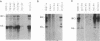Abstract
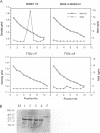
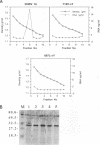
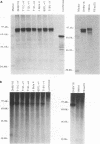
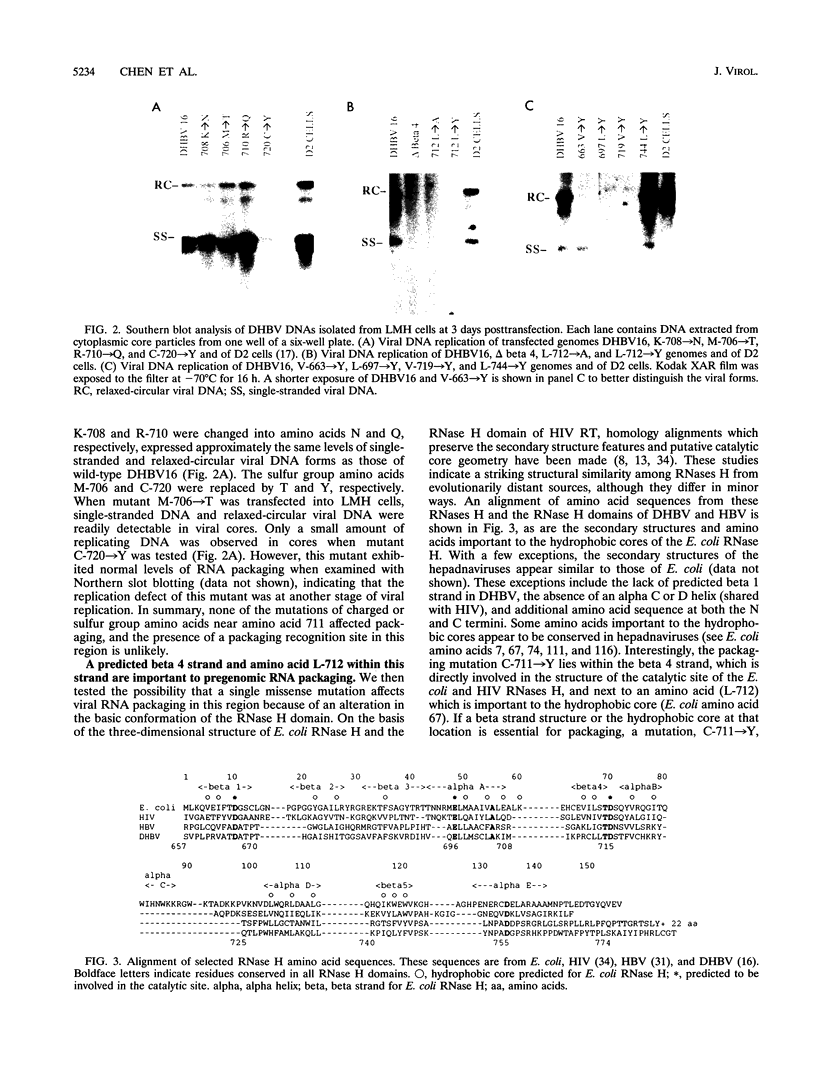
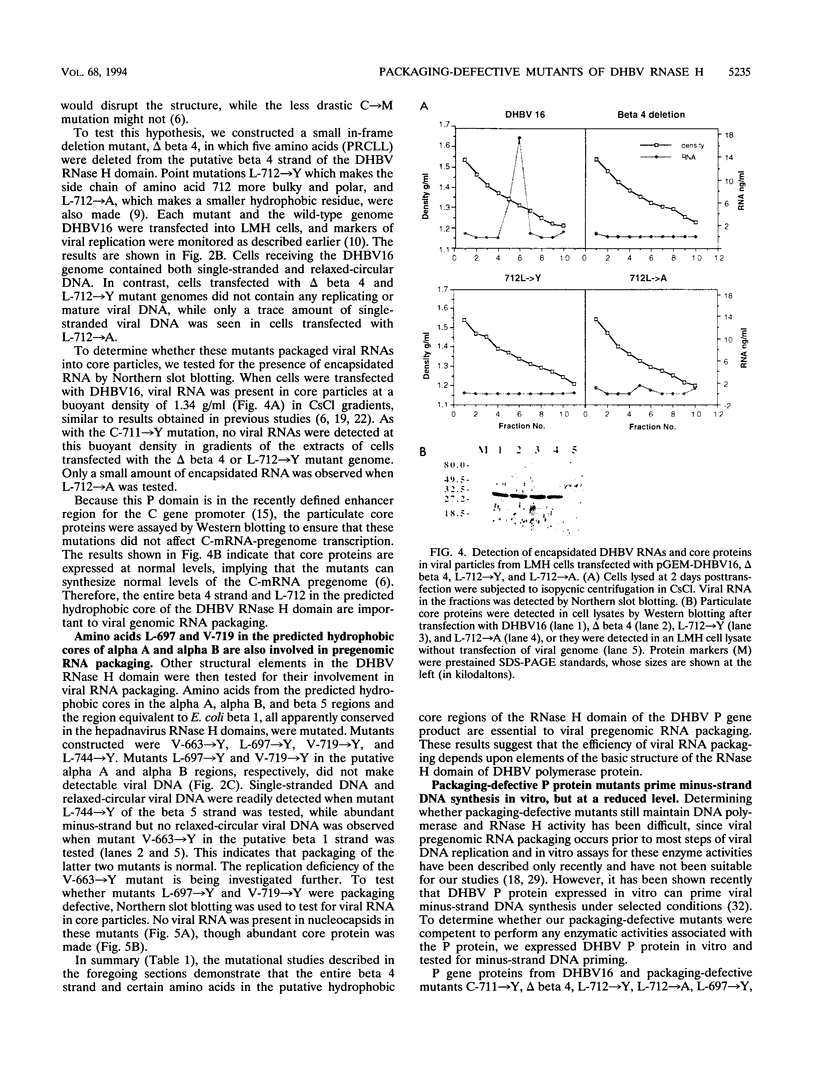
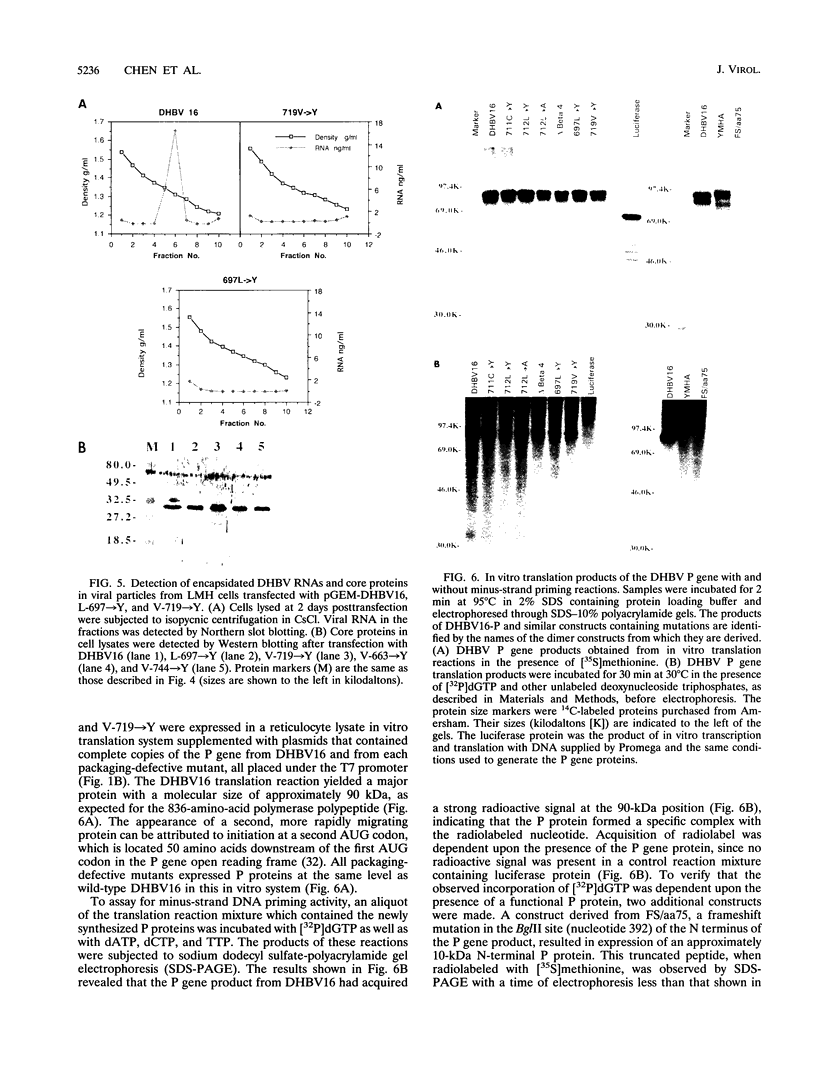
The genome of all hepadnaviruses has an open reading frame called the P gene, which encodes a polypeptide of 90 to 97 kDa. The product or products of this P gene are involved in multiple functions of the viral life cycle. These functions include a priming activity which initiates minus-strand DNA synthesis, a polymerase activity which synthesizes DNA by using either RNA or DNA templates (reverse transcriptase), a nuclease activity which degrades the RNA strand of RNA-DNA hybrids (RNase H), and involvement in packaging the RNA pregenome into nucleocapsids. In a previous study, we found that a single point mutation at position 711 in the duck hepatitis B virus (DHBV) P gene product RNase H domain prevented viral RNA packaging. In the present experiments, we have mutated additional conserved amino acids in the DHBV RNase H domain and examined the ability of viral genomes containing these mutations to package RNA and replicate viral DNA. Charged and sulfur group amino acids adjacent to Cys-711 were mutated. None of these mutants was defective in either RNA packaging or viral replication. We also tested a number of mutations on the basis of common elements in the crystal structures of Escherichia coli and human immunodeficiency virus reverse transcriptase RNase H enzymes and on the basis of the similarities of their amino acid sequences to those of the RNase H domains of DHBV and HBV. Our results revealed that the entire beta 4 strand and amino acids Leu-712, Leu-697, and Val-719 in the putative hydrophobic cores of the beta 4, alpha A, and alpha B regions, respectively, are involved in pregenomic RNA encapsidation. This suggests that the basic structure of the RNase H domain in the DHBV P gene product is required for viral RNA packaging. We used the in vitro DHBV minus-strand DNA priming system developed by Wang and Seeger (G.-H. Wang and C. Seeger, Cell 71:663-670, 1992) to test the effect of RNase H packaging mutations on P gene product enzymatic activity. While all packaging-defective mutants tested maintained DNA priming activity, levels were decreased 5- to 20-fold compared with that of the wild-type genome. This observation suggests that the hepadnavirus RNase H domain plays a role in optimizing priming of minus-strand DNA synthesis.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartenschlager R., Junker-Niepmann M., Schaller H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol. 1990 Nov;64(11):5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R., Schaller H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992 Sep;11(9):3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum H. E., Galun E., Liang T. J., von Weizsäcker F., Wands J. R. Naturally occurring missense mutation in the polymerase gene terminating hepatitis B virus replication. J Virol. 1991 Apr;65(4):1836–1842. doi: 10.1128/jvi.65.4.1836-1842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert J., Summers J. Two regions of an avian hepadnavirus RNA pregenome are required in cis for encapsidation. J Virol. 1994 Apr;68(4):2084–2090. doi: 10.1128/jvi.68.4.2084-2090.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L. J., Hirsch R. C., Ganem D., Varmus H. E. Effects of insertional and point mutations on the functions of the duck hepatitis B virus polymerase. J Virol. 1990 Nov;64(11):5553–5558. doi: 10.1128/jvi.64.11.5553-5558.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987 Aug;7(8):2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Robinson W. S., Marion P. L. Naturally occurring point mutation in the C terminus of the polymerase gene prevents duck hepatitis B virus RNA packaging. J Virol. 1992 Feb;66(2):1282–1287. doi: 10.1128/jvi.66.2.1282-1287.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condreay L. D., Aldrich C. E., Coates L., Mason W. S., Wu T. T. Efficient duck hepatitis B virus production by an avian liver tumor cell line. J Virol. 1990 Jul;64(7):3249–3258. doi: 10.1128/jvi.64.7.3249-3258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. F., 2nd, Hostomska Z., Hostomsky Z., Jordan S. R., Matthews D. A. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991 Apr 5;252(5002):88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- Eriksson A. E., Baase W. A., Zhang X. J., Heinz D. W., Blaber M., Baldwin E. P., Matthews B. W. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992 Jan 10;255(5041):178–183. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- Ganem D., Varmus H. E. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- Hirsch R. C., Lavine J. E., Chang L. J., Varmus H. E., Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as wel as for reverse transcription. Nature. 1990 Apr 5;344(6266):552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- Hirsch R. C., Loeb D. D., Pollack J. R., Ganem D. cis-acting sequences required for encapsidation of duck hepatitis B virus pregenomic RNA. J Virol. 1991 Jun;65(6):3309–3316. doi: 10.1128/jvi.65.6.3309-3316.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayanagi K., Miyagawa M., Matsushima M., Ishikawa M., Kanaya S., Ikehara M., Matsuzaki T., Morikawa K. Three-dimensional structure of ribonuclease H from E. coli. Nature. 1990 Sep 20;347(6290):306–309. doi: 10.1038/347306a0. [DOI] [PubMed] [Google Scholar]
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Liu C., Condreay L. D., Burch J. B., Mason W. Characterization of the core promoter and enhancer of duck hepatitis B virus. Virology. 1991 Sep;184(1):242–252. doi: 10.1016/0042-6822(91)90841-x. [DOI] [PubMed] [Google Scholar]
- Mandart E., Kay A., Galibert F. Nucleotide sequence of a cloned duck hepatitis B virus genome: comparison with woodchuck and human hepatitis B virus sequences. J Virol. 1984 Mar;49(3):782–792. doi: 10.1128/jvi.49.3.782-792.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraleda G., Wu T. T., Jilbert A. R., Aldrich C. E., Condreay L. D., Larsen S. H., Tang J. C., Colacino J. M., Mason W. S. Inhibition of duck hepatitis B virus replication by hypericin. Antiviral Res. 1993 Mar;20(3):235–247. doi: 10.1016/0166-3542(93)90023-c. [DOI] [PubMed] [Google Scholar]
- Oberhaus S. M., Newbold J. E. Detection of DNA polymerase activities associated with purified duck hepatitis B virus core particles by using an activity gel assay. J Virol. 1993 Nov;67(11):6558–6566. doi: 10.1128/jvi.67.11.6558-6566.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J. H., Bell K. D. Comparative studies of hepatitis B virus precore and core particles. Virology. 1990 Jan;174(1):185–191. doi: 10.1016/0042-6822(90)90067-2. [DOI] [PubMed] [Google Scholar]
- Oyama F., Kikuchi R., Crouch R. J., Uchida T. Intrinsic properties of reverse transcriptase in reverse transcription. Associated RNase H is essentially regarded as an endonuclease. J Biol Chem. 1989 Nov 5;264(31):18808–18817. [PubMed] [Google Scholar]
- Paeratakul U., De Stasio P. R., Taylor M. W. A fast and sensitive method for detecting specific viral RNA in mammalian cells. J Virol. 1988 Apr;62(4):1132–1135. doi: 10.1128/jvi.62.4.1132-1135.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit M. A., Pillot J. HBc and HBe antigenicity and DNA-binding activity of major core protein P22 in hepatitis B virus core particles isolated from the cytoplasm of human liver cells. J Virol. 1985 Feb;53(2):543–551. doi: 10.1128/jvi.53.2.543-551.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh J. C., Yaginuma K., Koike K., Summers J. Duck hepatitis B virus (DHBV) particles produced by transient expression of DHBV DNA in a human hepatoma cell line are infectious in vitro. J Virol. 1988 Sep;62(9):3513–3516. doi: 10.1128/jvi.62.9.3513-3516.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radziwill G., Tucker W., Schaller H. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J Virol. 1990 Feb;64(2):613–620. doi: 10.1128/jvi.64.2.613-620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhury S., Faruqi A. F., Shih C. Pregenomic RNA encapsidation analysis of eleven missense and nonsense polymerase mutants of human hepatitis B virus. J Virol. 1991 Jul;65(7):3617–3624. doi: 10.1128/jvi.65.7.3617-3624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanese N., Telesnitsky A., Goff S. P. Abortive reverse transcription by mutants of Moloney murine leukemia virus deficient in the reverse transcriptase-associated RNase H function. J Virol. 1991 Aug;65(8):4387–4397. doi: 10.1128/jvi.65.8.4387-4397.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavis J. E., Ganem D. Expression of functional hepatitis B virus polymerase in yeast reveals it to be the sole viral protein required for correct initiation of reverse transcription. Proc Natl Acad Sci U S A. 1993 May 1;90(9):4107–4111. doi: 10.1073/pnas.90.9.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesnitsky A., Goff S. P. RNase H domain mutations affect the interaction between Moloney murine leukemia virus reverse transcriptase and its primer-template. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1276–1280. doi: 10.1073/pnas.90.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. H., Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. J Virol. 1993 Nov;67(11):6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. H., Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992 Nov 13;71(4):663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- Yang W., Hendrickson W. A., Crouch R. J., Satow Y. Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein. Science. 1990 Sep 21;249(4975):1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]