Abstract
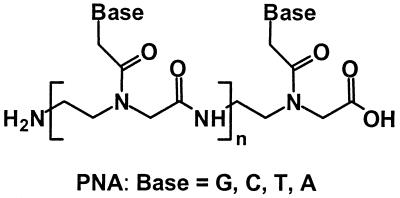
Peptide nucleic acids (PNA) are mimics with normal bases connected to a pseudopeptide chain that obey Watson–Crick rules to form stable duplexes with itself and natural nucleic acids. This has focused attention on PNA as therapeutic or diagnostic reagents. Duplexes formed with PNA mirror some but not all properties of DNA. One fascinating aspect of PNA biochemistry is their reaction with enzymes. Here we show an enzyme reaction that operates effectively on a PNA/DNA hybrid duplex. A DNA oligonucleotide containing a cis, syn-thymine [2+2] dimer forms a stable duplex with PNA. The hybrid duplex is recognized by photolyase, and irradiation of the complex leads to the repair of the thymine dimer. This finding provides insight into the enzyme mechanism and provides a means for the selective repair of thymine photodimers.
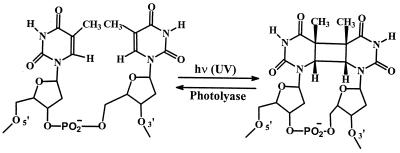
UV light damages nucleic acids primarily by causing dimerization of adjacent pyrimidines. This damage is repaired enzymatically by DNA photolyase, which binds selectively to cis, syn-cyclobutane dimers and, when this complex is activated by light, reforms the monomeric pyrimidines (1, 2). These reactions are outlined in Scheme S1 for thymines. Thymine photodimerization and its repair is an appealing system for examination of parallels in the properties of peptide nucleic acids (PNA), Scheme S2, and DNA. Photolyase is a site-selective, rather than a sequence-selective, enzyme. It binds to thymine dimers whether they are incorporated in superhelical, circular, linear, or single-stranded DNA. It appears that the only additional requirement for recognition by the enzyme is phosphate groups on the dimer-containing strand (3, 4). For these reasons, we suspected that photolyase might retain its ability to recognize and repair thymine dimers contained on the DNA strand of a DNA/PNA hybrid duplex (5–8). We carried out a series of experiments to assess this possibility.
Scheme 1.
Scheme 2.
MATERIALS AND METHODS
Melting Temperature (Tm) Measurements.
Absorbance versus temperature curves were measured at 260 nm in 10 mM phosphate buffer at pH 7 for solutions containing 2 μM of each strand of PNA or DNA shown in Scheme S3. The melting temperature, Tm, is assigned as the peak of the first derivative plot.
Scheme 3.
Gel Mobility Assay.
The oligonucleotides DNA(1) and DNA(3) were labeled with 32P at the 5′ end by using standard techniques (9). Separate samples of the radiolabeled DNA (2,500 cpm) were mixed with complementary DNA(2) (5 μM), PNA(1), or PNA(4) (5 μM) in 10 μl of 10 mM phosphate buffer at pH 7. The hybridization was carried out by heating to 90°C for 5 min and then cooling the solution to room temperature for 2 h. The samples were dried with a Speedvac at low heat for 2 h, and 5 μl of nondenaturing loading buffer was added. The analysis was carried out by electrophoresis in a 20% polyacrylamide gel (29:1 acrylamide and bisacrylamide) followed by autoradiography. The gel was run with TBE buffer containing 89 mM Tris-borate and 1 mM EDTA, pH 8.3, at 450 V for 5 h.
Analysis of [2+2] Thymine Dimer Repair by Photolyase.
The 32P 5′ end radiolabeled oligonucleotide (2,500 cpm) was mixed with various amounts of complementary PNA and DNA in 9 μl of 10 mM phosphate buffer containing enzyme assay buffer (3, 4) (50 mM Tris⋅HCl/10 mM NaCl/1.7 mM DTT/1 mM EDTA, pH 7.4). Hybridization was carried out by heating the solution at 90°C and then cooling to room temperature for 2 h. Photolyase (1 μl of a 1-μM solution) was added, and the samples were incubated in the dark for 30 min and then irradiated in a Rayonet (Southern New England Ultraviolet, Branford, CT) reactor equipped with 350-nm lamps. The samples were denatured by adding 1 μl (100 μM) of cold DNA(1) and heating at 90°C for 3 min followed by rapid cooling on ice. The thymine sequencing by means of KMnO4 used a modification of the standard procedure. The samples were added to centrifuge tubes, and 1 μl (0.5 mM) of calf-thymus DNA, 1 μl of 100 mM phosphate buffer, and 7.5 μl of water were added and mixed by vortexing for 5 s and then centrifuged for 5 s at 12,000 rpm. A freshly prepared solution of KMnO4 (0.5 μl of 0.5 M) was added to the samples and allowed to react for 45 s, and the reaction was quenched by adding DNA-precipitating buffer. The precipitated DNA was washed with 80% ethanol, dried, and subjected to piperidine treatment (100 μl of 1 mM piperidine for 1 h at 90°C). Loading buffer was added and the samples were analyzed on a 20% polyacrylamide (19:1 acrylamide and bisacrylamide) gel by electrophoresis run with TBE buffer containing 89 mM Tris-borate and 2 mM EDTA (pH 8.3) at 1,500 V for 3 h, followed by autoradiography.
RESULTS
The series of oligonucleotides and PNA derivatives used in these experiments are shown in Scheme S3. Irradiation of DNA(1) at >280 nm results in the expected absorbance decrease at 260 nm, which is characteristic of the formation of a thymine photodimer (10). The product mixture was separated by HPLC, and the major product was collected. It was shown to be DNA(3), which contains the expected dimer {TT}, based on the photoreversibility of the reaction and, more convincingly, by its rapid reformation of DNA(1) when it is treated with photolyase and irradiated with near-UV light (350 nm).
PNA (1), shown in Scheme S3 written in its N → C direction, is designed to form hybrid duplexes with DNA(1) and DNA(3) in the less stable parallel (N/5′) orientation. PNA(4), written in the C → N direction, is expected to form duplexes with DNA(1) and DNA(3) in the more stable antiparallel (N/3′) orientation (11). We determined the melting behavior (see Table 1) of these complexes in 10 mM phosphate buffer to verify formation of the duplexes and to assess their stability. The Tm for the parallel, PNA(1)/DNA(1), hybrid duplex is ca. 10°C greater than its all-DNA analog DNA(1)/DNA(2), and the Tm of the antiparallel structure PNA(4)/DNA(1) is 26°C above duplex DNA. PNA/DNA hybrids generally have greater stability than their natural analogs presumably because of counterion release (12) and reduced Coulombic repulsion (6). Significantly, the melting behavior for the duplexes containing thymine dimers in the DNA strand, DNA(3), have Tm values that are only a few degrees below those formed with DNA(1). This is consistent with previous conclusions that the formation of a thymine dimer destabilizes the duplex structure (13). However, the relatively small decrease in Tm indicates that there is only a minimal perturbation caused by dimerization of the thymines.
Table 1.
Melting temperatures (Tm, °C)* for DNA/DNA and DNA/PNA hybrid duplexes
| Oligonucleotide | DNA(1) | DNA(3) |
|---|---|---|
| DNA(2) | 57 | 53 |
| PNA(1) | 67 | 64 |
| PNA(4) | 83 | 78 |
| PNA(5) | 71 |
Absorption changes with temperature were measured at 260 nm in 10 mM sodium phosphate buffer at pH 7.
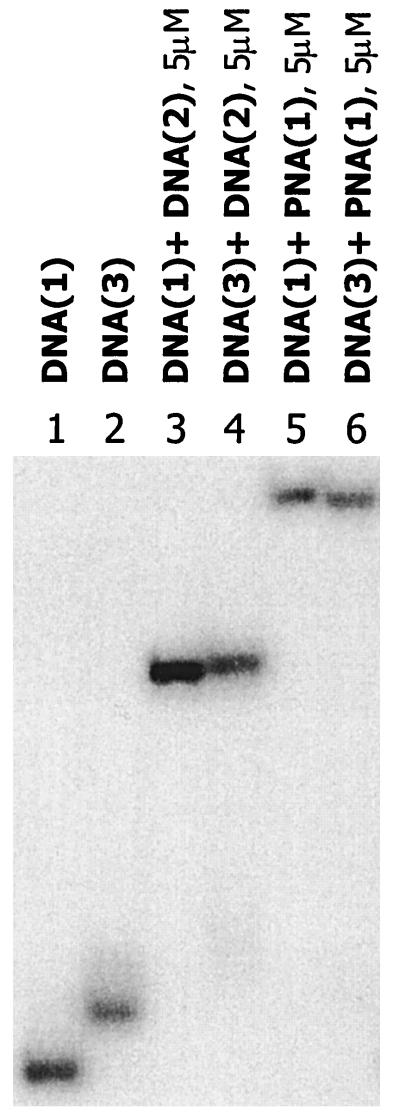
Further unambiguous evidence for the formation and stability of the duplex structures was obtained by analysis of gel retardation assays (14). The DNA oligomers were labeled with 32P at the 5′ end by using standard techniques (9). Fig. 1 shows the relative mobilities of single strands, duplex DNA, and DNA/PNA hybrids determined by electrophoresis on a 20% polyacrylamide gel under nondenaturing conditions. Interestingly, DNA(3) migrates more slowly than DNA(1). This probably is related to structural distortion resulting from dimerization. Fig. 1 also reveals that hybrid PNA/DNA duplexes migrate slower than their all-DNA analogs. This common observation undoubtedly is due to the reduced charge of the hybrid duplex caused by replacement of the sugar diphosphate backbone with a neutral pseudopeptide. The gel retardation experiments clearly show that complementary DNA(2) and PNA(1) give stable duplex structures with DNA(1) and DNA(3). Similar results are obtained with PNA(4). Control experiments show that there is no reduction in the mobility of DNA(1) or DNA(3) when excess noncomplementary PNA is present. Also, addition of excess complementary PNA to DNA(3) does not result in any change in the mobility of the hybrid duplex. These experiments confirm that formation of the hybrid duplexes is both specific and complete under these conditions.
Figure 1.
Gel mobility-shift assay demonstrating duplex formation by PNA with thymine [2+2] dimer. For comparison, duplex formation with DNA also was examined. Lanes: 1, DNA(1); 2, DNA(3); 3, DNA(1)+DNA(2); 4, DNA(3)+DNA(2); 5, DNA(1)+PNA(1); 6, DNA(3)+PNA(1).
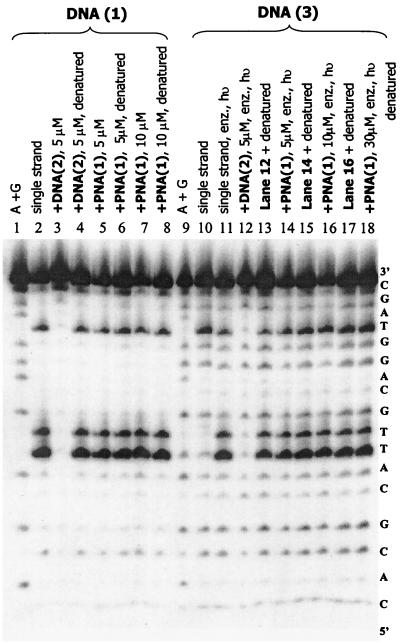
We developed a very convenient assay for thymine dimers in DNA. Reaction of DNA with KMnO4 is used regularly to locate thymines since this reaction causes cleavage uniquely at that base for single-stranded structures after treatment with piperidine (16). Interestingly, duplex DNA is not cleaved at thymine by KMnO4, but thymine in PNA/DNA hybrids is cleaved. Of most relevance to the current investigation, we discovered that the cis, syn-[2+2] thymine dimers in DNA are not cleaved by KMnO4 either as single strands, in DNA duplexes, or in PNA/DNA hybrids. A full description of this assay procedure will be presented elsewhere (16); here we use it to probe the formation and repair of thymine dimers. An illustration of this application is shown in Fig. 2. Lane 2 shows cleavage at each of the three thymines of a single-stranded sample of 5′-32P-labeled DNA(1) that was treated first with KMnO4 and then with piperidine. Formation of the DNA(1)/DNA(2) duplex protects the thymines (Fig. 2, lane 3), but formation of the DNA(1)/PNA(1) hybrid (lane 5) does not. Similar treatment of a single-stranded sample of dimer-containing DNA(3) (lane 10) shows cleavage only at the isolated thymine, but repair of the dimer by treatment of DNA(3) with photolyase and irradiation (lane 11) restores the cleavage of the three thymines.
Figure 2.
Autoradiogram demonstrating DNA photodimer [DNA(3)] repair by DNA photolyase. Samples containing photolyase (100 nM in enzyme assay buffer), indicated as such by “enz,” were irradiated (indicated by “hν”) at 350 nm in a Rayonet reactor for 7 min at ca. 15–20°C.
The ability of photolyase to recognize and repair thymine dimers in PNA/DNA hybrid duplexes can be assessed with confidence since the melting temperature and gel retardation experiments show stable hybrid duplex formation, and the KMnO4 assay permits the certain identification of dimer formation and repair. As expected, efficient repair (Fig. 2, compare lanes 10 and 13) of the thymine dimer is seen when photolyase is added to a sample of DNA(2)/DNA(3) [the DNA(3) is 5′-32P-labeled] in enzyme assay buffer and the sample is irradiated at 350 nm. Remarkably, very similar results were obtained when related experiments were carried out on the hybrid duplex PNA(1)/DNA(3). Treatment of the hybrid duplex with photolyase and irradiation gives rapid repair of the thymine dimer (Fig. 2, lane 14). Dark control experiments with enzyme and DNA(3) and PNA(1) show no repair. As a control to ensure that the enzyme is not reacting with single-stranded DNA(3) in equilibrium with the hybrid duplex, the concentration of PNA(1) was increased stepwise up to 30 μM. There is no reduction in repair efficiency (lane 18). Clearly and remarkably, photolyase recognizes and efficiently repairs thymine dimers in the DNA strand of the parallel PNA(1)/DNA(3) hybrid duplex.
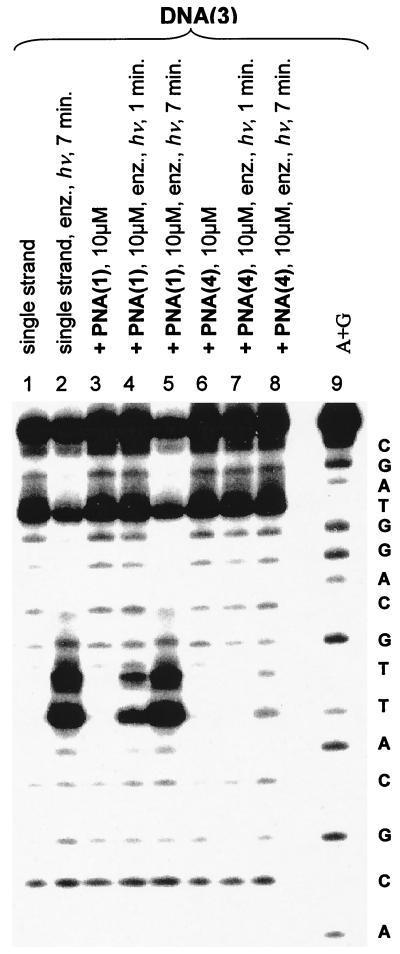
Similar experiments with the antiparallel duplex PNA(4)/DNA(3) give a different result. Fig. 3 shows the reaction of this hybrid with KMnO4. Both before (lane 6) and after treatment with photolyase (lanes 7 and 8, 1- and 7-min irradiation, respectively) only the undimerized thymine is detected. Control experiments show that all thymines in DNA(1)/PNA(4) are revealed by the KMnO4 treatment. Also, contemporaneous control experiments with the parallel PNA(1)/DNA(3) (Fig. 3, lanes 2–5) show efficient dimer repair. Clearly, photolyase recognizes and efficiently repairs cis, syn-thymine [2+2] dimers in parallel but not in antiparallel PNA/DNA hybrid duplexes.
Figure 3.
Autoradiogram demonstrating DNA photodimer [DNA(3)] response to DNA photolyase as hybrid duplexes with PNA(4) and PNA(1). Samples containing photolyase (100 nM in enzyme assay buffer) are indicated as such by “enz.” Samples were irradiated at 350 nm in a Rayonet reactor at ca. 15–20°C, except for the sequencing lane labeled A+G, for the times indicated.
PNA(5) has two fewer bases than DNA(3) because it lacks the two adenines that are opposite of the central thymines. The Tm of the antiparallel PNA(5)/DNA(1) hybrid duplex is 71°C, ca. 11°C below that of the full complement. This decrease in melting temperature results from the loss of hydrogen-bond stabilization. We expect that the central thymines on the complementary strand and the thymine dimer in PNA(5)/DNA(3) are “flipped out” in these hybrid duplexes. In contrast to the behavior of antiparallel PNA(4)/DNA(3), the thymine dimer in PNA(5)/DNA(3) is repaired by treatment with photolyase and irradiation. Evidently, destabilization of the duplex is sufficient to enable detectable operation of photolyase.
It is understood that recognition of thymine dimers by photolyase requires phosphate groups on the dimer-containing strand. Nevertheless, we explored the possibility that photolyase would recognize and repair thymine dimers on the PNA strand of a PNA/DNA hybrid duplex. We recently prepared and characterized PNA oligomers containing cis, syn-thymine [2+2] dimers (K. O’Shea, D.R., T.K., H.O., and G.B.S., unpublished data). Irradiation of PNA(2), λ > 280 nm, gives thymine dimer-containing PNA(3) (ca. 95% yield), which was characterized as the cis, syn-dimer structure by chemical transformations and by NMR spectral analysis. Treatment of PNA(3) with photolyase and irradiation, either as a single strand or as a hybrid duplex with its DNA complement, does not result in any reformation of PNA(1) as shown by HPLC analysis.
DISCUSSION
The structure of Escherichia coli photolyase has been determined by x-ray crystallography (2). The polypeptide chain is folded to bind a light-harvesting factor (5,10-methenyltetrahydrofolylpolyglutamate, MTHF) and a catalytic cofactor (FAD). The enzyme possesses a “hole” in the center of the helical domain containing the FAD, which matches the polarity of the thymine dimer. The mechanism for recognition and reaction follows from this structure. The thymine dimer is proposed to “flip out” from its duplex form and fit into the hole in the enzyme. Light absorbed by MTHF (or directly by FAD) generates the excited state of FAD, which transfers an electron to the bound dimer to form its radical anion. The dimer radical anion splits, an electron is eventually returned to FAD, and the repaired DNA is released from the enzyme, which completes a catalytic cycle.
The structure of the antiparallel PNA/DNA hybrid duplex in solution has been determined by NMR spectroscopy (18). It is a right-handed helix with a repeat of 13 bp. Its structural features are similar to those of natural nucleic acids. There is a major groove that is wide and deep, resulting in a comparatively narrow minor groove. A key conclusion from the structural analysis is that the PNA in a hybrid duplex conforms to that of its partner. This may be the key factor that controls the recognition of the hybrid duplex by photolyase. If the thymine dimer on the DNA strand on the hybrid duplex can flip out of the helix as is proposed for the natural structure, the enzyme “seeing” its normal substrate will bind and react in the usual way.
This model also accommodates the different behavior of the parallel and antiparallel hybrid duplexes. In the more stable antiparallel orientation (revealed by the increased Tm), the fraction of flipped out dimer will be much lower than in the natural substrate and lower than for the parallel structure. We suggest that photolyase fails to recognize the thymine dimer in the antiparallel structure because it is intrahelical. This view is strengthened by the results with antiparallel hybrid duplex PNA(5)/DNA(3), where the dimer is flipped out because of structural considerations. The thymine dimer is repaired in this duplex but not in PNA(4)/DNA(3). When the dimer is on the PNA strand of the duplex, even if it is flipped out, it is not recognized and cleaved by the enzyme, probably because it lacks flanking phosphate groups.
The recognition and reaction of PNA by enzymes is a new and important topic. Uhlmann found that PNA/DNA chimeras can serve as weak primers for some DNA polymerases, but Demidov (19) and Efimov (20) have shown that PNA is not a substrate for endonucleases or proteases. The findings reported in this work open a new chapter in the exploration of peptide nucleic acids. It is clear now that some enzymes will recognize and react with structures containing these synthetic constructs. This may be useful for structural modification to nucleic acids. For example, in the present case, inhibition of repair with antiparallel PNA will allow selective placement of thymine dimers in DNA. Further, it has been suggested that nonnucleotidic polymers could provide informational macromolecules relevant to the origin of life (21, 22). These results strengthen that case.
Acknowledgments
We thank Professor Aziz Sancar of the University of North Carolina for generously providing us with DNA photolyase. This work was supported by funding from the National Institutes of Health and from the National Science Foundation, for which we are grateful.
ABBREVIATION
- PNA
peptide nucleic acids
References
- 1. Sancar A. Biochemistry. 1994;33:2–9. doi: 10.1021/bi00167a001. [DOI] [PubMed] [Google Scholar]
- 2.Park H-W, Kim S-T, Sancar A, Deisenhofer J. Science. 1995;268:1866–1872. doi: 10.1126/science.7604260. [DOI] [PubMed] [Google Scholar]
- 3.Sancar G B, Smith F W, Sancar A. Biochemistry. 1985;24:1849–1855. doi: 10.1021/bi00329a007. [DOI] [PubMed] [Google Scholar]
- 4.Kim S-T, Sancar A. Biochemistry. 1991;30:8623–8630. doi: 10.1021/bi00099a019. [DOI] [PubMed] [Google Scholar]
- 5.Nielsen P E, Egholm M, Berg R H, Buchardt O. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 6.Egholm M, Buchardt O, Christensen L, Behrens C, Freier S M, Driver D A, Berg R H, Kim S K, Nordén B, Nielsen P E. Nature (London) 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 7.Armitage B, Ly D, Koch T, Frydenlund H, Orum H, G, Batz B H, Schuster G B. Proc Natl Acad Sci USA. 1997;94:12320–12325. doi: 10.1073/pnas.94.23.12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wittung P, Kim S K, Buchardt O, Nielsen P E, Nordèn B. Nucleic Acids Res. 1994;22:5371–5377. doi: 10.1093/nar/22.24.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 10.Banerjee S K, Christensen R B, Lawrence C W, LeCleric J E. Proc Natl Acad Sci USA. 1988;85:8141–8145. doi: 10.1073/pnas.85.21.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egholm, M., Behrens, C., Christensen, L., Berg, R. H., Nielsen, P. E. & Buchardt, O. (1993) J. Chem. Soc. Chem. Comm., 800–801.
- 12.Tomac S, Sarkar M, Ratilainen T, Wittung P, Nielsen P E, Nordén B, Gräslund A. J Am Chem Soc. 1996;117:5544–5552. [Google Scholar]
- 13.Taylor J-S, Garrett D S, Brockie I R, Svoboda D L, Tesler J. Biochemistry. 1990;29:8858. doi: 10.1021/bi00489a049. [DOI] [PubMed] [Google Scholar]
- 14.Bigey P, Sönnichsen S H, Meunier B, Nielsen P E. Bioconjugate Chem. 1997;8:267–270. doi: 10.1021/bc9700190. [DOI] [PubMed] [Google Scholar]
- 15.Hayatsu H, Ukita T. Biochem Biophys Res Commun. 1967;29:556–561. doi: 10.1016/0006-291x(67)90521-9. [DOI] [PubMed] [Google Scholar]
- 16.Ramaiah D, Koch T, Ørum H, Schuster G B. Nucleic Acids Res. 1998;26:3940–3944. doi: 10.1093/nar/26.17.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubin C M, Schmid C W. Nucleic Acids Res. 1980;8:4613–4619. doi: 10.1093/nar/8.20.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erikksson M, Nielsen P E. Nat Struct Biol. 1996;3:410–413. doi: 10.1038/nsb0596-410. [DOI] [PubMed] [Google Scholar]
- 19.Demidov V V, Potman V N, Frank-Kamenskii M D, Egholm M, Buchard O, Sonnichesen S H, Nielsen P E. Biochem Pharmacol. 1994;48:1310–1313. doi: 10.1016/0006-2952(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 20.Efimov V A, Choob M V, Buryakova A A, Kalinkina A L, Chakhmakhcheva O G. Nucleic Acids Res. 1998;26:566–575. doi: 10.1093/nar/26.2.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keefe A D, Miller S L. J Mol Evol. 1995;41:693–702. doi: 10.1007/BF00173147. [DOI] [PubMed] [Google Scholar]
- 22.Lutz M J, Benner S A, Hein S, Breipohl G, Uhlmann E. J Am Chem Soc. 1997;119:3177–3178. [Google Scholar]