Abstract
Although human immunodeficiency virus type 1 (HIV-1) specific CD4 T-helper cells are vital in mediating antiviral defence, little is known concerning the influence of HIV-1 antigenic variation on CD4 T-cell responses. In this study, the amino acid sequences of 5 synthetic HIV-1 envelope peptides used for in vitro stimulation (T2, P18 MN, P18 IIIB, T1 and TH4.1) were compared to the corresponding amino acid sequences of the gp 160 region of viruses isolated from HIV-1 infected children to determine whether variation in the envelope region of HIV-1 was associated with the ability to detect Env-specifice T-helper cell responses. Although the T2 region appeared to offer some evidence as to the role antigenic variation may have played in class II-restricted CD4 T-cell responses between those children who showed a detectable Env-stimulated T-helper cell response and those who did not, the other regions studied showed no evidence of being more conserved among those children who showed detectable responses. The combined amino acid variation across the specific peptide reions studied was not associated with peripheral levels of HIV-1, nor did the degree of amino acid variation dictate the clinical category into which the children had been classified, although there was a tendency towards HIV-1 isolates from the younger children showing a greater degree of amino acid variation than isolates from the older children. These results suggest that HIV-1 specific CD4 responses may be somewhat tolerant of viral variation, although further studies are required to fully elucidate the effect of antigenic variation on immune recognition.
Keywords: CD4 T-helper cells, gp160, HIV-1 infected children
Introduction
During infections with viruses and other pathogens, CD4 T-helper cells coordinate effector immune responses [1,2], which involves both the induction and maintenance of B-cell functions [3,4] as well as cytotoxic T-lymphocyte (CTL) responses [5-7]. While being key participants in the host response to infection, CD4 T-lymphocytes are also the principal target for human immunodeficiency virus type 1 (HIV-1) infection, and in the absence of virus suppression, newly activated CD4 T-cells may become infected, which leaves the virus-specific immune response without an essential component and unable to perform efficiently [8]. It appears that most HIV-1 infected subjects lose their HIV-1-specific CD4 T-cell responses very early during the course of infection, presumably due to the killing of developing CD4 memory/effector T-cells, either by direct HIV-1 infection [9], and/or by HIV-1-associated apoptosis [10,11]. Consequently, in most chronically infected adults, HIV-1-specific CD4 T-helper cell responses appear to be weak or absent [12,13]. Vigorous HIV-1-specific T-helper cell responses have however been observed among long-term non-progressors [8,14-16], and inversely correlated with viral load [8,14], suggesting that CD4 T-cells are critical in mediating an effective antiviral defence.
Major histocompatibility complex (MHC) molecules bind peptides that are recognised by antigen-specific receptors on T-lymphocytes, and under physiological conditions, these MHC-peptide assemblages are extremely stable. Whether a peptide will be stably bound by an MHC molecule is however dependent on the peptides interaction with a series of pockets on the groove surface of MHC, and a peptide must possess certain amino acid residues at certain positions to be able to bind to a particular MHC molecule. The positions of amino acid substitutions are an important determinant in recognition, as switches at certain positions can influence the anchoring of peptides on the MHC molecule. The combination of high viral turnover and error-prone replication generates high levels of genetic variation in the virus population, a consequence of which may be mutations within CTL and T-helper cell epitopes which affect binding to the presenting HLA molecule or affect interaction of the T-cell receptor with the peptide-HLA complex [17].
Although the role of antigenic variation in HLA class I-restricted epitopes has been widely studied [18-21], work on the influence of HIV-1 antigenic variation on HLA class II-restricted CD4 T-cell responses has been limited, probably due to the difficulty of detecting these responses in HIV-1 infected individuals at any stage of infection. The objective of this study was to determine whether differences in the amino acid sequences of envelope glycoproteins from HIV-1 isolates of infected children would influence the ability to detect Env-specific T-helper cell responses. This was done by comparing the amino acid sequences of the Env peptides used for in vitro stimulation to the corresponding amino acid sequences of the gp160 region of viruses isolated from the HIV-1 infected children. Understanding immunological factors that may contribute to the inhibition of disease progression in HIV-1 infected children could be used in developing strategies for intervention and prevention of disease.
Materials and Methods
Patient Samples
A group of HIV-1 vertically infected children attending Chris Hani Baragwanath Hospital, Johannesburg were enrolled for a study to evaluate the integrity of T-helper cell reactivity to various stimuli including a cocktail of HIV-1 envelope peptides [22]. In vitro T-helper cell reactivity was measured for a total of 52 children using a sensitive culture supernatant titration assay based on IL-2 dependent proliferation of a continuous T-lymphocyte (CTLL) mouse cell line as previously described [23]. The children in the original study ranged in age from 3 months to 11 years, and were selected to include a similar number of age-matched children with “mild” and “severe” clinical presentation of HIV-1 disease. For the current study, viral gp160 sequences were determined for isolates obtained from 16 of these HIV-1 infected children (Table 1). These 16 children were selected to include eight children, four with mild and four with severe disease who had detectable Env-stimulated T-helper cell responses when originally tested, and a further eight age-matched children (four with mild disease and four with severe disease) who had not responded to the Env stimulus. The clinical category, age, HIV-1 viral load, and CD4 T-cell counts for the 16 HIV-1 infected children selected for this study are shown in Table 1, while the peptide sequences and the gp160 amino acid residues of the peptides used for the HIV-1 stimulus are shown in Table 2.
Table 1.
Clinical category, age, HIV-1 viral load, and CD4 T-cell counts of the 16 HIV-1 infected children selected for viral gp160 sequence analysis
| Patient ID | Clinical category | Age (months) | Viral load log RNA copies/ml | CD4 T-cell count |
|---|---|---|---|---|
| T-helper cell Env responders | ||||
| LT10 | Mild | 46 | 4.62 | 744 |
| LT17 | Mild | 86 | 5.63 | 647 |
| LT40 | Mild | 78 | 5.60 | 53 |
| LT42 | Mild | 92 | 4.37 | 416 |
| LT1 | Severe | 17 | 5.56 | 1,045 |
| LT25 | Severe | 134 | 4.62 | 186 |
| LT38 | Severe | 43 | 4.42 | 844 |
| LT50 | Severe | 50 | 5.54 | 312 |
| T-helper cell Env non-responders | ||||
| LT5 | Mild | 43 | 3.03 | 866 |
| LT21 | Mild | 90 | 3.82 | 546 |
| LT39 | Mild | 75 | 4.09 | 760 |
| LT45 | Mild | 83 | 3.74 | 410 |
| LT15 | Severe | 127 | 4.86 | 168 |
| LT18 | Severe | 14 | 5.98 | 975 |
| LT28 | Severe | 53 | 5.73 | Not done |
| LT36 | Severe | 37 | 5.83 | 185 |
Table 2.
Synthetic HIV-1 envelope peptides used to stimulate IL-2 production
| HIV-1 gp160 peptides | Peptide sequences | Amino acid residues* |
|---|---|---|
| Conserved regions | ||
| T1 | KQIINMWQEVGKAMYA | aa 421–436 |
| T2 | HEDIISLWDQSLK | aa 105–117 |
| TH4.1 | DRVIEWQGAYRAIR | aa 827–841 |
| Hypervariable regions | ||
| P18 MN | RIHIGPGRAFYTTKN | aa 317–331 |
| P181IIIB | RIQRGPGRAFVTIGK | aa 317–331 |
Position on the HIV-1 HXB2 genome (Genebank accession number K03455).
Sequencing the gp160 Gene Region of HIV-1 Isolates
HIV-1 viral RNA was isolated from cell culture supernatant samples obtained from the co-culture of patient peripheral blood mononuclear cells (PBMC) with phytohaemagglutinin (PHA)-activated PBMC from healthy blood donors. In most cases, RNA was extracted from day 8 culture samples, except in those instances where p24 antigen levels were determined to be very low at this time interval, in which case a later culture time point was selected. Viral RNA was extracted using the QIAamp Viral RNA Mini Kit (Qiagen Inc., Valencia, CA, USA), according to the manufacturer’s instructions. Following extraction of the viral RNA, an initial reverse transcription step was performed with the env N primer [24] using Expand reverse transcriptase (RT). The cDNA was then subjected to a nested PCR amplification using env A and env N (outer primers) and env B and env M (inner primers) as described previously [24]. The PCR products were purified using the High Pure PCR Product Purification kit (Roche Applied Science, Indianapolis, IN, USA), and the complete gp160 regions were sequenced with a panel of 16 primers, using the ABI PRISM BigDye Terminator version 3.0 Cycle Sequencing kit, according to the manufacturer’s instructions. The sequencing reaction products were purified using isopropanol precipitation, and the clean sequencing reaction products were resuspended in Hi-Di formamide (10 ml/well) and run on an ABI PRISM 3100 automated sequencer. Sequence data was edited using the Sequencing analysis 3.3 program (ABI), and the full-length gp160 sequences were assembled using Sequencher version 4.0. A multiple alignment of the full-length gp160 sequences with references from HIV-1 subtypes A–K, CRF01_AE and CRF02_AG were generated in Clustal X, and phylogenetic trees were constructed for subtyping (Papathanasopoulos et al. pers. Comm.). The nucleotide sequences were translated to amino acid sequences using DNASIS version 3.5, aligned in Clustal X, and viewed in Genedoc.
Results
Analysis of the Viral gp160 Amino Acid Sequences and Comparison to the Amino Acid Sequences of the Peptides Used for in vitro Stimulation
In this study viral gp160 sequences were determined for isolates obtained from 16 HIV-1 infected children (Table 1). Eight of these children, four each in the mild and severe disease groups, showed detectable Env-stimulated T-helper cell responses, and a further eight age-matched children (four with mild disease and four with severe disease) who did not respond to the Env stimulus were selected. Phylogenetic analysis of the gp160 sequences to determine virus genotype showed that all the viral isolates belonged to the HIV-1 subtype C clade (data not shown).
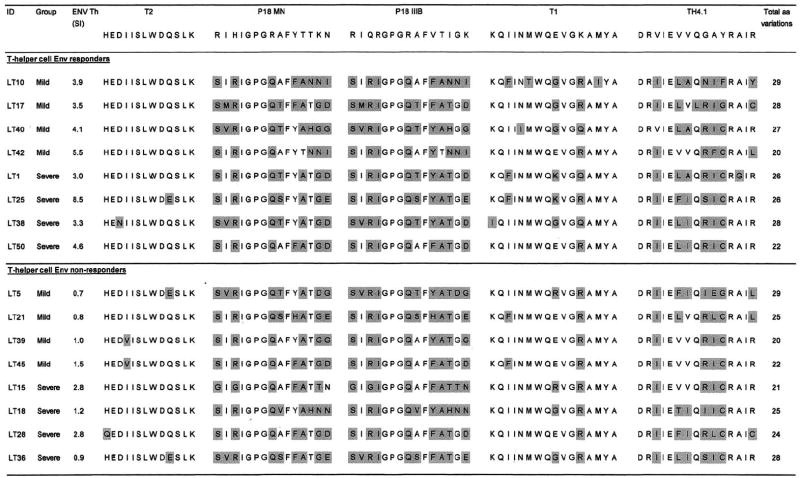
The full gp160 nucleotide sequences of all the viral isolates were translated to amino acid sequences to allow comparison of the Env peptide sequences used for in vitro stimulation to the corresponding virus sequences. Fig. 1 shows the amino acid sequences of the peptides used for in vitro stimulation and the corresponding regions of the 16 viral isolates. Differences between the peptide sequences and viral sequences are highlighted and the total number of amino acid variations are indicated.
Fig. 1.
Comparison of the amino acid sequences of the 5 synthetic HIV-1 Env peptides used for in vitro stimulation to the corresponding amino acid sequences of the gp160 region of viruses isolated from the HIV-1 infected children. Amino acid differences between the Env peptides and the corresponding viral epitopes are shaded. In vitro T-helper cell reactivity to the Env peptides are expressed as SI’s, which are the ratio of counts per minute (cpm) from Env-stimulated cultures to that of unstimulated cultures. All SI’s >3 were considered as a positive response to the Env stimulus.
T2 was the most conserved region of the five studied, with 9 of the 16 isolates having 100% amino acid homology to the T2 peptide, which was 13 amino acids in length. Of these isolates, six belonged to the group who showed a positive T-helper cell response to the Env peptides while three were isolates from children who showed no T-helper reactivity to the Env cocktail. The two children who showed T-helper cell reactivity to the Env cocktail and who showed differences in the T2 region were LT25 (Q → E switch at position 10) and LT38 (D → N switch at position 3). Two of the children who did not respond to the Env peptides also showed the Q → E substitution at position 10 (LT5 and LT36), while in the non-responder group, a H → Q switch at position 1 (LT28) and I → V substitution at position 4 was also observed in two children. The P18 MN and P18 IIIMB sequences on the V3 loop of gp160 showed considerable amino acid variation with most isolates showing between 6 and 8 amino acid differences from the peptides used for in vitro stimulation. The GPGQ motif on the tip of the V3 loop was highly conserved however, with all isolates having this sequence. The T1 region was fairly well conserved, with most isolates showing differences of between 1 and 3 amino acids from the T1 peptide sequence. Most variation occurred at positions 3 (I → F) and 9 (E → G/K/R), while all isolates showed a K → Q or R switch at position 12. As with P18 MN and P18 IIIMB, the TH4.1 region also showed considerable amino acid variation, with isolates showing between 4 and 7 amino acid differences from the TH4.1 peptide used for in vitro stimulation. Although positions 1, 2, 4, 5, 12, and 14 of TH4.1 were conserved for all isolates, most variation occurred at positions 3 (V → I), 6 (V → L/F/T), 7 (V → A/I), and a cluster at positions 9, 10, and 11.
Association between the Combined Amino Acid Variation across the Five Specific Peptide Regions and the Age, Clinical Category, and HIV-1 Viral Load of the HIV-1 Infected Children
In addition to looking at the effect of viral amino acid sequence changes on the ability to detect Env-stimulated T-helper cell responses in the HIV-1 infected children, the combined number of amino acid variations for the five regions analysed in this study was calculated for each isolate (Fig. 1). For those individuals who showed a positive T-helper cell response to the Env stimulus, the mean number of amino acid variations did not differ compared to those individuals who had no detectable Env-stimulated response.
We also investigated whether the total degree of variation observed across these specific peptide regions was associated with the age and clinical category of the HIV-1 infected children or with the amount of circulating virus in these children. The combined amino acid variation across the specific peptide regions studied was not associated with peripheral levels of HIV-1, nor did the degree of amino acid variation dictate the clinical category into which the children had been classified. There was a tendency towards HIV-1 isolates from the younger children showing a greater degree of amino acid variation than isolates from the older children (Spearman rank correlation coefficient ρ = −0.43; p = 0.096).
Discussion
In evolutionary terms, HIV-1 has been a human pathogen for a relatively short time, during which it has evolved into distinct clades that are widely spread throughout the world. The rapid replication kinetics and the low-fidelity RT enzyme of HIV-1 allow a high mutation rate, which enables the virus to evade host immune surveillance. Studies with CTLs have shown that mutations, even single amino acid changes, of viral sequences that specify immunogenic epitopes can lead to escape from CD8 T-cell recognition [18-21,25], impairing the ability of the host to control infection. Little data exists however on the influence of HIV-1 antigenic variation on HLA class II-restricted CD4 T-cell responses. During the early stages of the HIV-1 epidemic, Siliciano et al. [26] analysed the effect of HIV-1 genomic heterogeneity on T-cell recognition using T-cell clones specific for HIV-1 gp120. They observed that the interaction of gp120 epitopes with CD4 T-cell receptors and MHC is precise and poorly cross-reactive, with even one conservative substitution drastically reducing recognition. In another study, by Harcourt et al. [27], PBMC from asymptomatic HIV-1 infected patients were assayed for proliferative responses to HIV-1 antigens. Two individuals who showed positive responses were further analysed, and a dominant CD4 T-lymphocyte response was localised to p17 Gag and p24 Gag epitopes, with proviral DNA sequencing revealing substantial sequence variation within both these peptide epitopes. Although in general, epitope variation did not diminish binding to class II molecules, several of the viral variants failed to stimulate both fresh PBMC and cultured CD4 T-lymphocyte cell lines, indicating that variant antigens arise in HIV-1 infected patients that fail to stimulate the T-cell antigen receptor of HLA classII-restricted CD4 T-lymphocytes, even though the peptide epitopes can be presented on the cell surface. These authors postulate that these naturally occurring variants that fail to stimulate the circulating T-cell repertoire may curtail helper responses at sites where variant viruses predominate such as the lymph nodes [27].
In our current study, the amino acid sequences of HIV-1 envelope peptides used for in vitro stimulation were compared to the corresponding peptide sequences of the gp160 region of viruses isolated from HIV-1 infected children, to determine whether variation in the gp160 region of the infecting viruses was associated with the ability to detect CD4 T-helper cell responses. A limited understanding of the extent of sequence variability that can be tolerated by class II molecules before immune reactivity to defined epitopes is abolished currently exists, and this is the first study to compare Env viral sequences to functional T-helper cell responses in HIV-1 subtype C infected children. The selection of the HIV-1 stimulus used in this study was based on previous reports which had identified these peptides to be broadly immunogenic across MHC haplotypes [28-30], and which have documented T-helper cell responses to these peptides in several, independent populations of HIV-1 exposed, uninfected individuals [31-34]. The P18 MN and P18 IIIB variable regions of the virus isolates showed considerable amino acid variation from the peptides used for in vitro stimulation and it seems unlikely that these peptides would have been recognised. Regarding the TH4.1 and T1 conserved regions, a similar degree of amino acid variation was observed between those children who showed a detectable Env-stimulated T-helper cell response and those who did not. T2 was the most conserved region of the five studied, with 9 of the 16 isolates having 100% amino acid homology to the T2 peptide sequence. Of these isolates, six belonged to the group that showed a positive T-helper cell response to the Env peptides while the remaining three were isolates from children who showed no T-helper reactivity to the Env cocktail.
Although the T2 region appeared to offer some evidence as to the role antigenic variation may have played in class II-restricted CD4 T-cell responses between those children who showed a detectable Env-stimulated T-helper cell response and those who did not, the other regions studied showed no evidence of being more conserved among those children who showed detectable responses. These results suggest, in contrast to other reports that have shown a negative effect of viral variation on CD4 T-cell recognition [26,27], that CD4 responses may be somewhat tolerant of viral variation. T-helper cells may have been reactive to more than one of the peptides in the cocktail, and although limitations of small blood volumes from the HIV-1 infected children in this study did not allow peptide responses to be tested individually, this approach would be important in future studies to provide more insight into immunogenic epitopes.
HIV-1-specific T-helper cells are critical in mediating an effective antiviral defence, as shown in long-term non-progressing HIV-1 infected patients [8,14-16]. We have previously shown, in the same cohort of HIV-1 infected children tested in this current study, that HIV-1 specific CD4 T-cell responses may play a role in containing progression of HIV-1 infection, where children (with a severe clinical profile) who lacked Env peptide-stimulated T-helper cell responses were 6-fold more likely to die during the year after enrollment than those who displayed an Env-specific T-helper cell response [22]. Mutations that enable HIV-1 to evade immune surveillance have been postulated to be harmful in that they would render the host’s immune system unable to control infection. This is supported by data from Moore et al. [25], showing that HIV-1 infected patients harbouring virus with escape mutations had a level of viraemia that was approximately a log higher than individuals infected with HIV-1 that lacked such mutations. In addition, it has been shown that cells expressing mutated HIV-1 peptides can even block the lysis of cells infected with unmutated virus [18]. In our current study however, the combined amino acid variation across the specific peptide regions studied was not associated with peripheral levels of HIV-1.
The interaction between host immune responses and HIV-1 is complex and there are a number of host and viral factors which could influence the ability of leukocytes to mount an effective response. Current knowledge of the precise epitopes of different HIV-1 proteins targeted by CD4 T-cells is limited, as is an understanding of the extent of sequence variability that can be tolerated by class II molecules before immune reactivity to defined epitopes is abolished. In this study, the only region that appeared to offer some evidence as to the role antigenic variation may have played in the ability to detect class II-restricted CD4 T-cell responses was T2. Given the pivotal role of CD4 T-cell help in CTL and antibody responses, delineation of major T-helper cell recognition sites of HIV-1 proteins, and further elucidation of the effect of antigenic variation on immune recognition would provide vital information for the design of an effective HIV-1 vaccine.
Nucleotide sequence accession numbers
| Our number | Accession number |
|---|---|
| 99ZALT1 | AY522721 |
| 99ZALT10 | AY522722 |
| 99ZALT15 | AY522723 |
| 99ZALT17 | AY522724 |
| 99ZALT18 | AY522725 |
| 99ZALT21 | AY522726 |
| 99ZALT25 | AY522727 |
| 99ZALT28 | AY522728 |
| 99ZALT36 | AY522729 |
| 99ZALT38 | AY522730 |
| 99ZALT39 | AY522731 |
| 99ZALT40 | AY522732 |
| 99ZALT42 | AY522733 |
| 99ZALT45 | AY522734 |
| 99ZALT5 | AY522735 |
| 99ZALT50 | AY522736 |
Acknowledgments
This study was supported in part by the South African AIDS Vaccine Initiative (SAAVI) and by grants from NICHD (HD 42402, HD 36177). This study formed part of a Ph.D. thesis, by S. Meddows-Taylor, approved for that degree by the University of the Witwatersrand, Johannesburg, South Africa.
References
- 1.Kalams SA, Walker BD. J Exp Med. 1998;188:2199–2204. doi: 10.1084/jem.188.12.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maloy KJ, Burkhart C, Freer G, Rulicke T, Pircher H, Kono DH, Theofilopoulos AN, Ludewig B, Hoffman-Rohrer U, Zinkernagel RM, Hengartner H. J Immunol. 1999;162:2867–2874. [PubMed] [Google Scholar]
- 3.McMichael AJ, Phillips RE. Annu Rev Immunol. 1997;15:271–296. doi: 10.1146/annurev.immunol.15.1.271. [DOI] [PubMed] [Google Scholar]
- 4.Letvin NL. J Clin Invest. 1998;102:1643–1644. doi: 10.1172/JCI5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matloubian M, Concepcion RJ, Ahmed R. J Virol. 1994;68:8056–8063. doi: 10.1128/jvi.68.12.8056-8063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romieu R, Baratin M, Kayibanda M, Guillet JG, Viguer M. Int Immunol. 1998;10:1273–1279. doi: 10.1093/intimm/10.9.1273. [DOI] [PubMed] [Google Scholar]
- 7.Kalams SA, Buchbinder SP, Rosenberg ES, Billingsley JM, Colbert DS, Jones NG, Shea AK, Trocha AK, Walker BD. J Virol. 1999;73:6715–6720. doi: 10.1128/jvi.73.8.6715-6720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenberg ES, Billingsley JM, Caliendo AM, Boswell SL, Sax PE, Kalams SA, Walker BD. Science. 1997;278:1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- 9.Levy JA. Microbiol Rev. 1993;57:183–289. doi: 10.1128/mr.57.1.183-289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groux H, Torpier G, Monte D, Mouton Y, Capron A, Ameisen JC. J Exp Med. 1992;175:331–340. doi: 10.1084/jem.175.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clerici M, Sarin A, Berzofsky JA, Landay AL, Kessler HA, Hashemi F, Hendrix CW, Blatt SP, Rusnak J, Dolan MJ, Coffman RL, Henkart PA, Shearer GM. AIDS. 1996;10:603–611. doi: 10.1097/00002030-199606000-00005. [DOI] [PubMed] [Google Scholar]
- 12.Pontesilli O, Carlesimo M, Varani AR, Ferrara R, Guerra EC, Bernadi ML, Ricci G, Mazzone AM, D’Offizi G, Aiuti F. Clin Exp Immunol. 1995;100:419–424. doi: 10.1111/j.1365-2249.1995.tb03716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenberg ES, Walker BD. AIDS Res Hum Retroviruses. 1998;14:S143–S147. [PubMed] [Google Scholar]
- 14.Pitcher CJ, Quittner C, Peterson DM, Connors M, Koup RA, Maino VC, Picker LJ. Nat Med. 1999;5:518–525. doi: 10.1038/8400. [DOI] [PubMed] [Google Scholar]
- 15.Pontesilli O, Carotenuto P, Kerkhof-Garde SR, Roos MT, Keet IP, Coutinho RA, Goudsmit J, Miedema F. AIDS Res Hum Retroviruses. 1999;15:973–981. doi: 10.1089/088922299310485. [DOI] [PubMed] [Google Scholar]
- 16.Alatrakchi N, Di Martino V, Thibault V, Autran B. AIDS. 2002;16:713–717. doi: 10.1097/00002030-200203290-00006. [DOI] [PubMed] [Google Scholar]
- 17.Goulder PJ, Rowland-Jones SL, McMichael AJ, Walker BD. AIDS. 1999;13:S121–S136. [PubMed] [Google Scholar]
- 18.Klenerman P, Rowland-Jones S, McAdam S, Edwards J, Daenke S, Lalloo D, Koppe B, Rosenberg W, Boyd D, Edwards A, Giangrande P, Phillips RE, McMichael AJ. Nature. 1994;369:403–407. doi: 10.1038/369403a0. [DOI] [PubMed] [Google Scholar]
- 19.Price DA, Goulder PJ, Klenerman P, Sewell AK, Easterbrook PJ, Troop M, Bangham CR, Phillips RE. Proc Natl Acad Sci USA. 1997;94:1890–1895. doi: 10.1073/pnas.94.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael AJ, Rowland-Jones S. Nat Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 21.Kelleher AD, Long C, Holmes EG, Alien RL, Wilson J, Conlon C, Workman C, Shaunak S, Olson K, Goulder PJ, Brander C, Ogg G, Sullivan JS, Dyer W, Jones I, McMichael AJ, Rowland-Jones S, Phillips RE. J Exp Med. 2001;193:375–386. doi: 10.1084/jem.193.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn L, Meyers TM, Meddows-Taylor S, Simmank K, Sherman GG, Tiemessen CT. J Infect Dis. 2001;184:691–698. doi: 10.1086/322988. [DOI] [PubMed] [Google Scholar]
- 23.Clerici M, Stocks NI, Zajac RA, Boswell RN, Bernstein DC, Mann DL, Shearer GM, Berzofsky JA. Nature. 1989;339:383–385. doi: 10.1038/339383a0. [DOI] [PubMed] [Google Scholar]
- 24.Gao F, Morrison SG, Robertson DL, Thornton CL, Craig S, Karlsson G, Sodroski J, Morgado M, Galvao-Castro B, von Briesen H, Beddows S, Weber J, Sharp PM, Shaw GM, Hahn B. J Virol. 1996;70:1651–1667. doi: 10.1128/jvi.70.3.1651-1667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 26.Siliciano RF, Lawton T, Knall C, Karr RW, Berman P, Gregory T, Reinherz EL. Cell. 1988;54:561–575. doi: 10.1016/0092-8674(88)90078-5. [DOI] [PubMed] [Google Scholar]
- 27.Harcourt GC, Garrard S, Davenport MP, Edwards A, Phillips RE. J Exp Med. 1998;188:1785–1793. doi: 10.1084/jem.188.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cease KB, Margalit H, Cornette JL, Putney SD, Robey WG, Ouyang C, Streicher HZ, Fischinger PJ, Gallo RC, DeLisi C, Berzofsky JA. Proc Natl Acad Sci USA. 1987;84:4249–4253. doi: 10.1073/pnas.84.12.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hale PM, Cease KB, Houghten RA, Ouyang C, Putney S, Javaherian K, Margalit H, Cornette JL, Spouge JL, DeLisi C. Int Immunol. 1989;1:409–415. doi: 10.1093/intimm/1.4.409. [DOI] [PubMed] [Google Scholar]
- 30.Berzofsky JA, Pendleton CD, Clerici M, Ahlers J, Lucey DR, Putney SD, Shearer GM. J Clin Invest. 1991;88:876–884. doi: 10.1172/JCI115389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clerici M, Berzofsky JA, Shearer GM, Tacket CO. J Infect Dis. 1991;164:178–182. doi: 10.1093/infdis/164.1.178. [DOI] [PubMed] [Google Scholar]
- 32.Clerici M, Giorgi JV, Chou CC, Gudeman VK, Zack JA, Gupta P, Ho HN, Hishanian PG, Berzofsky JA, Shearer GM. J Infect Dis. 1992;165:1012–1019. doi: 10.1093/infdis/165.6.1012. [DOI] [PubMed] [Google Scholar]
- 33.Clerici M, Levin JM, Kessler HA, Harris A, Berzofsky JA, Landay AL, Shearer GM. J Am Med Assoc. 1994;271:42–46. [PubMed] [Google Scholar]
- 34.Mazzoli S, Trabattoni D, Lo Caputo S, Piconi S, Ble C, Meacci F, Ruzzante S, Salvi A, Semplici F, Longhi R, Fusi ML, Tofani N, Biasin M, Villa ML, Mazzotta F, Clerici M. Nat Med. 1997;3:1250–1257. doi: 10.1038/nm1197-1250. [DOI] [PubMed] [Google Scholar]