Abstract
Communication between the 5′ and 3′ ends is a common feature of several aspects of eukaryotic mRNA metabolism. In the nucleus, the pre-mRNA 5′ end is bound by the nuclear cap binding complex (CBC). This RNA–protein complex plays an active role in both splicing and RNA export. We provide evidence for participation of CBC in the processing of the 3′ end of the message. Depletion of CBC from HeLa cell nuclear extract strongly reduced the endonucleolytic cleavage step of the cleavage and polyadenylation process. Cleavage was restored by addition of recombinant CBC. CBC depletion was found to reduce the stability of poly(A) site cleavage complexes formed in nuclear extract. We also provide evidence that the communication between the 5′ and 3′ ends of the pre-mRNA during processing is mediated by the physical association of the CBC/cap complex with 3′ processing factors bound at the poly(A) site. These observations, along with previous data on the function of CBC in splicing, illustrate the key role played by CBC in pre-mRNA recognition and processing. The data provides further support for the hypothesis that pre-mRNAs and mRNAs may exist and be functional in the form of “closed-loops,” due to interactions between factors bound at their 5′ and 3′ ends.
The biosynthesis of most eukaryotic nuclear mRNAs requires the modification of the 5′ end of the RNA by the cotranscriptional addition of an m7G(5′)ppp(5′)N cap structure (1, 2), the removal of introns by splicing (3), and the modification of the 3′ end by endonucleolytic cleavage and poly(A) addition (4, 5).
Polyadenylation in vertebrates requires two cis-acting RNA sequence elements which straddle the cleavage site and embody the core poly(A) site: the AAUAAA hexamer 10–30 nucleotides upstream of the cleavage site, and an amorphous U- or G+U-rich element downstream of the cleavage site. Six factors, comprised of at least 13 proteins, are required for pre-mRNA cleavage and polyadenylation (4, 5). Cleavage and polyadenylation specificity factor (CPSF) (6, 7) binds the pre-mRNA upon recognition of the AAUAAA hexamer, while cleavage stimulatory factor (CstF) (8) binds the downstream element. Together, CPSF and CstF form a relatively stable pre-mRNA–protein complex (9) that allows for the recruitment of cleavage factors Im and IIm (10), and poly(A) polymerase (11, 12). Following the endonucleolytic cleavage of the pre-mRNA, poly(A) addition requires both CPSF and poly(A) polymerase (7, 13). Poly(A) binding protein II, however, confers both processivity and tail length control to the poly(A) addition reaction (14, 15).
The 5′ cap structure has been shown to influence the efficiency of 3′ processing in vitro (16–18). The addition of the cap analog m7GpppG to HeLa cell nuclear extract resulted in a reduction in poly(A) site cleavage, although even at high levels of the analog, processing was not completely abolished (16, 18). Uncapped pre-mRNAs were found to be poorly processed in nuclear extract, and have been shown to compete less efficiently for 3′ processing factors than capped pre-mRNAs (17, 18). Although not essential for 3′ end formation in vitro, the impact of the cap upon processing in crude nuclear extract suggested that the cap may indeed contribute to processing within the complex milieu of the nucleus. A similar picture has emerged from the investigation of pre-mRNA splicing, in which the 5′ cap, although likewise not essential for processing, does contribute to the removal of the cap proximal intron (19–22).
In the nucleus, the 5′ cap is bound by the nuclear cap-binding complex (CBC) that in human cells is comprised of two proteins: CBP20 and CBP80 (23–27). This RNA–protein complex plays an active role in both splicing (19–21, 24) and RNA export (25, 28). We now show that CBC participates in a second mRNA processing reaction. Depletion of CBC from HeLa cell nuclear extract diminished poly(A) site cleavage, and subsequent addition of recombinant CBC restored processing. Poly(A) site cleavage complex stability was found to be reduced in CBC-depleted nuclear extracts, and the physical interaction of the CBC/cap complex with the 3′ processing complex was demonstrated by coimmunoprecipitation.
The 5′ and 3′ ends of eukaryotic mRNAs communicate at nearly every stage in the life of the message. Nuclear export (29), translation (30–32), and mRNA decay (33) all appear to involve the coordinate recognition of RNA–protein complexes at both termini. We now present evidence that CBC bound at the 5′ end of the pre-mRNA participates in the actual biosynthesis of the mRNA 3′ end.
METHODS
Preparation of HeLa Nuclear Extracts.
HeLa cell nuclear extracts were prepared as described (34), with the following exceptions. All buffers contained Tris (pH 7.9) at the concentrations indicated for Hepes⋅KOH. Buffer C contains pepstatin at 0.7 mg/ml and leupeptin at 0.4 mg/ml. The high salt buffer C contained 800 mM (NH4)2SO4 instead of NaCl, and the low salt buffer C contained KCl instead of NaCl. The nuclear extract was centrifuged for 60 min at 95,000 × g and used undialyzed for in vitro assays.
Depleted Nuclear Extracts.
Depletions were conducted as described (24), with the following exceptions. The HeLa nuclear extract was adjusted to 500 mM (NH4)2SO4 and then clarified by centrifugation at 14,000 rpm in a microfuge for 5 min at 4°C prior to filtration through a 0.45 μm syringe filter. Eluates from filtration were recycled over protein-A Sepharose antibody columns (either the rabbit preimmune or the polyclonal anti-CBP80), which had been crosslinked as described (24). CBC levels were monitored by enhanced chemiluminescence (Amersham) Western blot analysis, using a polyclonal CBP80 antiserum. Depleted extracts were dialyzed into buffer D (100 mM KCl/20 mM Hepes, pH 7.9/20% glycerol/0.2 mM EDTA/0.5 mM DTT/0.1 mM phenylmethylsulfonyl fluoride).
Preparation of Recombinant CBC.
rCBC was prepared as described (25). Briefly, human CBP20 was expressed in Escherichia coli (1-l culture) as a histidine-tagged protein, and human CBP80 was expressed in E. coli (6-l culture) as an untagged protein. Histidine-tagged CBP20 was purified on a nickel-nitrilotriacetic acid column as previously described (25). 2 ml of purified CBP20 along with 0.5 ml nickel-nitrilotriacetic acid-agarose was added to a CBP80 expressing lysate and incubated at room temperature for 1 hr. The heterodimeric CBC complex was then purified via the histidine tag resident on CBP20. Purification of CBC was monitored by gel mobility shift assays using capped (m7GpppG) or uncapped (pppG) 32P-labeled RNAs. Gel shift assays were conducted as described (23).
Generation of 32P-Labeled RNA Substrates.
L3 RNA substrates for in vitro cleavage reactions were prepared as described (9). The RNA previously termed AAU + ds (9) is referred to here as L3. All of the substrates were transcribed using SP6 polymerase (Epicentre Technologies, Madison, WI). Transcription reactions contained 0.5 mM ATP, CTP, UTP, 15 μM GTP, 0.1 μM [α-32P]GTP with a specific activity of 3,000 Ci/mmol (DuPont; 1 Ci = 37 GBq), and 0.25 mM cap analog. Cap analogs were obtained from the following sources: m7GpppG (Epicentre Technologies) and Gm2′pppG7m (Pharmacia). RNA transcripts were gel purified and stored at −20°C in H2O.
In Vitro 3′ Cleavage Assays.
Cleavage reactions were carried out essentially as described (17), with the following exceptions. 2.9 × 10−14 mol of 32P-labeled pre-mRNA (100,000 dpm) were incubated in a total reaction volume of 25 μl containing 4–40% (vol/vol) depleted HeLa nuclear extract, 68 mM KCl, 29 mM Tris (pH 7.8), 5.8% glycerol, 0.12 mM EDTA, 0.5–1.0 mM 3′dATP, 2% polyvinylalcohol, and 0.5 μg tRNA for 10–40 min at 30°C. Complementation of CBC-depleted nuclear extract with recombinant CBC was carried out as described above for depleted nuclear extract cleavages, with the following modifications: all components excluding 32P-labeled pre-mRNA and 3′dATP were added together on ice and preincubated at 30°C for 10 min. The reactions were then chilled on ice prior to the addition of 32P-labeled substrate and 3′dATP. rCBC complementation reactions were returned to 30°C for 40 min. All cleavage reactions were stopped by the addition of 170 μl ETS (10 mM EDTA, 10 mM Tris, 0.5% SDS) and 5 μl proteinase K (20 mg/ml) and incubated at 37°C for at least 10 min. Reactions were phenolized and ethanol precipitated, followed by denaturing 10% PAGE.
In Vitro Polyadenylations.
32P-labeled pre-mRNA (2.9 × 10−14 mol) (100,000 dpm) was incubated in a total reaction volume of 25 μl containing 4–40% (vol/vol) CBC- or mock-depleted nuclear extract, 68 mM KCl, 29 mM Tris (pH 7.8), 5.8% glycerol, 0.12 mM EDTA, 0.7 mM ATP, 0.5 mM MgCl2, 2% polyvinylalcohol, and 0.5 μg tRNA for 10 min at 30°C. Reactions were treated with ETS/proteinase K as described above for 15 min at 37°C, phenolized, ethanol precipitated, and electrophoresed on a denaturing 5% polyacrylamide gel.
3′ Processing Complex Analysis.
3′ processing complex stability assays were conducted essentially as described (35). 30 μl reactions contained 12.5% (vol/vol) of depleted nuclear extract, 1% polyvinyl alcohol, 0.75 μg tRNA, 9 μg total yeast RNA, 75 mM KCl, 15 mM Tris (pH 7.9), 7.5% glycerol, 20 mM EDTA, 50 mM DTT, 10 mM phenylmethylsulfonyl fluoride, and 4.3 × 10−14 mol (100,000 dpm) of 32P-labeled HIV-1 pre-mRNA (use/CPS full-length for cleavage complexes or use/CPS precleaved for polyadenylation complexes) (35). Reactions were incubated for 10 min at 30°C, and a 5-μl aliquot was removed from each sample as time point 0, transferred to ice and treated with 5 mg/ml heparin. To the remaining 25 μl reaction, 4.0 × 10−12 mol of unlabeled capped L3 pre-mRNA was added. Incubation of the reaction was continued at 30°C and 5 μl aliquots were removed from each reaction at 5, 10, 20, and 40 min, placed on ice and treated with 5 mg/ml of heparin. Reactions were electrophoresed on a nondenaturing 3% polyacrylamide (80:1) gel in 25 mM Tris, 25 mM boric acid, and 1 mM EDTA at 300 V for 1.5 hr at 4°C. The gel was preelectrophoresed for 30 min prior to loading.
RNase H/Immunoprecipitations.
RNA–protein complexes were assembled as follows: 125 μl reactions contained 12.5% (vol/vol) of nuclear extract (preadsorbed with protein A-Sepharose to remove nonspecific binding proteins), 1% polyvinyl alcohol, 2.5 μg tRNA, 30 μg total yeast RNA, 75 mM KCl, 15 mM Tris (pH 7.9), 7.5% glycerol, 20 mM EDTA, 50 mM DTT, 10 mM phenylmethylsulfonyl fluoride, 1 mM MgCl2, and 2.2 × 10−13 mol (500,000 dpm) of 32P-labeled HIV-1 pre-mRNA (use/CPS full-length; ref. 35). The HIV-1 pre-mRNA containing a wild-type hexamer (AAUAAA) was either capped (m7GpppG) or uncapped (pppG), and an HIV-1 pre-mRNA containing a mutant hexamer (AAUAAA → CCACCC) was capped. Reactions were incubated for 10 min at 30°C. For RNase H digestion, 1.3 mM of the DNA oligo (5′-GTACAGGCAAAA-3′), complementary to a sequence between the 5′ end of the RNA and the poly(A) site, was added to the reaction. After incubation of the reaction at 30°C for 10 min, EDTA was added to 2.5 mM to block further cleavage by RNase H. 25 μl aliquots were removed from the reaction and added to 20 μl of protein A-Sepharose/antibody preparation or protein G-Sepharose/antibody preparation (50% slurry). Protein G-Sepharose beads or preimmune serum/protein A and αCBP80/protein A crosslinked preparations were incubated in the presence of 100 μg of total yeast RNA in NET-2 buffer (150 mM NaCl/50 mM Tris (pH 7.9)/0.05% Nonidet P40) at room temperature for 30 min. The beads or bead/antibody preparations were then washed twice in 10 volumes of NET-2. After washing, the beads and bead/antibody preparations were resuspended in NET-2 buffer to a final 50% slurry. Immunoprecipitations with the α64k (Clinton MacDonald, Texas Tech, Lubbock, TX) and the α-2,2,7trimethylguanosine (αTMG) (Oncogene Science) antibodies were performed as follows. To the 20 μl of blocked protein G beads, either 10 μl of purified αTMG or 10 μl of α64k hybridoma culture supernatant was added, followed by the 25 μl reaction aliquot. Immunoprecipitations using the crosslinked αCBP80 or crosslinked preimmune serum also received a 25 μl aliquot of the reaction mixture along with 10 μl of NET-2 buffer. Immunoprecipitation reactions were incubated on ice for 45 min with frequent but gentle mixing. The reactions were centrifuged (pulse in microfuge) and the supernatant was removed. The pellets were washed four times with 10 volumes of NET-2 buffer. After the final wash, 195 μl of ETS and 5 μl of proteinase K were added to the beads and incubated at 37°C for 15 min. Reactions were phenolized, ethanol precipitated, and electrophoresed on a denaturing 10% polyacrylamide gel.
RESULTS
Depletion of CBC from HeLa Cell Nuclear Extract Inhibits Poly(A) Site Cleavage.
Previous studies have shown that a 5′ cap structure (m7GpppG) augmented mRNA 3′ processing in HeLa cell nuclear extract (16–18). Addition of exogenous m7GpppG, but not ApppG, to nuclear extract reduced, but did not abolish poly(A) site cleavage, suggesting that a titratable factor mediated the enhancement of 3′ processing (16, 18). In the nucleus, the m7GpppG cap is bound by the heterodimeric CBC composed of CBP20 and CBP80 (23–27). CBC was considered the most likely candidate for the m7GpppG-titratable factor involved in the enhancement of poly(A) site cleavage. We therefore depleted CBC from HeLa nuclear extract using a polyclonal CBP-80 antibody. It has been determined that immunodepletion with anti-CBP80 coimmunodepletes CBP20 (24).
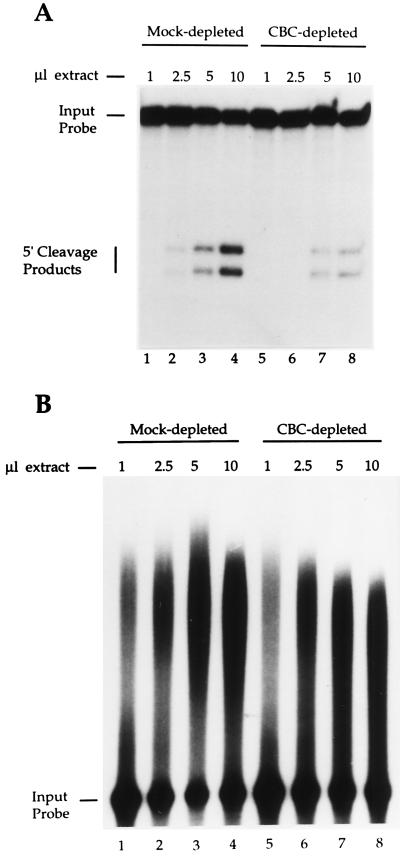
A comparison of the poly(A) site cleavage activity of the CBC-depleted and mock-depleted HeLa cell nuclear extract revealed a 4-fold reduction in the depleted extract (Fig. 1A, compare lanes 4 and 8). The difference in poly(A) site cleavage activity in the mock-depleted vs. the CBC-depleted extract was equivalent to the difference in cleavage activity between m7GpppG-capped and uncapped pre-mRNAs, in HeLa cell nuclear extract (data not shown).
Figure 1.
Inhibition of poly(A) site cleavage by CBC depletion. (A) An m7GpppG-capped pre-mRNA containing the L3 poly(A) site was incubated with increasing amounts of either a mock-depleted (lanes 1–4) or a CBC-depleted (lanes 5–8) HeLa cell nuclear extract in the presence of 3′ dATP. Products of the cleavage reaction were resolved on a denaturing 10% polyacrylamide gel. (B) An m7GpppG-capped precleaved RNA (extending only to the poly(A) cleavage site) containing the HIV-1 poly(A) site was incubated with increasing amounts of either mock-depleted (lanes 1–4) or CBC-depleted (lanes 5–8) HeLa cell nuclear extract in the presence of ATP. The reaction products were resolved on a denaturing 5% polyacrylamide gel.
We also examined the impact of CBC-depletion upon the second step of 3′ processing, poly(A) addition, using a precleaved RNA substrate [an RNA extending only to the poly(A) cleavage site]. In contrast to poly(A) site cleavage, the efficiency of poly(A) addition to the precleaved RNA was only slightly reduced (≈10%) by CBC depletion (Fig. 1B).
Recombinant CBC Restores Poly(A) Site Cleavage Activity to a CBC-Depleted HeLa Cell Nuclear Extract.
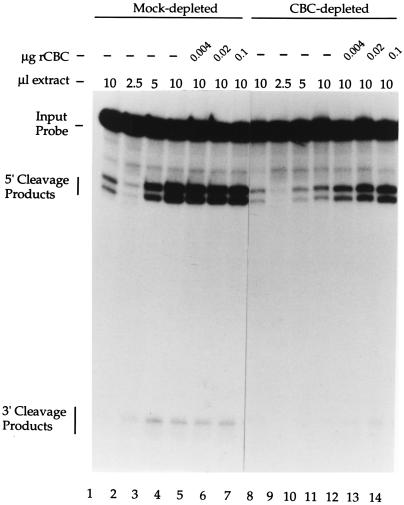
To determine whether the absence of CBC alone was responsible for the reduction in poly(A) site cleavage activity in depleted extract, purified recombinant CBC (rCBC) was added back to the depleted extract. Fig. 2 (lanes 2–4 and 9–11) illustrates the level of processing of capped pre-mRNA with increasing concentrations of mock- and CBC-depleted extract, respectively. Fig. 2, lanes 1 and 8, illustrate the level of processing of uncapped pre-mRNA in mock- and CBC-depleted extract, respectively, at the highest extract concentration. As illustrated in Fig. 2 (lanes 12–14), rCBC was sufficient for the restoration of efficient poly(A) site cleavage activity to the CBC-depleted nuclear extract. The addition of rCBC had no detectable impact upon poly(A) site cleavage when added to the mock-depleted nuclear extract (Fig. 2, lanes 5–7). Thus, rescue of cleavage activity was specific, and not due to nonspecific stimulation of processing by the addition of rCBC. Furthermore, the inability of rCBC to enhance poly(A) site cleavage of an uncapped pre-mRNA over a wide range of concentrations (data not shown) suggests that the actual binding of CBC to the pre-mRNA is required for efficient 3′ processing. CBC is therefore essential for the cap-dependent enhancement of poly(A) site cleavage in HeLa cell nuclear extract.
Figure 2.
rCBC restores poly(A) site cleavage efficiency to a CBC-depleted HeLa cell nuclear extract. Uncapped (pppG, lanes 1 and 8) or capped (m7GpppG, lanes 2–7 and 9–14) pre-mRNAs containing the L3 poly(A) site were incubated in mock-depleted (lanes 1–7) or CBC-depleted (lanes 8–14) HeLa cell nuclear extract in the presence of 3′ dATP. The extracts were supplemented with the indicated amount of recombinant CBC (lanes 5–7 and 12–14). The reaction products were resolved on a denaturing 10% polyacrylamide gel.
3′ Processing Complex Stability Is Diminished in the Absence of CBC.
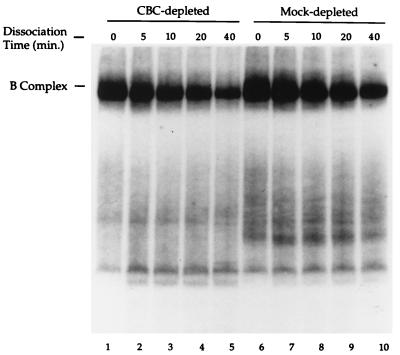
To address the mechanism by which CBC participates in the cap-dependent enhancement of poly(A) site cleavage in nuclear extract we examined the impact of CBC depletion upon 3′ processing complex stability. The efficiency of 3′ processing has been shown (4, 5, 36–40) to correlate with the stability of processing complexes formed on the pre-mRNA in vitro. A capped 32P-labeled pre-mRNA containing the HIV-1 poly(A) site was incubated at 30°C in each extract to allow for complex formation. After a 10-min incubation, an aliquot was withdrawn, placed on ice, and treated with heparin (0 time point). A 100-fold molar excess of unlabeled capped L3 pre-mRNA was then added to the reaction and the incubation was continued at 30°C. Aliquots were withdrawn at 5, 10, 20, and 40 min, placed on ice and treated with heparin.
As illustrated in Fig. 3, the stability of poly(A) site cleavage complexes (“B” complexes—containing both CPSF and CstF; ref. 9) was reduced in the CBC-depleted extract relative to the mock-depleted extract (Fig. 3, compare lanes 1–5 and 6–10). Quantitation revealed a two-fold reduction in the B complex half-life in the CBC-depleted extract. In a set of five independent experiments, the mean reduction in B complex half-life in the CBC-depleted extract was 3.6-fold (SD = 1.5). It should be noted, however, that in crude extract we are unable to distinguish between CPSF/CstF/RNA complexes and higher order complexes containing additional 3′ processing factors by gel mobility shift. The possibility therefore remains that CBC may affect the association of additional 3′ processing factors with the CPSF/CstF/RNA complex.
Figure 3.
CBC contributes to B complex stability. A 32P-labeled m7GpppG-capped pre-mRNA containing the HIV-1 poly(A) site was incubated with either CBC-depleted (lanes 1–5) or mock-depleted (lanes 6–10) nuclear extract. 3′ processing complexes were allowed to form for 10 min at 30°C. An aliquot was removed, denoted time 0, and treated with heparin at 0°C. A 100-fold molar excess of unlabeled m7GpppG-capped pre-mRNA was added to the remaining reaction and aliquots were removed after an additional incubation for 5, 10, 20, and 40 min. The RNA/protein complexes were resolved on a nondenaturing 3% polyacrylamide gel. The B complex contains both CPSF and CstF (9).
The stability of poly(A) addition complexes (“A” complexes—containing CPSF, but not CstF; ref. 9) formed on a precleaved HIV-1 pre-mRNA was also examined. Quantitation of the A complexes formed in CBC- and mock-depleted extracts (data not shown) revealed no significant difference in A complex stability. These results were consistent with our observations that the efficiency of poly(A) addition in the two extracts was nearly equivalent (Fig. 1B).
Physical Interaction Between the CBC/Cap Complex and the 3′ Processing Complex.
The impact of CBC on cleavage complex stability in HeLa cell nuclear extract suggested a potential physical interaction between these two RNA–protein complexes. To investigate the existence of protein-mediated interactions of the 5′ end of the RNA and the poly(A) site, we utilized an RNase H/immunoprecipitation assay. Our strategy was to form 3′ processing complexes on capped pre-mRNA in a HeLa cell nuclear extract and then cleave the RNA between the cap and the poly(A) site using a complementary DNA oligonucleotide and the endogenous RNase H activity of the extract. The RNase H digestion was stopped after 10 min of incubation by the addition of EDTA. The potential interaction between the 5′ and 3′ RNase H digestion products was then examined by the immunoprecipitation of the RNA–protein complexes with antibody directed against CBP80 (anti-CBP80) or against the 64-kDa subunit of CstF (anti-64k). If CBC and the 3′ processing complex physically interact, each antibody should be capable of immunoprecipitating both the 5′ and 3′ RNase H cleavage products. Two other antibodies were used: (i) rabbit preimmune serum, as a control, and (ii) an anti-TMG mAb. The anti-TMG antibody recognizes a set of small nuclear RNAs via their unique TMG cap, as confirmed by the 3′ end labeling of anti-TMG immunoprecipitated HeLa cell nuclear extract RNAs (data not shown).
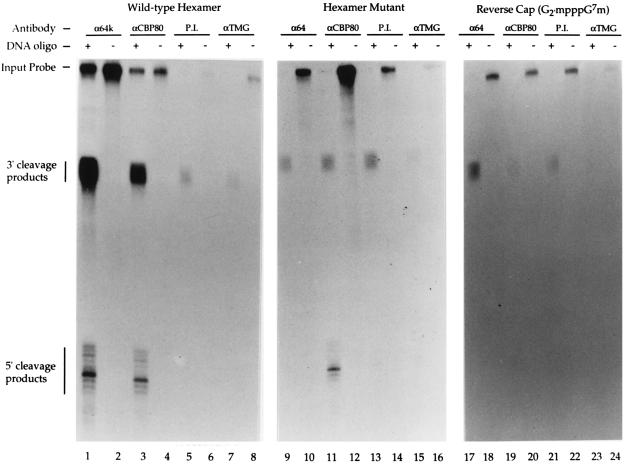
Three RNAs were examined: (i) an m7GpppG-capped RNA containing a wild-type poly(A) site (Fig. 4, lanes 1–8); (ii) an m7GpppG-capped RNA containing a mutant hexamer (AAUAAA to CCACCC) (Fig. 4, lanes 9–16) that does not support 3′ processing complex formation (data not shown); and (iii) a Gm2′pppG7m-capped (“reverse” capped) RNA containing a wild-type poly(A) site (Fig. 4, lanes 17–24). CBC does not detectably bind to RNAs carrying this reverse cap structure in vitro (data not shown). The HIV-1 pre-mRNA was again used for these experiments, since it was more amenable to RNase H cleavage than the L3 pre-mRNA (data not shown). As shown in Fig. 4, immunoprecipitation of RNA–protein complexes formed on the m7GpppG-capped RNA, containing a wild-type poly(A) site, with either the anti-64k or anti-CBP80 antibodies resulted in the coisolation of the 5′ and 3′ RNase H cleavage products (Fig. 4, lanes 1–4). The preimmune and anti-TMG antibodies immunoprecipitated background levels of these cleavage products as well as the full-length RNA (Fig. 4, lanes 5–8). The amount of RNA isolated following the immunoprecipitation of complexes formed on the m7GpppG-capped hexamer mutant RNA with the anti-64k antibody was similar to that of the preimmune antibody control (Fig. 4, compare lanes 9 and 10 to 13 and 14). Although the amount of full-length RNA in these immunoprecipitates was high, the amount of 5′ and 3′ RNase H cleavage products were dramatically reduced relative to the wild-type pre-mRNA (Fig. 4, lanes 1 and 2). Immunoprecipitation of complexes formed on the m7GpppG-capped hexamer mutant RNA with the anti-CBP80 antibody resulted in the isolation of significant levels of both full-length and 5′ RNase H cleavage product (Fig. 4, lanes 11 and 12). Importantly, however, immunoprecipitation of the 3′ cleavage products with this antibody was reduced to a level comparable to that of the preimmune control immunoprecipitation (Fig. 4, compare lanes 11 and 13). Immunoprecipitation of complexes formed on the reverse-capped (Gm2′pppG7m) RNA, containing a wild-type poly(A) site, with the anti-64k antibody resulted in the isolation of an increased level of 3′ products relative to the level of 3′ products isolated by immunoprecipitation with the preimmune serum (Fig. 4, compare lanes 17 and 21). No 5′ products were detectable. The level of 3′ products recovered from the reverse-capped RNA following immunoprecipitation with the anti-64k antibody was reduced, however, relative to the m7GpppG-capped capped RNA (Fig. 4, compare lanes 1 and 17). This reduction in the recovery of 3′ products is consistent with the reduced stability of cleavage complexes formed in the absence of CBC (Fig. 3). The amount of full-length, 5′, and 3′ products isolated by immunoprecipitation by the anti-CBP80 antibody was approximately equivalent to that isolated by immunoprecipitation with the preimmune serum (Fig. 4, compare lanes 19, 20 and 21, 22).
Figure 4.
Physical interaction of the CBC/cap and 3′ processing complexes. m7GpppG-capped pre-mRNAs containing either a wild-type HIV-1 poly(A) site (lanes 1–8) or an HIV-1 poly(A) site containing a hexamer mutation (AAUAAA to CCACCC), and a Gm2′pppG7m-capped (reverse capped) RNA containing a wild-type HIV-1 poly(A) site (lanes 17–24) were incubated in HeLa cell nuclear extract in the presence (odd-numbered lanes) or absence (even-numbered lanes) of a DNA oligonucleotide complementary to a region between the cap and poly(A) site. After 10 min at 30°C, EDTA was added to block the endogenous extract RNase H activity. Immunoprecipitations were then conducted with the following antibodies: anti-64k CstF (lanes 1, 2, 9, 10, 17, and 18), anti-CBP80 (lanes 3, 4, 11, 12, 19, and 20), preimmune (P.I.) serum (lanes 5, 6, 13, 14, 21, and 22), and anti-TMG (lanes 7, 8, 15, 16, 23, and 24). The RNA was extracted from the immunoprecipitates and electrophoresed on a denaturing 10% polyacrylamide gel.
No evidence was obtained for an association of small nuclear ribonucleoproteins (snRNPs) with either the CBC/cap or the 3′ processing complexes (Fig. 4, lanes 7, 8, 15, 16, 23, and 24). Taken together, our results provide strong evidence of a physical interaction between the CBC/cap complex and 3′ processing factors bound at the poly(A) site.
DISCUSSION
Eukaryotic pre-mRNA 3′ processing requires a complex array of proteins that serve in the recognition of an extraordinarily limited set of conserved sequence elements (4, 5). An understanding of the mechanism of poly(A) site recognition is emerging in which multiple weak RNA:protein and protein:protein interactions contribute to the assembly of 3′ processing complexes at authentic poly(A) sites. The demonstration that CBC enhances poly(A) site cleavage adds yet another component to the network of interactions responsible for poly(A) site recognition. We propose that the participation of CBC in 3′ processing reflects a fundamental role of the CBC/cap complex in the targeting of nascent RNA polymerase II transcripts for processing.
Early work on mRNA 3′ processing demonstrated that the presence of an m7GpppG cap structure on the pre-mRNA enhanced poly(A) site cleavage efficiency in HeLa cell nuclear extract (17). Dinucleotide cap analog competition experiments indicated a requirement for a titratable cap-binding factor for efficient processing (16, 18). We have now demonstrated that CBC is required for the enhancement of 3′ processing by the 5′ cap. Immunodepletion of CBC from HeLa cell nuclear extract reduced the poly(A) site cleavage efficiency of a capped pre-mRNA to the level of an uncapped pre-mRNA. Addition of recombinant CBC to the depleted extract restored efficient processing. In every case examined, the impact of the structure of the 5′ end of the pre-mRNA on 3′ processing correlated directly with the binding of CBC (data not shown). The physical interaction of the CBC/cap complex with the processing complex assembled at the poly(A) site, as demonstrated by RNase H/coimmunoprecipitation experiments, supports a mechanism for 3′ processing in which the 5′ and 3′ ends of the pre-mRNA are juxtaposed by protein:protein interactions. An analysis of 3′ processing complexes indicated that CBC increased the stability of poly(A) site cleavage complexes formed in HeLa cell nuclear extract.
The impact of CBC on 3′ processing complex stability and its influence on endonucleolytic cleavage at the poly(A) site parallels the role of CBC in pre-mRNA splicing. In each case, CBC is not essential for processing, but rather it stabilizes the binding of processing factors to the pre-mRNA and enhances processing efficiency. Work in both mammalian (20) and yeast (19, 21) systems has demonstrated that CBC participates in an early step in spliceosome assembly. CBC influences the initial binding of U1 snRNP to the cap proximal 5′ splice site and serves to stabilize the E complex (HeLa cells) or commitment complex (Saccharomyces cerevisiae). In a similar manner, we have shown that CBC enhances the first step in 3′ processing, endonucleolytic cleavage. CBC enhanced the stability of the CPSF/CstF/RNA (B) complex formed in HeLa cell nuclear extracts. The efficiency of 3′ processing has previously been shown to correlate with the stability of processing complexes formed on the pre-mRNA in vitro (4, 5, 36–40). CBC depletion had little influence, however, on the efficiency of poly(A) addition. An analysis of poly(A) addition complexes indicated that CBC had no detectable impact on the stability of CPSF/RNA (A) complexes formed on a precleaved RNA. Since cleavage is required for poly(A) addition in vivo, an increase in cleavage efficiency would be expected to increase the overall efficiency of 3′ processing, and thus of mRNA production.
The splicing of an upstream intron has been shown to enhance 3′ processing both in vivo and in vitro (41–43). The cooperative interaction of the splicing and 3′ processing machineries is a prediction of the exon definition model of pre-mRNA processing (44). According to the exon definition model, an internal exon is defined through the interaction of RNA–protein complexes that reside at the splice junctions flanking the exon. The initial establishment of contacts across exons, which in vertebrates are relatively short (average size, 137 nucleotides), then facilitates the assembly of spliceosomes across much larger introns. The participation of CBC in the definition of the 5′ terminal exon is suggested by its role in the assembly of splicing complexes at the 5′ splice site of the cap proximal intron (19–21). In an analogous manner, cooperative interactions between complexes at the last 3′ splice site and the poly(A) site may serve in the definition of the 3′ terminal exon. The 3′ splice site has recently been shown to functionally replace the cap for both efficient 3′ processing (18) and splicing (20) in vitro. The mechanism by which the 3′ splice site enhances 3′ processing may therefore be similar to that of the CBC/cap complex. The roles of CBC and splicing in 3′ processing are unlikely to be completely redundant, however. The 3′ processing of a capped pre-mRNA has been shown to be stimulated up to 5-fold by the addition of an upstream intron in vitro (43). In addition, we observe an interaction of CBC with the 3′ processing complex in the absence of ATP—conditions that do not support spliceosome assembly. The cooperative interaction of both CBC and splicing factors with the 3′ processing complex may serve to meet the more stringent requirements imposed by communication across 3′ terminal exons, which are often the largest exon in a vertebrate gene (average length, ≈600 nucleotides). We propose that the CBC/cap complex participates, along with the assembly of splicing complexes on the pre-mRNA, in poly(A) site selection and definition of the 3′ terminal exon.
Both the functional and physical interaction of 5′ and 3′ RNA/protein complexes described here appear to require an, as yet, unidentified intermediate(s). Recombinant CBC, together with partially purified CPSF and CstF were not sufficient to support the coimmunoprecipitation of both RNA products following RNase H digestion (data not shown). This observation is consistent with the loss of cap-dependent enhancement of poly(A) site cleavage in reactions reconstituted with partially-purified processing factors (data not shown). The impact of CBC on the binding of U1 snRNP at the 5′ splice site, along with the enhancement of 3′ processing by an upstream intron, suggests a potential role for splicing factors in the interaction between CBC and the 3′ processing complex. The U1A protein, a component of the U1 snRNP, has been shown to modulate 3′ processing (45–48), and the assembly of splicing factors on the calcitonin 3′ processing enhancer (49) has been shown to participate in alternative poly(A) site selection. However, we have found no evidence for the participation of snRNPs in the interaction between the cap and poly(A) site. An anti-TMG antibody, capable of recognizing a set of small nuclear RNAs, was unable to immunoprecipitate 3′ processing complexes assembled on a capped pre-mRNA in HeLa cell nuclear extract. We also examined the potential involvement of SR proteins, which were shown to provide an alternative to CBC in splicing activation when added in excess (20). The addition of the recombinant SR protein SC35, however, had no impact on 3′ processing in CBC-depleted nuclear extract (data not shown). Interestingly, the carboxyl-terminal domain of the large subunit of RNA polymerase II has recently been implicated in 3′ processing. The demonstration that the carboxyl-terminal domain is not only associated with 3′ processing factors, but is required for 3′ processing in vivo (50), suggests that the CBC/cap complex and the 3′ processing complex might be juxtaposed through interactions with RNA polymerase II. Interactions between CBC and 3′ processing factors could supercede those involving the polymerase carboxyl-terminal domain once the nascent transcript has been capped and CBC has bound to it. Alternatively, interaction with CBC might only become possible once the 3′ processing factors have dissociated from the carboxyl-terminal domain and bound to the 3′ processing signals on the pre-mRNA. In either scenario the result would be to facilitate the formation and maintenance of a closed loop structure on the pre-mRNA.
The interaction of CBC with the 3′ processing complex may be important not only in poly(A) site selection, but in the correct pairing of mRNA termini for their coordinate recognition in subsequent transactions. Electron micrographs of Balbiani ring premessenger RNP particles in Chironomus tentans indicates that nuclear mRNAs can assemble into a ring structure in which the 5′ and 3′ ends of the transcript are juxtaposed (29). Subsequent docking of the mRNP at the nuclear pore complex involves a specific orientation of the RNA ends, with the 5′ end being translocated through the pore first, while the 3′ end appears to tether the RNP via the inner annulus. CBC remains associated with the 5′ cap until passage through the nuclear pore complex is completed (51). Within the cytoplasm, functional interaction of the 5′ and 3′ ends has also been observed in secondary cap methylation (31), translation initiation (30, 52), and mRNA turnover (33, 53). The coordinate recognition of both termini in each of these processes suggests that an mRNA may indeed function as a “closed-loop” (54). The participation of the mRNA 5′ end in 3′ processing may therefore represent only the first of a series of interactions between the mRNA termini that appear to play a role in nearly every aspect of the life of a eukaryotic message.
Acknowledgments
We thank Brent Graveley for critical reading of the manuscript and for generously providing purified recombinant SC35. This work was supported by grants from the National Institutes of Health (GM46624) to G.M.G. and the Lucille P. Markey Charitable Trust. P.F. and E.I. were supported by a postdoctoral fellowship and a project grant (to I.W.M.) from the Human Frontiers Science Programme Organisation, respectively.
ABBREVIATIONS
- CBC
cap binding complex
- CPSF
cleavage and polyadenylation specificity factor
- CstF
cleavage stimulatory factor
- rCBC
recombinant CBC
- TMG
trimethylguanosine
- RNP
ribonucleoprotein
- snRNP
small nuclear RNP
References
- 1.Rasmussen E B, Lis J T. Proc Natl Acad Sci USA. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salditt-Georgieff M M, Harpold M, Che-Kiang S, Darnell J. Cell. 1980;19:69–78. doi: 10.1016/0092-8674(80)90389-x. [DOI] [PubMed] [Google Scholar]
- 3.Sharp P A. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 4.Keller W. Cell. 1995;81:829–832. doi: 10.1016/0092-8674(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 5.Wahle E. Biochim Biophys Acta. 1995;1261:183–194. doi: 10.1016/0167-4781(94)00248-2. [DOI] [PubMed] [Google Scholar]
- 6.Bienroth S, Wahle E, Suter-Crazzolara C, Keller W. J Biol Chem. 1991;266:19768–19776. [PubMed] [Google Scholar]
- 7.Murthy K G K, Manley J L. J Biol Chem. 1992;267:14804–14811. [PubMed] [Google Scholar]
- 8.Takagaki Y, Manley J L, MacDonald C C, Wilusz J, Shenk T. Genes Dev. 1990;4:2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- 9.Gilmartin G M, Nevins J R. Genes Dev. 1989;3:2180–2189. doi: 10.1101/gad.3.12b.2180. [DOI] [PubMed] [Google Scholar]
- 10.Takagaki Y, Ryner L C, Manley J L. Genes Dev. 1989;3:1711–1724. doi: 10.1101/gad.3.11.1711. [DOI] [PubMed] [Google Scholar]
- 11.Raabe T, Bollum F J, Manley J L. Nature (London) 1991;353:229–234. doi: 10.1038/353229a0. [DOI] [PubMed] [Google Scholar]
- 12.Wahle E. J Biol Chem. 1991;266:3131–3139. [PubMed] [Google Scholar]
- 13.Bienroth S, Keller W, Wahle E. EMBO J. 1993;12:585–594. doi: 10.1002/j.1460-2075.1993.tb05690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheets M D, Wickens M. Genes Dev. 1989;3:1401–1412. doi: 10.1101/gad.3.9.1401. [DOI] [PubMed] [Google Scholar]
- 15.Wahle E. J Biol Chem. 1995;270:2800–2808. doi: 10.1074/jbc.270.6.2800. [DOI] [PubMed] [Google Scholar]
- 16.Hart R P, McDevitt M A, Nevins J R. Cell. 1985;43:677–683. doi: 10.1016/0092-8674(85)90240-5. [DOI] [PubMed] [Google Scholar]
- 17.Gilmartin G M, McDevitt M A, Nevins J R. Genes Dev. 1988;2:578–587. doi: 10.1101/gad.2.5.578. [DOI] [PubMed] [Google Scholar]
- 18.Cooke C, Alwine J C. Mol Cell Biol. 1996;16:2579–2584. doi: 10.1128/mcb.16.6.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colot H V, Stutz F, Rosbash M. Genes Dev. 1996;10:1699–1708. doi: 10.1101/gad.10.13.1699. [DOI] [PubMed] [Google Scholar]
- 20.Lewis J D, Izaurralde E, Jarmolowski A, Mcguigan C, Mattaj I W. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 21.Lewis J D, Gorlich D, Mattaj I W. Nuc Acids Res. 1996;24:3332–3336. doi: 10.1093/nar/24.17.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohno M, Sakamoto H, Shimura Y. Proc Natl Acad Sci USA. 1987;84:5187–5191. doi: 10.1073/pnas.84.15.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izaurralde E, Stepinski J, Darzynkiewicz E, Mattaj I W. J Cell Biol. 1992;118:1287–1295. doi: 10.1083/jcb.118.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izaurralde E, Lewis J, Mcguigan C, Jankowska M, Darzynkiewicz E, Mattaj I W. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 25.Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, Mcguigan C, Mattaj I W. Nature (London) 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 26.Kataoka N, Ohno M, Moda I, Shimura Y. Nucleic Acids Res. 1995;23:3638–3641. doi: 10.1093/nar/23.18.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohno M, Kataoka N, Shimura Y. Nucleic Acids Res. 1990;18:6989–6995. doi: 10.1093/nar/18.23.6989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Görlich D, Kraft R, Kostka S, Vogel F, Hartmann E, Laskey R A, Mattaj I W, Izaurralde E. Cell. 1996;87:21–32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- 29.Mehlin H, Daneholt B, Skoglund U. Cell. 1992;69:605–613. doi: 10.1016/0092-8674(92)90224-z. [DOI] [PubMed] [Google Scholar]
- 30.Gallie D R. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 31.Kuge H, Richter J D. EMBO J. 1995;14:6301–6310. doi: 10.1002/j.1460-2075.1995.tb00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarun S Z, Sachs A B. EMBO J. 1996;24:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 33.Beelman C A, Stevens A, Caponigro G, Lagrandeur T E, Hatfield L, Fortner D M, Parker R. Nature (London) 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- 34.Wahle E, Keller W. In: RNA Processing: A Practical Approach. Higgins S J, Hames B D, editors. II. Oxford: Oxford Univ. Press; 1994. pp. 1–33. [Google Scholar]
- 35.Gilmartin G M, Fleming E S, Oetjen J, Graveley B R. Genes Dev. 1995;9:72–83. doi: 10.1101/gad.9.1.72. [DOI] [PubMed] [Google Scholar]
- 36.Ahmed Y F, Gilmartin G M, Hanly S M, Nevins J R, Greene W C. Cell. 1991;64:727–737. doi: 10.1016/0092-8674(91)90502-p. [DOI] [PubMed] [Google Scholar]
- 37.Gilmartin G M, Fleming E S, Oetjen J. EMBO J. 1992;11:4419–4428. doi: 10.1002/j.1460-2075.1992.tb05542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss E A, Gilmartin G M, Nevins J R. EMBO J. 1991;10:215–219. doi: 10.1002/j.1460-2075.1991.tb07938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prescott J, Falck-Pedersen E. J Biol Chem. 1992;267:8175–8181. [PubMed] [Google Scholar]
- 40.Prescott J, Falck-Pedersen E. Mol Cell Biol. 1994;14:4682–4693. doi: 10.1128/mcb.14.7.4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nesic D, Cheng J, Maquat L E. Mol Cell Biol. 1993;13:3359–3369. doi: 10.1128/mcb.13.6.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nesic D, Maquat L E. Genes Dev. 1994;8:363–375. doi: 10.1101/gad.8.3.363. [DOI] [PubMed] [Google Scholar]
- 43.Niwa M, Rose S D, Berget S M. Genes Dev. 1990;4:1522–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 44.Berget S M. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 45.Gunderson S I, Vagner S, Polycarpou-Schwarz M, Mattaj I W. Genes Dev. 1997;11:761–773. doi: 10.1101/gad.11.6.761. [DOI] [PubMed] [Google Scholar]
- 46.Boelens W C, Jansen E J R, Venrooij W J V, Stripecke R, Mattaj I W, Gunderson S I. Cell. 1993;72:881–892. doi: 10.1016/0092-8674(93)90577-d. [DOI] [PubMed] [Google Scholar]
- 47.Gunderson, S. I., Beyer, K., Martin, G., Keller, W., Boelens, W. C. & Mattaj, I. W. (1994) Cell, 531–541. [DOI] [PubMed]
- 48.Lutz C S, Murthy K G K, Schek N, Oconnor J P, Manley T L, Alwine J C. Genes Dev. 1996;10:325–337. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- 49.Lou H, Gagel R F, Berget S M. Genes Dev. 1996;10:208–219. doi: 10.1101/gad.10.2.208. [DOI] [PubMed] [Google Scholar]
- 50.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G H, Greenblatt J, Patterson S D, Wickens M, Bentley D L. Nature (London) 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 51.Visa N, Izaurralde E, Ferreira J, Daneholt B, Mattaj I W. J Cell Biol. 1996;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tarun S Z, Sachs A B. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 53.Caponigro G, Parker R. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 54.Jacobson A. In: Translational Control. Hershey J W B, Matthews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 451–480. [Google Scholar]