell lysates harvested at 72 hours. Plates were coated with 500 ng of Galanthus nivalis lectin, washed and blocked with PBS containing 0.5% tween 20 and 5% non-fat milk. H77 E1E2 containing cell lysates were allowed to bind overnight and washed. Subject serum specimens or pooled normal human AB serum (NHS; negative control) were serially diluted in PBS. Diluted samples were added to their respective wells and incubated at 37° C for 1 hour. Wells were washed in PBS-tween prior to addition of horseradish peroxidase conjugated anti-human IgG (BD Pharmingen, San Diego, CA). Wells were washed and ABTS substrate (KPL, Gaithersburg, MD) added. The substrate reaction was stopped after 30 min by addition of 1% SDS and absorbance was read at 405 nm. To determine the positive cut-off, 2 times the mean absorbance of the negative control wells was used.
Statistical Analysis
Antibody measurements were performed blinded with respect to the GM and KM phenotype status of the subjects, and the results were provided to independent genetic epidemiologists (YL, YU, and RCE) who conducted analyses according to standard statistical methods. Because of almost absolute linkage disequilibrium between particular GM alleles in a given race, data were analyzed as a group (phenotypes), rather than the presence or absence of individual markers. Furthermore, significant linkage disequilibrium may be the result of certain selective evolutionary advantages (immunity to pathogens), making the analysis by phenotypes biologically more meaningful. Subjects with very unusual GM phenotypes and those whose frequency was <10% were combined as “other” for statistical analyses, so as not to have a test with too many degrees of freedom. Antibody titers were log10 transformed to obtain residual homoscedasticity. Student’s t test and analysis of variance were used to determine the significance of the differences in antibody titers associated with different phenotypic groups. Statistical significance was defined as p < 0.05. One-sided p values are presented, as we a priori expected a relationship between particular phenotypes and higher antibody levels, based on the observations in our previous study [1]. It is important, however, to note that associations with the GM phenotypes and antibody responsiveness would remain significant even if a two-sided test were used.
RESULTS
The distribution of GM and KM phenotypes in relation to the endpoint titers of antibodies to HCV E1E2 proteins is given in Table 1. Of the three most frequent GM phenotypes present in this study population, GM 1,17 5,13 is most likely homozygous for the common Negroid haplotype GM 1,17 5, 13. Presence of GM 21 in GM 1,17 5,13,21 and that of GM 3 and 23 in GM 1,3,17 23 5,13 indicates Caucasian admixture. These subjects are most probably heterozygous for the common Negroid haplotype GM 1,17 5, 13 and common Caucasoid haplotypes GM 1,17 21 and 3 23 5,13, respectively. The majority (52%) of the subjects were homozygous for the most common KM allele, KM3, the rest being KM 1,3 heterozygotes (33%) and KM 1 homozygotes (15%). (Three alleles—KM1, KM1,2 and KM3—segregate at the KM locus. Over 98% of subjects positive for KM1 are also positive for KM2; the KM1 allele is extremely rare. In this—and in most other investigations involving KM allotypes—positivity for KM1 includes both KM1 and KM1,2 alleles.)
TABLE 1.
Distribution (number of subjects) of GM and KM phenotypes in relation to endpoint titers of antibodies to HCV E1E2 proteins
| Phenotypes | Endpoint E1E2 titers
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 25 | 50 | 200 | 400 | 800 | 1600 | 3200 | 6400 | 12800 | 25600 | |
| GM 1,17 5,13,21 | 2 | 2 | 4 | 1 | 5 | 3 | 3 | 4 | ||
| GM 1,3,17 23 5,13 | 1 | 2 | 5 | 6 | 5 | 3 | 1 | |||
| GM 1,17 5,13 | 3 | 6 | 4 | 5 | 2 | |||||
| Other GM | 2 | 1 | 1 | 4 | 3 | 6 | 10 | 4 | 2 | |
|
| ||||||||||
| KM 1 | 1 | 2 | 4 | 4 | 3 | 1 | ||||
| KM 1,3 | 1 | 1 | 4 | 5 | 5 | 4 | 7 | 6 | ||
| KM 3 | 3 | 1 | 3 | 5 | 5 | 14 | 14 | 2 | 4 | 1 |
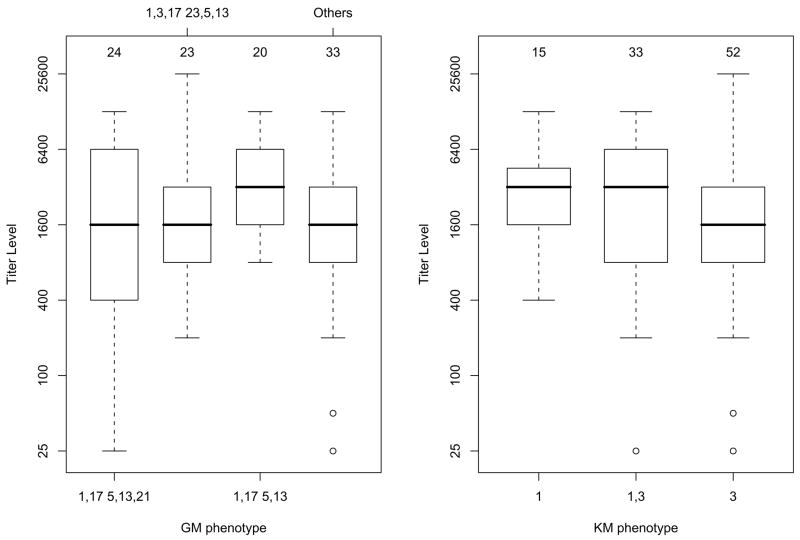
In our previous study, prevalence of subjects with the GM 1,17 5,13 phenotype and those who were KM 1-carriers (KM 1,1 homozygotes or KM 1,3 heterozygotes) was higher in the group that cleared the HCV infection than in those who were persistently infected [1]. Most importantly, the combined effect of these phenotypes showed a highly significant association with spontaneous clearance of HCV. These results led us to hypothesize that one mechanism underlying this association may be that subjects with GM 1,17 5,13 and KM 1-carrier phenotypes have higher levels of anti-HCV antibodies than those lacking these phenotypes. Therefore, we compared the antibody levels of subjects with these phenotypes to those lacking these phenotypes. As shown in Figure 1, the median antibody titers associated with GM 1,17 5,13 were twofold higher than those associated with other phenotypes (3200 vs 1600; p = 0.008). Likewise, KM 1-carriers had significantly higher levels of anti-E1E2 antibodies than the non-KM 1-carriers (3200 vs 1600; p = 0.041).
Figure 1.
Boxplots of anti-E1E2 IgG antibody titers relative to each GM (left panel) and KM (right panel) phenotype. Bold horizontal lines represent the median titers corresponding to the GM and KM phenotypes, while the boxes are indicative of the interquartile (IQR) ranges. The dashed lines outside the boxes extend 1.5 IQR from the median; values outside the 1.5 IQR are represented by open dots. The number of subjects with each phenotype is given above each box.
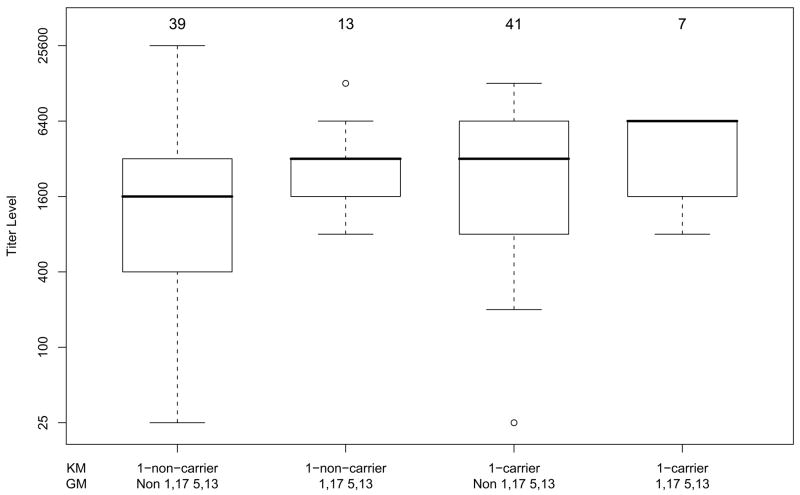
We also investigated the joint effects of these GM and KM phenotypes on the levels of anti-E1E2 antibody titers. It can be seen from the medians in Figure 2 that the joint action of GM 1,17 5,13 and KM 1-carrier phenotypes on antibody level is additive on the log scale, i.e. multiplicative (p = 0.48 for the interaction term in a regression model on the log scale, thus not rejecting multiplicative interaction on the raw scale). Specifically, within non-KM 1 carriers, there is a twofold increase in the median antibody levels for GM 1,17 5,13-carriers compared to non-carriers (3200 vs 1600, p = 0.007). The same twofold increase is observed within KM 1-carriers (6400 vs 3200), but it did not reach statistical significance, possibly due to the small sample size in one of the comparison cells. We also analyzed the data taking into account possible confounding factors such as sex and the duration of HCV infection (measured as years since first infection). Duration of infection, which ranged from 7 to 23 years in this sample, was not significantly associated with anti-E1E2 titers by itself or in any model with other variables. Females had slightly higher antibody levels; however, with GM and KM phenotypes in the model, sex was not significantly associated with antibody responsiveness to E1E2 proteins.
Figure 2.
Boxplots showing joint effects of GM 1,17 5,13 and KM 1-carrying phenotypes on anti-E1E2 IgG antibody levels. Bold horizontal lines represent the median antibody titers corresponding to the particular GM and KM phenotype combinations, while the boxes are indicative of the interquartile (IQR) ranges. The dashed lines outside the boxes extend 1.5 IQR from the median; values outside the 1.5 IQR are represented by open dots. The number of subjects with each phenotype is given above each box.
DISCUSSION
The results presented here show that HCV-infected subjects with GM 1,17 5,13 and KM 1-carrier phenotypes have a fourfold higher anti-E1E2 antibody concentration than those lacking these phenotypes. At least two mechanisms—that are not mutually exclusive—could explain the involvement of GM and KM allotypes in humoral immunity to HCV. These determinants, when present in the membrane-bound form of IgG (mIgG), could directly influence antibody responsiveness. Memory B cells, which predominantly express mIgG show enhanced response to antigen stimulation than cells expressing IgM on their surface [26,27]. Perhaps mIgG molecules expressing GM 1,17 5,13 and KM 1 determinants are more compatible receptors for HCV E1E2 epitopes and thus provoke a strong humoral immunity, whereas the mIgG molecules with other GM/KM allotypes form less compatible receptors for these proteins. GM determinants could directly influence the conformation of the Ig V regions involved in antigen binding and thus cause changes in antibody specificity. Contrary to the current dogma in immunology that the V region of Ig is the sole determinant of antibody specificity, several studies have shown that structural variation in the C region affects the expression of certain idiotypes and causes variation in the specificity of V-region-identical Ig molecules [14,28–31]. The C-region structural changes due to amino acid substitutions associated with GM 1,17 5,13 and KM 1-carrier phenotypes could impose conformational changes in the V regions, resulting in variation in antibody specificity to the E1 and E2 epitopes.
In addition to their direct contribution to antibody specificity, C-region allotypic determinants could indirectly influence antibody responsiveness through linkage disequilibrium with the relevant V-region sequences. A report of Ig mimicry by HCV envelope protein E2 lends support to this mechanism of involvement of GM and KM allotypes in antibody responses to HCV [32]. These investigators have shown that the N-terminal region of E2 is antigenically and structurally similar to the conserved V regions of Ig κchains and, to a lesser extent, to the V regions of heavy chains and the T-cell receptors. A good match between the E2 sequences and the host Ig V regions will render the host non-responsive to HCV due to tolerance mechanisms, whereas a poor match will result in a vigorous immune response. KM alleles are known to be in linkage disequilibrium with particular Vκ sequences [33]. Although not yet investigated, it is possible that particular GM alleles, too, are in linkage disequilibrium with certain Vγ sequences. If E2 is a molecular mimic of these Vκ and Vγsequences, it follows that anti-HCV antibody responses are likely to be associated with GM and KM alleles that are in linkage disequilibrium with these V-region determinants.
The observed multiplicative effect of GM and KM phenotypes on antibody responsiveness suggests that the association of γ and κ chains in IgG antibodies directed against E1 and E2 glycoproteins may not be random. Only γ and κ chains carrying specific GM and KM allotypes might form a paratope with the necessary quaternary structure for an effective recognition of the E1E2 epitopes. Nonrandom pairing of heavy and light chains has been reported in experimental animals [34,35]. It may be relevant to note that joint effects of GM 1,17 5,13 and KM 1 phenotypes on antibody responsiveness to the Epstein-Barr virus antigens have been observed [6].
Association of GM 1,17 5,13 and KM 1-carrier phenotypes with high antibody responsiveness to E1 and E2 epitopes could explain, at least in part, their association with HCV clearance observed in our previous study [1]. As mentioned earlier, antibodies to E1E2 could contribute to HCV clearance by influencing its cell entry and neutralization of its infectious epitopes. In addition, anti-E2 IgG antibodies mediate ADCC against HCV-infected cells, which could also be modulated by GM allotypes. IgG-mediated ADCC is triggered upon ligation of FcγR to the Fc of IgG molecules [36]. It follows that genetic variation in FcγR and Fc could contribute to the inter-individual differences in ADCC, which links the specific humoral responses to the vigorous innate cytotoxic effector responses. Like the Fc (GM) of IgG, activating receptors FcγRIIa and Fc RIIIa are genetically polymorphic and certain alleles are more efficient in causing ADCC than others [37,38]. Determinants expressed on Fc and FcγR are probably some of the most likely ligand-receptor candidate pairs for gene-gene interactions in the human genome. Thus, for instance, anti-HCV-E2 IgG antibodies with Fc of a particular GM genotype could preferentially associate with the FcγRIIIa (expressed on natural killer cells) of a particular genotype [39] and influence the destruction of HCV-infected liver cells through ADCC. This would be analogous to the reported interaction between particular killer cell immunoglobulin-like receptors (KIR) and their HLA-C ligand in the resolution of HCV infection [40].
It is difficult to adequately examine the relationship between Ig allotypes and antibody responsiveness in subjects who have cleared the HCV infection, for the antibody levels drop precipitously after clearance. We examined the anti-E1E2 antibody titers in 50 subjects who had cleared the HCV infection. In about half of these subjects the antibody titers were below 50. No associations between GM and KM allotypes and anti-E1E2 antibody responsiveness were found, possibly a reflection of low antibody levels and small sample size (unpublished observations). In future studies, we plan to allotype the subjects and quantitate the anti-HCV antibody responses in the acute phase of HCV infection, and perform follow-up studies to monitor their influence on the long-term viremia status—resolved vs chronic infection—and the pathologies associated with this viral infection.
The interplay between the host and viral factors leading to the resolution or persistence of HCV infection is not understood. We, and others, have reported an association between viral complexity and HCV persistence [41,42]. There also is strong evidence that humoral immune responses increase viral diversity, especially those to envelope epitopes. For example, substantially less HCV envelope diversity accumulates in humans with impaired B cell responses and in chimpanzees that fail to produce anti-envelope antibodies [43,44]. However, it remains unclear whether viral diversity causes or is a consequence of viral persistence and what role humoral immune responses play in this process.
The bulk of published data suggest that the vigor and breadth of adaptive T cell responses contribute substantially to recovery from HCV infection. However, the mechanisms underlying immune failure—despite the presence of cellular immune responses against HCV epitopes—remain poorly understood [45,46]. Future studies involving GM allotypes and cellular immunity to HCV may shed some light on this paradox, as these markers—either individually or epistatically with certain HLA and KM alleles—are associated with several cell-meditated immune functions [47–49].
Acknowledgments
This work was supported in part by NIH grants DK070877, GM28356, DK068555, DA012568, and DA13324. We thank Mr. Keith Rocca for expert technical assistance.
ABBREVIATIONS
- HCV
hepatitis C virus
- Ig
Immunoglobulin
- C
constant
- V
variable
- ADCC
antibody dependent cellular cytotoxicity
- mIgG
membrane-bound form of IgG
- KIR
killer cell immunoglobulin-like receptors
- FcγR
Fcgamma Receptor
- bDNA
branched DNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pandey JP, Astemborski J, Thomas DL. Epistatic effects of immunoglobulin GM and KM allotypes on outcome of infection with hepatitis C virus. J Virol. 2004;78:4561–4565. doi: 10.1128/JVI.78.9.4561-4565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vejbaesya S, Tanwandee T, Pandey JP. Immunoglobulin GM and KM genotypes in hepatitis C virus infection. J Med Virol. 2004;73:384–386. doi: 10.1002/jmv.20102. [DOI] [PubMed] [Google Scholar]
- 3.Muratori P, Sutherland SE, Muratori L, Granito A, Guidi M, Pappas G, et al. Immunoglobulin GM and KM allotypes and prevalence of anti-LKM1 autoantibodies in patients with hepatitis C virus infection. J Virol. 2006;80:5097–5099. doi: 10.1128/JVI.80.10.5097-5099.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Namboodiri AM, Budkowska A, Nietert PJ, Pandey JP. Fcγ receptor-like hepatitis C virus core protein binds differentially to IgG of discordant Fc (GM) genotypes. Mol Immunol. 2007;44:3805–3808. doi: 10.1016/j.molimm.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pandey JP, Montes-Cano MA, Aguilar-Reina J, Gonzalez-Escribano MF. Interactive effects of immunoglobulin gamma and human leucocyte antigen genotypes on clearance and persistence of infection with hepatitis C virus. Clin Exp Immunol. 2007;150:518–522. doi: 10.1111/j.1365-2249.2007.03519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biggar RJ, Pandey JP, Henle W, Nkrumah FK, Levine PH. Humoral immune response to Epstein-Barr virus antigens and immunoglobulin allotypes in African Burkitt lymphoma patients. Int J Cancer. 1984;33:577–580. doi: 10.1002/ijc.2910330505. [DOI] [PubMed] [Google Scholar]
- 7.Hyöty H, Pandey JP, Lehtinen M, Kulomaa P, Leinikki P. Immunoglobulin allotypes and virus antibodies in Finnish type 1 diabetic patients. Exp Clin Immunogenet. 1988;5:52–59. [PubMed] [Google Scholar]
- 8.Pandey JP. Immunoglobulin GM genes and IgG antibodies to cytomegalovirus in patients with systemic sclerosis. Clin Exp Rheumatol. 2004;22:S35–S37. [PubMed] [Google Scholar]
- 9.Netski DM, Mosbruger T, Depla E, Maertens G, Ray SC, Hamilton RG, et al. Humoral immune response in acute hepatitis C virus infection. Clin Infect Dis. 2005;41:667–675. doi: 10.1086/432478. [DOI] [PubMed] [Google Scholar]
- 10.Chen M, Sallberg M, Sonnerborg A, Weiland O, Mattsson L, Jin L, et al. Limited humoral immunity in hepatitis C virus infection. Gastroenterology. 1999;116:135–143. doi: 10.1016/s0016-5085(99)70237-4. [DOI] [PubMed] [Google Scholar]
- 11.Lal RB, Buckner C, Khabbaz RF, Kaplan JE, Reyes G, Hadlock K, et al. Isotypic and IgG subclass restriction of the humoral immune responses to human T-lymphotropic virus type-1. Clin Immunol Immunopathol. 1993;67:40–49. doi: 10.1006/clin.1993.1043. [DOI] [PubMed] [Google Scholar]
- 12.Skvaril F. IgG subclasses in viral infections. Monogr Allergy. 1986;19:134–143. [PubMed] [Google Scholar]
- 13.Sundqvist VA, Linde A, Kurth R, Werner A, Helm EB, Popovic M, et al. Restricted IgG subclass responses to HTLV-III/LAV and to cytomegalovirus in patients with AIDS and lymphadenopathy syndrome. J Infect Dis. 1986;153:970–973. doi: 10.1093/infdis/153.5.970. [DOI] [PubMed] [Google Scholar]
- 14.Torres M, Fernández-Fuentes N, Fiser A, Casadevall A. The immunoglobulin heavy chain constant region affects kinetic and thermodynamic parameters of antibody variable region interactions with antigen. J Biol Chem. 2007;282:13917–13927. doi: 10.1074/jbc.M700661200. [DOI] [PubMed] [Google Scholar]
- 15.Pestka JM, Zeisel MB, Blaser E, Schurmann P, Bartosch B, Cosset FL, et al. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc Natl Acad Sci USA. 2007;104:6025–6030. doi: 10.1073/pnas.0607026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartosch B, Cosset FL. Cell entry of hepatitis C virus. Virology. 2006;348:1–12. doi: 10.1016/j.virol.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 17.Zhang P, Wu CG, Mihalik K, Virata-Theimer ML, Yu MY, Alter HJ, et al. Hepatitis C virus epitope-specific neutralizing antibodies in Igs prepared from human plasma. Proc Natl Acad Sci USA. 2007;104:8449–8454. doi: 10.1073/pnas.0703039104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nattermann J, Schneiders AM, Leifeld L, Langhans B, Schulz M, Inchauspé G, et al. Serum antibodies against the hepatitis C virus E2 protein mediate antibody-dependent cellular cytotoxicity (ADCC) J Hepatol. 2005;42:499–504. doi: 10.1016/j.jhep.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 19.Vlahov D, Anthony JC, Muñoz A, Margolik J, Celentano DD, Solomon L, et al. The ALIVE Study: a longitudinal study of HIV-1 infection in intravenous drug users: description of methods. J Drug Issues. 1991;21:759–776. [PubMed] [Google Scholar]
- 20.Thomas DL, Astemborski J, Rai RM, Anania FA, Schaeffer M, Galai N, et al. The natural history of hepatitis C virus infection: host, viral, and environmental factors. JAMA. 2000;284:450–456. doi: 10.1001/jama.284.4.450. [DOI] [PubMed] [Google Scholar]
- 21.Vyas GN, Fudenberg HH, Pretty HM, Gold ER. A new rapid method for genetic typing of human immunoglobulins. J Immunol. 1968;100:274–279. [PubMed] [Google Scholar]
- 22.Schanfield MS, van Loghem E. Human immunoglobulin allotypes. In: Weir DM, editor. Handbook of Experimental Immunology (Volume 3): Genetics and Molecular Immunology. Blackwell; Boston: 1986. pp. 94.1–94.18. [Google Scholar]
- 23.Pandey JP. Genetics of immunoglobulins. In: Virella G, editor. Medical Immunology. Marcel Dekker; New York: 2007. pp. 73–81. [Google Scholar]
- 24.Dard P, Lefranc MP, Osipova L, Sanchez-Mazas A. DNA sequence variability of IGHG3 alleles associated to the main G3m haplotypes in human populations. Eur J Hum Genet. 2001;9:765–772. doi: 10.1038/sj.ejhg.5200700. [DOI] [PubMed] [Google Scholar]
- 25.Grubb R. Advances in human immunoglobulin allotypes. Exp Clin Immunogenet. 1995;12:191–197. doi: 10.1159/000424871. [DOI] [PubMed] [Google Scholar]
- 26.Martin SW, Goodnow CC. Burst-enhancing role of the IgG membrane tail as a molecular determinant of memory. Nat Immunol. 2002;3:182–188. doi: 10.1038/ni752. [DOI] [PubMed] [Google Scholar]
- 27.Wakabayashi C, Adachi T, Wienands J, Tsubata T. A distinct signaling pathway used by the IgG-containing B cell antigen receptor. Science. 2002;298:2392–2395. doi: 10.1126/science.1076963. [DOI] [PubMed] [Google Scholar]
- 28.Morahan G, Berek C, Miller JFAP. An idiotypic determinant formed by both immunoglobulin constant and variable regions. Nature. 1983;301:720–722. doi: 10.1038/301720a0. [DOI] [PubMed] [Google Scholar]
- 29.Pritsch O, Hudry-Clergeon G, Buckle M, Petillot Y, Bouvet JP, Gagnon J, et al. Can immunoglobulin CH1 constant region domain modulate antigen binding affinity of antibodies? J Clin Invest. 1996;98:2235–2243. doi: 10.1172/JCI119033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper LJN, Robertson D, Granzow R, Greenspan NS. Variable domain-identical antibodies exhibit IgG subclass-related differences in affinity and kinetic constants as determined by surface plasmon resonance. Mol Immunol. 1994;31:577–584. doi: 10.1016/0161-5890(94)90165-1. [DOI] [PubMed] [Google Scholar]
- 31.McLean GR, Torres M, Elguezabal N, Nakouzi A, Casadevall A. Isotype can affect the fine specificity of an antibody for a polysaccharide antigen. J Immunol. 2002;169:1379–1386. doi: 10.4049/jimmunol.169.3.1379. [DOI] [PubMed] [Google Scholar]
- 32.Hu YW, Rocheleau L, Larke B, Chui L, Lee B, Ma M, et al. Immunoglobulin mimicry by Hepatitis C Virus envelope protein E2. Virology. 2005;332:538–549. doi: 10.1016/j.virol.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 33.Moxley G, Gibbs RS. Polymerase chain reaction-based genotyping for allotypic markers of immunoglobulin kappa shows allelic association of Km with kappa variable segment. Genomics. 1992;13:104–108. doi: 10.1016/0888-7543(92)90208-a. [DOI] [PubMed] [Google Scholar]
- 34.Czerwinski M, Siemaszko D, Siegel DL, Spitalnik SL. Only selected light chains combine with a given heavy chain to confer specificity for a model glycopeptide antigen. J Immunol. 1998;160:4406–4417. [PubMed] [Google Scholar]
- 35.Primi D, Drapier AM, Cazenave PA. Highly preferential VH-VL pairing in normal B cells results in antigen-independent selection of the available repertoire. J Immunol. 1987;138:1607–1612. [PubMed] [Google Scholar]
- 36.Nimmerjahn F, Ravetch JV. Antibodies, Fc receptors and cancer. Curr Opin Immunol. 2007;19:239–245. doi: 10.1016/j.coi.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 37.van der Pol W, van de Winkel JG. IgG receptor polymorphisms: risk factors for disease. Immunogenetics. 1998;48:222–232. doi: 10.1007/s002510050426. [DOI] [PubMed] [Google Scholar]
- 38.Stockmeyer B, Valerius T, Repp R, Heijnen IA, Buhring HJ, Deo YM, et al. Preclinical studies with FcγR bispecific antibodies and granulocyte colony-stimulating factor-primed neutrophils as effector cells against HER-2/neu overexpressing breast cancer. Cancer Res. 1997;57:696–701. [PubMed] [Google Scholar]
- 39.Pandey JP. Genetic polymorphism of Fc. Science. 2006;311:1376–1377. doi: 10.1126/science.311.5766.1376d. [DOI] [PubMed] [Google Scholar]
- 40.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 41.Ray SC, Wang YM, Laeyendecker O, Ticehurst JR, Villano SA, Thomas DL. Acute hepatitis C virus structural gene sequences as predictors of persistent viremia: hypervariable region 1 as a decoy. J Virol. 1999;73:2938–2946. doi: 10.1128/jvi.73.4.2938-2946.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder JC, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 43.Ray SC, Mao Q, Lanford RE, Bassett S, Laeyendecker O, Wang YM, Thomas DL. Hypervariable region 1 sequence stability during hepatitis C virus replication in chimpanzees. J Virol. 2000;74:3058–3066. doi: 10.1128/jvi.74.7.3058-3066.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Booth JC, Kumar U, Webster D, Monjardino J, Thomas HC. Comparison of the rate of sequence variation in the hypervariable region of E2/NS1 region of hepatitis C virus in normal and hypogammaglobulinemic patients. Hepatology. 1998;27:223–227. doi: 10.1002/hep.510270134. [DOI] [PubMed] [Google Scholar]
- 45.Cox AL, Mosbruger T, Lauer GM, Pardoll D, Thomas DL, Ray SC. Comprehensive analyses of CD8+ T cell responses during longitudinal study of acute human hepatitis C. Hepatology. 2005;42:104–112. doi: 10.1002/hep.20749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox AL, Mosbruger T, Mao Q, Liu Z, Wang XH, Yang HC, et al. Cellular immune selection with hepatitis C virus persistence in humans. J Exp Med. 2005;201:1741–1752. doi: 10.1084/jem.20050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Legrand L, Rivat-Perran L, Huttin C, Dausset J. HLA-and Gm-linked genes affecting the degradation rate of antigens (sheep red blood cells) endocytized by macrophages. Hum Immunol. 1982;4:1–13. doi: 10.1016/0198-8859(82)90045-3. [DOI] [PubMed] [Google Scholar]
- 48.Wachsmuth RR, Pandey JP, Fedrick JA, Nishimura Y, Sasazuki T. Interactive effect of Gm and Km allotypes on cellular immune responses to streptococcal cell wall antigen. Exp Clin Immunogenet. 1987;4:163–166. [PubMed] [Google Scholar]
- 49.Schmidt W, Königstedt B, Zipprich B, Scheibe E, Nilius R. Immunoglobulin allotypes and immunoreactivity in chronic liver disease. Hepatogastroenterology. 1987;34:206–211. [PubMed] [Google Scholar]