Abstract
It has been suggested that the tethering caused by binding of the N-terminal region of smooth muscle caldesmon (CaD) to myosin and its C-terminal region to actin contributes to the inhibition of actin-filament movement over myosin heads in an in vitro motility assay. However, direct evidence for this assumption has been lacking. In this study, analysis of baculovirus-generated N-terminal and C-terminal deletion mutants of chicken-gizzard CaD revealed that the major myosin-binding site on the CaD molecule resides in a 30-amino acid stretch between residues 24 and 53, based on the very low level of binding of CaDΔ24–53 lacking the residues 24–53 to myosin compared with the level of binding of CaDΔ54–85 missing the adjacent residues 54–85 or of the full-length CaD. As expected, deletion of the region between residues 24 and 53 or between residues 54 and 85 had no effect on either actin-binding or inhibition of actomyosin ATPase activity. Deletion of residues 24–53 nearly abolished the ability of CaD to inhibit actin filament velocity in the in vitro motility experiments, whereas CaDΔ54–85 strongly inhibited actin filament velocity in a manner similar to that of full-length CaD. Moreover, CaD1–597, which lacks the major actin-binding site(s), did not inhibit actin-filament velocity despite the presence of the major myosin-binding site. These data provide direct evidence for the inhibition of actin filament velocity in the in vitro motility assay caused by the tethering of myosin to actin through binding of both the CaD N-terminal region to myosin and the C-terminal region to actin.
It is well known that actin-activated ATP hydrolysis by myosin and cross-bridge cycling in smooth muscle cells are primarily regulated by the reversible phosphorylation of the 20-kDa regulatory light chains of myosin (for review, see refs. 1–3). However, myosin phosphorylation–dephosphorylation alone cannot explain the force maintenance and relaxation processes of smooth muscle; full tension can be maintained with reduced levels of myosin phosphorylation (4–7), and relaxation can occur even when myosin is phosphorylated (6, 8). Thus, thin-filament-mediated regulation has been postulated to complement the major regulatory mechanism (myosin phosphorylation and dephosphorylation) of smooth muscle contraction (for review, see refs. 9–11).
Caldesmon (CaD), a protein that is associated with the thin filaments in smooth muscle cells, has been shown to play a role in the regulation of smooth muscle contraction. The following evidence supports the regulatory role of CaD: (i) CaD inhibits the actin-activated ATP hydrolysis by phosphorylated myosin and this inhibition is reversed by calmodulin in the presence of Ca2+ (12–17), (ii) CaD inhibits the movement of actin filaments over smooth muscle heads in an in vitro motility assay (18–20), and (iii) a synthetic peptide containing the calmodulin-binding sequence of CaD enhances the basic tension of skinned smooth muscle fibers (21). Functionally, smooth muscle CaD can be divided into three domains: the N-terminal domain, which binds to the S1/S2 junction of myosin (22, 23); the C-terminal domain, which binds to actin, tropomyosin, tubulin, calmodulin, and other Ca2+-binding proteins and inhibits actomyosin ATPase activity (10, 13, 15, 24–26); and the central helical domain, which separates the N-terminal domain from the C-terminal domain (27). Low-affinity binding sites for actin (28, 29) and tropomyosin (28, 30) are also present in the N-terminal region of CaD, but the precise binding sites have not been identified.
The functional properties of the CaD C-terminal domain have been well-studied by using CaD peptides obtained from limited proteolysis, chemical cleavage, and bacterial expression systems, and the major functional motifs for actin-binding, calmodulin-binding, and inhibition of actomyosin ATPase activity on the CaD molecule have been localized to its C-terminal end (24, 27, 28, 31–34). Use of a series of CaD truncation and internal deletion mutants generated from a baculovirus expression system has enabled analyses of the functional domains on the CaD molecule while retaining the structural integrity of most of the molecule. Those studies revealed the presence of three functional motifs for actin-binding/actomyosin ATPase inhibition in the C-terminal domain of CaD (35–37). The actin-binding capacity and inhibitory activity can be enhanced by smooth muscle tropomyosin or released by the Ca2+-binding protein calmodulin (refs. 13–17; for review, see refs. 10 and 11).
Although the major myosin-binding site had been localized to the N-terminal domain of CaD (23, 38, 39), its exact location is unknown. Exact definition of the myosin-binding site is important because of its implication in promoting the assembly and stability of smooth muscle myosin filaments in the presence of Mg2+-ATP (40) and its involvement in cross-linking myosin with actin, an event that has been postulated to play a role in force maintenance in smooth muscle cells (refs. 22, 23, 41, and 42; for review, see ref. 10). Force maintenance in smooth muscle (termed “latch”), which requires a low level of myosin light chain phosphorylation (7) and a low cross-bridge cycling rate (43), is not understood.
In this study, we focused on the precise localization of the N-terminal myosin-binding site on chicken-gizzard smooth muscle CaD and the effect of CaD binding to myosin on the myosin–actin interaction. By using baculovirus-expressed internal deletion mutants that contained the entire structure of the molecule except the targeted sites to be tested, we demonstrated that (i) the major myosin-binding site on the CaD molecule is located in a 30-amino acid stretch between residues 24–53 and (ii) both N-terminal myosin-binding and C-terminal actin-binding sites are required for the CaD-mediated inhibition of actin-filament velocity, but only the C-terminal region of CaD is required for inhibition of actomyosin ATPase activity. These data demonstrate that tethering of myosin to actin by binding N-terminal and C-terminal regions of CaD to the S2 portion of myosin and to actin, respectively, inhibits the unloaded shortening velocity of actin filaments.
MATERIALS AND METHODS
Construction of Recombinant Baculovirus Transfer Vectors.
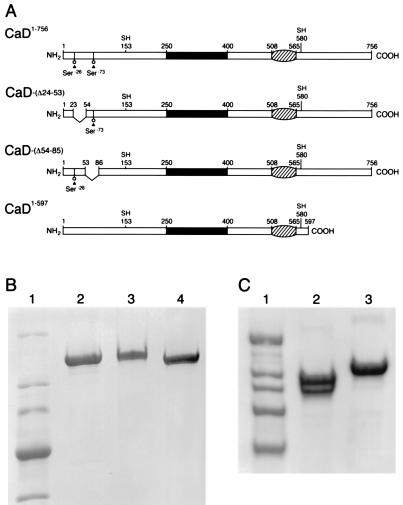
The cDNAs encoding the full-length CaD and CaD1–597 (Fig. 1) were constructed by the PCR cloning strategy as described (35). Recombinant pBluescript CaD plasmid containing the entire coding sequence of chicken-gizzard smooth muscle CaD (a gift from Joseph Bryan) was used as the template for PCR amplification. Both CaDΔ24–53 and CaDΔ54–85 cDNAs were generated by site-directed mutagenesis (36) using the following primers: CaDΔ24–53, 5′-GATACATCTCCTTCTTCCTCTGCTTCAAGGCGC-3′; and CaDΔ54–85, 5′-CCAACAATGCAGCTTCCTTTTGCCGCAGCCTTTCC-3′. The mutagenized CaD cDNAs were released by BamHI digestion and subcloned into the Pvl941 vector at a BamHI cloning site. The correct orientation and construction of each CaD mutant (Fig. 1) were confirmed by restriction mapping and DNA sequencing, respectively (35, 36).
Figure 1.
N-terminal and C-terminal deletion mutants of chicken-gizzard smooth muscle CaD. (A) Solid boxes indicate the central repetitive region consisting of a 13-amino acid sequence repeated eight times (27). V-shaped areas represent the region deleted in the CaD mutants. Numbers indicate the amino acid position. Hatched areas indicate regions of sequence homology between CaD and troponin T, as determined by amino acid sequence alignment (27). The two cysteine residues in the CaD molecule are indicated as SH. Ser-26 and Ser-73 represent the sites phosphorylated by either Ca2+/calmodulin-dependent kinase or casein kinase II (44, 45). (B) The SDS/PAGE profile of the purified full-length CaD and the CaD N-terminal deletion mutants. Lanes: 1, molecular mass standards (myosin, 200 kDa; β-galactosidase, 116.3 kDa; phosphorylase B, 97.4 kDa; BSA, 66.2 kDa; and ovalbumin, 45 kDa); 2–4, CaDΔ24–53, full-length CaD, and CaDΔ54–85, respectively. (C) The SDS/PAGE profile of CaD1–597. Lanes: 1, molecular mass standards as described above; 2 and 3, CaD1–597 and full-length CaD, respectively. Note that CaD1–597 shown in lane 2 migrated in SDS/PAGE as two bands. The lower band was caused by slight proteolytic degradation of CaD1–597. The cleaved site in CaD1–597 is either outside the myosin-binding site (residues 24–53) or in its C terminus, because the concentration of CaD1–597 required for myosin-binding saturation was similar to that of full-length CaD and increases in the concentration (up to 20 μM) of CaD1–597 did not increase the binding to myosin after binding of CaD1–597 reached saturation (data not shown).
Transfection and Isolation of Recombinant Baculovirus.
Spodoptera frugiperda (Sf9) insect cells were transfected as described (46). All recombinant viruses were isolated in plaque assays and amplified as described (46, 47).
Protein Purification.
Recombinant full-length CaD and CaD mutants were purified by using published procedures (46). The yields of CaDΔ24–53, CaDΔ54–85, and CaD1–597 ranged from 25 to 50 mg for 1 liter of cell culture (109 cells). Smooth muscle actin, CaD, myosin, and tropomyosin were prepared from chicken gizzard (48–50). Prior to purification, smooth muscle myosin was fully thiophosphorylated by using the endogenous kinase (50). Unphosphorylated myosin was prepared by incubating the myosin extract at 25°C for 10 min in 2 mM EGTA in the absence of ATP to allow dephosphorylation of the myosin light chains by the endogenous phosphatase (51). The extent of myosin light chain phosphorylation and dephosphorylation was determined from urea gel electrophoresis (49). The concentrations of full-length CaD and CaD mutants were determined by the Lowry method (52), and the concentrations of other proteins were measured spectrophotometrically by using the following extinction coefficients: actin, E2771% = 1.9; tropomyosin, E2771% = 1.9; and myosin, E2771% = 0.647.
Binding Assays for Smooth Muscle Myosin and Actin.
The full-length CaD and CaD mutants (Fig. 1A) were labeled with [14C]iodoacetamide (35) and used in both myosin-binding and acting-binding assays. Physical binding of full-length CaD and CaD mutants to phosphorylated myosin, unphosphorylated myosin, actin, and tropomyosin–actin from smooth muscle was determined by cosedimentation assays (35). The amounts of bound and unbound CaD mutants were estimated from the radioactivity in the pellets and supernatants, respectively, as described (35). All binding assays were done in triplicate. The apparent dissociation constants for myosin, actin, and actin–tropomyosin bindings were determined by Scatchard analysis (53) and by weighted nonlinear least-squares curve fitting as described (54).
ATPase Assay.
Actin-activated ATPase activity of smooth muscle myosin was determined as described (55). Specific assay conditions are given in the figures. ATPase assays for each experimental parameter were repeated three times with three different protein preparations. Data from a representative experiment are shown in each figure.
In Vitro Motility Assays.
Myosin with various degrees of light chain phosphorylation was obtained by mixing fully phosphorylated myosin with unphosphorylated myosin at the desired ratios. The concentrations of the myosin and CaD used in the in vitro motility assay were kept constant (smooth muscle myosin, 200 μg/ml; full-length CaD or each of CaD mutants, 0.2 μM). The concentrations of smooth muscle tropomyosin and rhodamine-phalloidine-labeled skeletal muscle actin were 0.025 μM and 0.1 μM, respectively. The in vitro motility assay was carried out with slight modification (18, 19) of the procedure published by Korn et al. (56).
RESULTS AND DISCUSSION
Effects of the CaD N-Terminal Mutants on Myosin Binding, Actin Binding, and Inhibition of Actomyosin ATPase Activity.
Two N-terminal internal deletion mutants of chicken-gizzard smooth muscle CaD, CaDΔ24–53 and CaDΔ54–85 (Fig. 1A), were constructed by using a site-directed mutagenesis method and overexpressed in a baculovirus expression system. Full-length CaD and CaD1–597 were prepared as described (35) and used as controls. As expected, CaDΔ24–53, CaDΔ54–85, and CaD1–597 migrated on the SDS/polyacrylamide gel faster than the full-length CaD (Fig. 1 B and C).
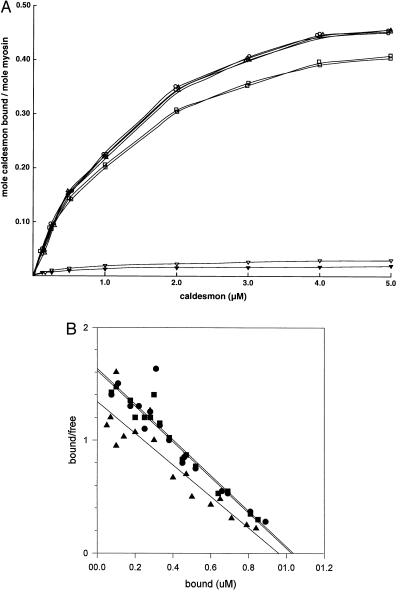
CaDΔ54–85 and CaD1–597 bound to phosphorylated myosin as strongly as did the full-length CaD (Fig. 2A), whereas CaDΔ24–53 bound to phosphorylated myosin less strongly. Saturation occurred at a stoichiometry of 0.45 mol of CaDΔ54–85 per mol of myosin and 0.4 mol of CaD1–597 per mol of myosin (Fig. 2A). The apparent Kd values of myosin-binding for CaDΔ54–85 and CaD1–597 were 0.7 ± 0.04 × 10−6 M and 0.84 ± 0.37 × 10−6 M, respectively (Fig. 2B). The apparent dissociation constant of full-length CaD was identical to that of CaDΔ54–85, whereas the very low-level binding of CaDΔ24–53 to myosin precluded any reliable determination of the apparent dissociation constant in this case. These studies strongly suggest that the major myosin-binding site on the CaD molecule is located in a 30-amino acid stretch between residues 24 and 53. One might argue that the inability of CaDΔ24–53 to bind to myosin is due to a conformational change caused by the disruption of the actual myosin-binding site upon deletion of the neighboring region (residues 24–53). Indeed, there is no direct evidence to rule out the possibility that the lack of myosin binding for CaDΔ24–53 is caused by a change in the conformation of the CaD molecule due to deletion of residues 24–53. However, if this is true, it is likely that the major myosin-binding site is located in the region upstream, but not downstream, of the sequence between residues 24 and 53 because deletion of the region between residues 54 and 85 (CaDΔ54–85) has no effect on myosin-binding. However, the region between residues 1 and 27 is probably not implicated in myosin binding (refs. 27 and 57; for review, see ref. 58). We also found that an N-terminal truncation mutant lacking the region between residues 1 and 23 bound to smooth muscle myosin as strongly as CaD1–756 did (data not shown). Therefore, it is unlikely that the region between residues 1 and 23 contains the major myosin-binding site. Thus, these data support our conclusion that the major myosin-binding site in the CaD molecule is located in the region between residues 24 and 53.
Figure 2.
Binding of CaD mutants to either phosphorylated or unphosphorylated smooth muscle myosin. (A) Fully phosphorylated (open symbols) and unphosphorylated (closed symbols) smooth muscle myosins at a constant concentration (2 μM) was mixed with various concentrations of 14C-labeled full-length CaD (○, •), CaDΔ24–53 (▿, ▾), CaDΔ54–85 (▵, ▴), and CaD1–597 (□, ▪) in 40 mM KCl, 5 mM MgCl2, 0.1 mM CaCl2, 2 mM DTT, 1 mM ATP, and 10 mM imidazole hydrochloride (pH 7.0) for 20 min at 27°C, and mixtures were cosedimented as described (35). Proteins in the supernatants and pellets were subjected to SDS/PAGE. After Coomassie blue staining, the protein bands corresponding to full-length CaD or each CaD mutant were excised and the radioactivity was measured in a liquid scintillation counter (35). Nonspecific binding was estimated by measuring radioactivity in gel slips excised from blank areas on the gels adjacent to the bands of full-length CaD or CaD mutants. (B) Scatchard analysis of full-length CaD (•), CaDΔ54–85 (▪), and CaD1–597 (▵) binding to phosphorylated smooth muscle myosin. The binding affinities of full-length and CaD mutants in theses experiments to unphosphorylated myosin were identical to those shown in this figure (data not shown). CaDΔ24–53 does not bind to myosin, so it is not shown.
The maximum binding of CaD to myosin in this study (Fig. 2), as well as in our previous study (46), was 0.5 mol of CaD per mol of myosin. The apparent Kd value of 0.7 ± 0.04 × 10−6 M obtained in this study is slightly higher than that reported by Hemric and Chalovich (41). The reported stoichiometry of CaD binding to myosin ranges from 1 to 3 mol of CaD per mol of myosin (for review, see ref. 10). The differences in the stoichiometry of CaD binding to myosin in different studies may be due to differences in assay conditions. Binding assays in this study were performed under the conditions of the ATPase assays, except for ATP concentration (2 mM for the ATPase assays vs. 1 mM for the binding assays).
The slight reduction in myosin binding to CaD1–597 (Fig. 2) suggests the existence of a weak myosin-binding motif between residues 1 and 23 or between residues 86 and 756 of chicken-gizzard smooth muscle CaD. A weak myosin-binding motif in the CaD C-terminal region has been implicated by the work of Matsumura and coworkers (59) who showed that a bacterially produced rat nonmuscle CaD peptide encompassing the region between residues 235 and 531 bound to myosin. Our finding that CaDΔ24–53 bound weakly to smooth muscle myosin (Fig. 2) is inconsistent with the observation of Huber et al. (60, 61) that a recombinant peptide of human nonmuscle CaD that contains the sequence-equivalent residues 476–737 of chicken-gizzard smooth muscle CaD bound to myosin with an affinity similar to that of intact CaD. Possibly, the deletion of a large region of CaD results in a significant conformational change that increases the affinity of the weak myosin-binding site located in the C-terminal region of CaD.
Because the tail region of smooth muscle myosin is bent toward the S1/S2 junction upon phosphorylation of the light chain (62, 63) and the CaD-binding site in the myosin molecule is also located in the S1/S2 junction (22), the different conformation of unphosphorylated myosin molecule might affect the CaD–myosin interaction. However, both full-length CaD and CaD mutants CaDΔ54–85 and CaD1–597 bound to phosphorylated myosin as tightly as to unphosphorylated myosin (Fig. 2). Our present study and those of others (64) suggest that the different conformation in unphosphorylated myosin has no effect on the CaD–myosin interaction.
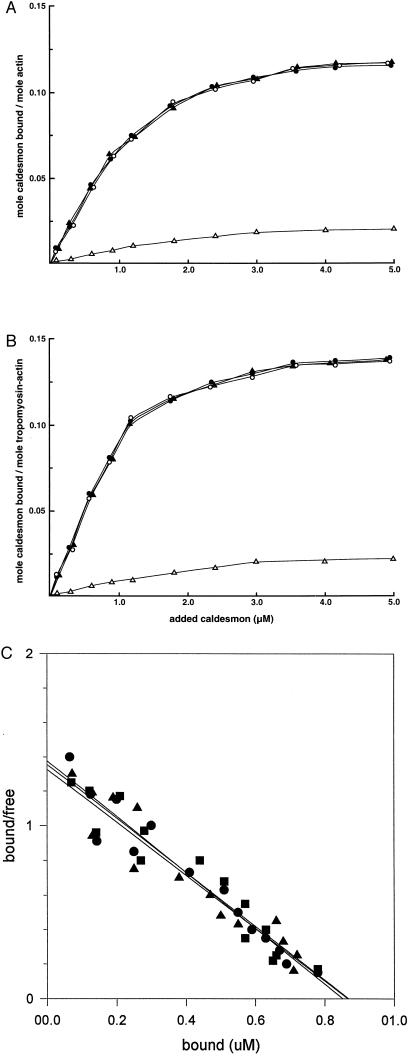
To determine whether the CaD sequence between residues 24 and 85 contributes to the binding of CaD to actin, we examined the binding of CaDΔ24–53 and CaDΔ54–85 to smooth muscle actin or tropomyosin–actin. As shown in Fig. 3A, both CaDΔ24–53 and CaDΔ54–85 bound to actin with a binding affinity indistinguishable from that of full-length CaD (Kd = 0.5 ± 0.03 × 10−6 M; Fig. 3C). By contrast, CaD1–597, lacking the region between residues 598 and 756, bound weakly to actin, consistent with our previous observation (35). The apparent Kd values for actin–tropomyosin binding of CaDΔ24–53 and CaDΔ54–85 were 0.51 ± 0.025 and 0.497 ± 0.02, respectively (Fig. 3C). The presence of smooth muscle tropomyosin enhanced the binding of full-length CaD and the CaD N-terminal mutants, CaDΔ24–53 and CaDΔ54–85, to actin and did not affect the actin binding of CaD1–597 (Fig. 3B).
Figure 3.
Binding of the CaD mutants to actin or tropomyosin-actin. Binding of full-length CaD (•), CaDΔ24–53 (○), CaDΔ54–85 (▴), and CaD1–597 (▵) to smooth muscle actin (A) or tropomyosin-actin (B) was measured as described (40). The actin concentration remained constant at 5 μM, and tropomyosin was mixed with actin at a 1:4 molar ratio. The binding assays were performed in 40 mM KCl, 5 mM MgCl2, 0.1 mM CaCl2, 2 mM DTT, 2 mM ATP, and 10 mM imidazole hydrochloride (pH 7.0). (C) Scatchard analysis of full-length CaD (•), CaDΔ24–53 (▪), and CaDΔ54–85 (▴) to smooth muscle tropomyosin–actin. The measured Kd values are shown in Results and Discussion. The binding of CaD1–597 was too weak to get meaningful data from Scatchard analysis.
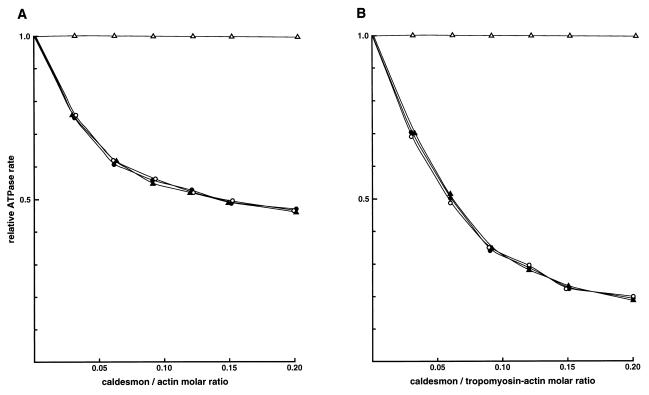
Deletion of residues 598–756 abolished the inhibition of actomyosin ATPase activity by CaD both in the absence and presence of smooth muscle tropomyosin (Fig. 4 A and B). However, CaDΔ24–53 and CaDΔ54–85, lacking 30 and 32 amino acid residues, respectively, inhibited the actin-activated myosin ATPase activity in a pattern identical to that of full-length CaD (Fig. 4A), and the presence of tropomyosin increased the inhibitory potential for full-length CaD, CaDΔ24–53, and CaDΔ54–85 (Fig. 4B).
Figure 4.
Inhibition of actin-activated myosin ATPase by the CaD mutants. Inhibition of actin- (A) or tropomyosin–actin- (B) activated myosin ATPase activities was assessed as described (55). Symbols are the same as in Fig. 3. The actin concentration remained constant at 25 μM, and tropomyosin was mixed with actin at a 1:4 molar ratio. The actin–tropomyosin-activated ATPase rate of smooth muscle myosin that is fully phosphorylated is 0.275 ± 0.021 μmol of Pi per mg per min. Conditions of the assays are as in Fig. 3.
Inhibition of Actin-Filament Velocity by Full-Length CaD and Its Mutants.
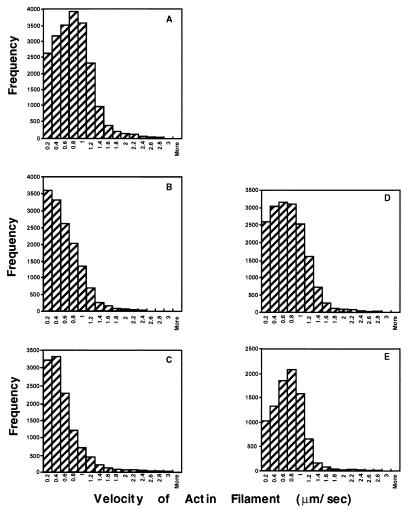
Several studies have reported that in the presence of tropomyosin, smooth muscle CaD inhibits the velocity of actin filaments sliding over smooth muscle or skeletal muscle myosin (18–20). We previously showed that smooth muscle CaD also inhibited actin filament sliding velocity over myosin with low levels of light chain phosphorylation (19). To determine whether the CaD N-terminal myosin-binding domain and C-terminal actin-binding domain are involved in CaD-mediated regulation of actin filament sliding velocity over myosin with different levels of light chain phosphorylation, we tested the effects of both CaD N-terminal and C-terminal deletion mutants on smooth muscle actin-filament velocity in the presence of smooth muscle tropomyosin in an in vitro motility assay. Maximal inhibition of actin-filament velocity occurs at 0.2 μM CaD (18); thus, full-length CaD or each CaD mutant was used at a constant concentration of 0.2 μM in this study. In the absence of CaD, most of the actin filaments moved at relatively high velocities (Fig. 5A), whereas the presence of full-length CaD shifted the velocity distribution to much lower velocities (Fig. 5B). Interestingly, CaDΔ24–53 did not inhibit actin-filament velocity (Fig. 5D), whereas the adjacent internal deletion mutant CaDΔ54–85 inhibited actin-filament velocity almost as strongly as did full-length CaD, as indicated by the similar velocity distributions (Fig. 5C, compare Fig. 5 C and B), consistent with the specificity of the sequence between residues 24 and 53 for myosin binding. The CaD C-terminal truncation mutant CaD1–597, which binds tightly to myosin, also did not inhibit actin filament velocity (Fig. 5E). The average velocity (in μm/sec) for actin filaments was 0.39 ± 0.01 for full-length CaD, 0.49 ± 0.03 for CaDΔ54–85, 0.83 ± 0.02 for CaDΔ24–53, and 0.8 ± 0.02 for CaD1–597. The average actin-filament velocity in the absence of CaD was 0.92 ± 0.01.
Figure 5.
Distribution of velocities of actin filament sliding over fully phosphorylated smooth muscle myosin in the presence of either full-length CaD or each CaD mutant. The velocity distribution of smooth muscle actin filament sliding either in the absence of CaD (A) or the presence of a constant concentration (0.2 μM) of full-length CaD or each CaD mutant (B, full-length; C, CaDΔ54–85; D, CaDΔ24–53; E, CaD1–597) was determined as described (18, 19). The assay conditions for the in vitro motility measurement were 20 mM KCl, 20 mM Mops (pH 7.2), 5 mM MgCl2, 0.1 mM EGTA, 1 mM ATP, 5 mM DTT, glucose at 2.5 mg/ml, glucose oxidase at 0.1 mg/ml, and catalase at 0.02 μg/ml. Actin filament velocity was measured at 30°C.
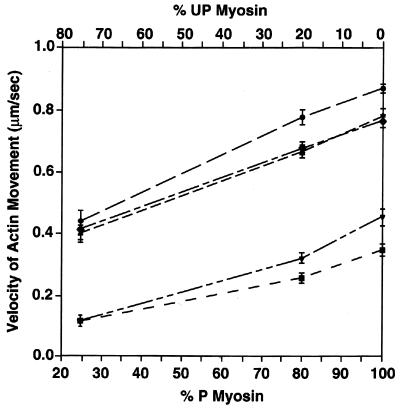
When either 80% or 25% phosphorylated smooth myosin was used, both full-length CaD and CaDΔ54–85 strongly inhibited actin filament velocity, whereas CaDΔ24–53 and CaD1–597 only slightly inhibited the actin filament velocity (Fig. 6). Note that when fully phosphorylated myosin or myosin with 80% phosphorylation was used, the inhibition of actin-filament velocity by CaDΔ54–85 was slightly lower than that by full-length CaD, and CaDΔ24–53 and CaD1–597 inhibited the actin-filament velocity at a very low level compared with that in the absence of CaD (Fig. 6).
Figure 6.
Effect of full-length CaD or each of CaD mutants on the velocity of actin filaments sliding over fully phosphorylated smooth muscle myosin and smooth muscle myosin at 80% or 25% phosphorylation. Phosphorylated and unphosphorylated myosins were mixed as described in Materials and Methods to obtain different levels of light chain phosphorylation while keeping the myosin concentration constant. The actin filament velocity was measured in the absence of CaD (•) or in the presence of full-length CaD (▪), CaDΔ24–53 (▴), CaDΔ54–85 (▾), and CaD1–597 (⧫), respectively.
One of the key properties of smooth muscle CaD is binding to myosin through its N-terminal myosin-binding domain (Fig. 2) and to actin through its C-terminal region (19, 38). The dual interaction of CaD has led to the hypothesis that CaD may play a role in the modulation of slow-cycling cross-bridges during force maintenance of smooth muscle cells, a phenomenon termed “latch” state by Murphy and colleagues (4, 5). This hypothesis is supported by recent observations that smooth muscle CaD inhibits actin-filament velocity in in vitro motility assays (18–20). In the present study, we found that full-length CaD significantly reduced the actin-filament velocity in the presence of smooth muscle tropomyosin (Figs. 5 and 6), consistent with previous reports (18, 19), and that deletion of either the myosin-binding domain between residues 24 and 53 or the CaD C-terminal actin-binding region between residues 598 and 756 nearly abolished the ability of CaD to inhibit actin-filament velocity (Figs. 5 and 6). In contrast, CaDΔ54–86, which has the same myosin-binding capacity as full-length CaD, strongly inhibited actin-filament velocity. Apparently, inhibition of actin-filament velocity by CaD requires both the C-terminal actin-binding site and the N-terminal myosin-binding site, through which the CaD tethers myosin to actin and thus inhibits the movement of actin filaments.
We also found that actin-filament velocity increases upon raising the level of light chain phosphorylation of smooth muscle myosin (Fig. 6). However, the extent of inhibition of actin-filament velocity by full-length CaD or CaDΔ54–85 did not depend on the phosphorylation level of smooth muscle myosin (Fig. 6), because both full-length CaD and CaDΔ54–85 were extensively inhibitory even at a very low level of phosphorylated myosin (Fig. 6). We previously showed that mixing phosphorylated myosin with myosin rods lacking heads but containing the S2 regions also decreases the movement of actin filaments (19). These data imply that the decrease in chemical energy due to a low ATP hydrolysis rate may not affect the ability of CaD to inhibit the movement of actin filaments. The observation that the regulation of actin-filament velocity by CaD is not dependent upon the phosphorylation level of myosin may, in part, explain the regulatory role for CaD in the in vivo force maintenance at low levels of myosin phosphorylation (4–7) and slow cross-bridge cycling (43). The direct evidence for a relationship between tethering and in vitro motility of actin filaments and the requirement for a very low level of phosphorylation to facilitate the decreased velocity strongly suggests a role for CaD during force maintenance, a condition characterized by low levels of myosin phosphorylation (7) and low ATPase turnover rate (43). Further studies using a skinned fiber system and the CaD N-terminal mutants or synthetic peptides containing the major myosin-binding or actin-binding motif should help to reveal the physiological functions of CaD.
Acknowledgments
We are grateful to R. Rasmus and the Tyson Company for providing fresh chicken gizzards, Marina Hoffman for editorial assistance, Dr. James R. Seller for critical reading of our manuscript, and Dr. Joseph Bryan for the chicken smooth muscle caldesmon cDNA that was used originally to construct the recombinant baculovirus vector. Photographic work was done by Jamie Hayden (Bio-Graphics). This work was supported by National Institutes of Health Grants DK 39740 and DK 47514.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviation: CaD, caldesmon.
References
- 1.Kamm K E, Stull J T. Annu Rev Pharmacol. 1985;25:593–620. doi: 10.1146/annurev.pa.25.040185.003113. [DOI] [PubMed] [Google Scholar]
- 2.Sellers J R, Adelstein R S. In: The Enzymes. Boyer P D, Krebs E G, editors. Orlando, FL: Academic; 1987. pp. 381–418. [Google Scholar]
- 3.Hartshorne D J. In: Physiology of the Gastrointestinal Tract. 2nd Ed. Johnson L R, editor. New York: Raven; 1987. pp. 423–482. [Google Scholar]
- 4.Dillon P F, Aksoy M O, Driska S P, Murphy R A. Science. 1981;211:495–497. doi: 10.1126/science.6893872. [DOI] [PubMed] [Google Scholar]
- 5.Aksoy M O, Mras S, Kamm K E, Murphy R A. Am J Physiol. 1983;245:C255–C270. doi: 10.1152/ajpcell.1983.245.3.C255. [DOI] [PubMed] [Google Scholar]
- 6.Tansey M G, Hori M, Karaki H, Kamm K E, Stull J T. FEBS Lett. 1990;270:219–221. doi: 10.1016/0014-5793(90)81272-p. [DOI] [PubMed] [Google Scholar]
- 7.Dillon P F, Murphy R A. Am J Physiol. 1982;242:C102–C108. doi: 10.1152/ajpcell.1982.242.1.C102. [DOI] [PubMed] [Google Scholar]
- 8.Fischer W, Pfitzer G. FEBS Lett. 1989;258:59–62. doi: 10.1016/0014-5793(89)81615-1. [DOI] [PubMed] [Google Scholar]
- 9.Trybus K M. Cell Motil Cytoskeleton. 1991;18:81–85. doi: 10.1002/cm.970180202. [DOI] [PubMed] [Google Scholar]
- 10.Marston S B, Redwood C S. Biochem J. 1991;279:1–16. doi: 10.1042/bj2790001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobue K, Sellers J R. J Biol Chem. 1991;266:12115–12118. [PubMed] [Google Scholar]
- 12.Sobue K, Muramoto Y, Fujita M, Kakiuchi S. Proc Natl Acad Sci USA. 1981;78:5652–5655. doi: 10.1073/pnas.78.9.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sobue K, Takahash K, Wakabayash I. Biochem Biophys Res Commun. 1985;132:645–651. doi: 10.1016/0006-291x(85)91181-7. [DOI] [PubMed] [Google Scholar]
- 14.Marston S B, Smith C W. J Muscle Res Cell Motil. 1985;6:669–708. doi: 10.1007/BF00712237. [DOI] [PubMed] [Google Scholar]
- 15.Horiuchi K Y, Miyata H, Chacko S. Biochem Biophys Res Commun. 1986;136:962–968. doi: 10.1016/0006-291x(86)90426-2. [DOI] [PubMed] [Google Scholar]
- 16.Lash J A, Sellers J R, Hathaway D R. J Biol Chem. 1986;261:16155–16160. [PubMed] [Google Scholar]
- 17.Velaz L, Hemric M E, Benson C E, Chalovich J M. J Biol Chem. 1989;264:9602–9610. [PubMed] [Google Scholar]
- 18.Shirinsky V P, Biryukov K G, Hettasch J M, Sellers J R. J Biol Chem. 1992;267:15886–15892. [PubMed] [Google Scholar]
- 19.Horiuchi K, Chacko S. J Muscle Res Cell Motil. 1995;16:11–19. doi: 10.1007/BF00125306. [DOI] [PubMed] [Google Scholar]
- 20.Fraser I D C, Marston S B. J Biol Chem. 1995;270:19688–19693. doi: 10.1074/jbc.270.34.19688. [DOI] [PubMed] [Google Scholar]
- 21.Katsuyama H, Wang C L A, Morgan K G. J Biol Chem. 1992;267:14555–14558. [PubMed] [Google Scholar]
- 22.Ikebe M, Reardon S. J Biol Chem. 1988;263:3055–3058. [PubMed] [Google Scholar]
- 23.Southerland C, Walsh M P. J Biol Chem. 1989;264:578–583. [PubMed] [Google Scholar]
- 24.Szpacenko A, Dabrowska R. FEBS Lett. 1986;202:182–186. doi: 10.1016/0014-5793(86)80683-4. [DOI] [PubMed] [Google Scholar]
- 25.Marston S B, Lehman W, Woody C J, Smith C W J. In: Advances in Protein Phosphatase II. Merlevede W, Lens H, Disalvo J, editors. Leuven, Belgium: Leuven Univ. Press; 1985. pp. 171–189. [Google Scholar]
- 26.Ishikawa R, Kagami O, Hayashi C, Kohama K. Cell Motil Cytoskeleton. 1992;23:244–251. doi: 10.1002/cm.970230404. [DOI] [PubMed] [Google Scholar]
- 27.Wang C L A, Wang L W C, Xu S, Lu R C, Saavedra-Alanis V, Bryan J. J Biol Chem. 1991;266:9166–9172. [PubMed] [Google Scholar]
- 28.Redwood C S, Marston S B. J Biol Chem. 1993;268:10969–10976. [PubMed] [Google Scholar]
- 29.Zhuang S, Wang C L A. Biophys J. 1997;72:188A. (abstr.). [Google Scholar]
- 30.Tsuruda T S, Watson M H, Foster D B, Lin J J-C, Mak A S. Biochem J. 1995;309:951–957. doi: 10.1042/bj3090951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dabrowska R, Gouch A, Galazkiewicz B, Osinska H. Biochim Biophys Acta. 1985;842:70–75. doi: 10.1016/0304-4165(85)90295-8. [DOI] [PubMed] [Google Scholar]
- 32.Horiuchi K Y, Samuel M, Chacko S. Biochemistry. 1991;30:712–717. doi: 10.1021/bi00217a019. [DOI] [PubMed] [Google Scholar]
- 33.Dobrowolski Z, Borovikov Y S, Nowak E, Galazkiewicz B, Dabrowska R. Biochem Biophys Acta. 1988;956:140–150. doi: 10.1016/0167-4838(88)90260-9. [DOI] [PubMed] [Google Scholar]
- 34.Riseman V M, Lynch W P, Nefsky B, Bretscher A. J Biol Chem. 1989;264:2869–2875. [PubMed] [Google Scholar]
- 35.Wang Z, Horiuchi K Y, Chacko S. J Biol Chem. 1996;271:2234–2242. doi: 10.1074/jbc.271.4.2234. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, Chacko S. J Biol Chem. 1996;271:25707–25714. doi: 10.1074/jbc.271.41.25707. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z, Yang Z Q, Chacko S. J Biol Chem. 1997;272:16896–16903. doi: 10.1074/jbc.272.27.16896. [DOI] [PubMed] [Google Scholar]
- 38.Velaz L, Ingraham R H, Chalovich J M. J Biol Chem. 1990;265:2929–2934. [PubMed] [Google Scholar]
- 39.Katayama E, Horiuchi K Y, Chacko S. Biochem Biophys Res Commun. 1989;160:1316–1322. doi: 10.1016/s0006-291x(89)80147-0. [DOI] [PubMed] [Google Scholar]
- 40.Katayama E, Scoot-Woo G, Ikebe M. J Biol Chem. 1995;270:3919–3925. doi: 10.1074/jbc.270.8.3919. [DOI] [PubMed] [Google Scholar]
- 41.Hemric M E, Chalovich J M. J Biol Chem. 1990;265:19672–19678. [PMC free article] [PubMed] [Google Scholar]
- 42.Adam L P, Haeberle J R, Hathaway D R. J Biol Chem. 1989;264:7698–7703. [PubMed] [Google Scholar]
- 43.Siegman M J, Butler T M, Mooers S U, Davies R E. J Gen Physiol. 1980;76:609–629. doi: 10.1085/jgp.76.5.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikebe M, Reardon S. J Biol Chem. 1990;265:17607–17612. [PubMed] [Google Scholar]
- 45.Bogatcheva N V, Vorotnikov A V, Birykov K G, Shirinsky V P. Biochem J. 1993;290:437–442. doi: 10.1042/bj2900437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Horiuchi K Y, Jacob S S, Gopalakurup S, Chacko S. J Muscle Res Cell Motil. 1994;15:646–658. doi: 10.1007/BF00121072. [DOI] [PubMed] [Google Scholar]
- 47.Wang Z, Myles G M, Brandt C S, Lioubin M N, Rohrschneider L. Mol Cell Biol. 1993;13:5348–5357. doi: 10.1128/mcb.13.9.5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horiuchi K Y, Chacko S. Biochemistry. 1989;28:9111–9116. doi: 10.1021/bi00449a023. [DOI] [PubMed] [Google Scholar]
- 49.Chacko S. Biochemistry. 1981;20:702–707. doi: 10.1021/bi00507a005. [DOI] [PubMed] [Google Scholar]
- 50.Heaslip R J, Chacko S. Biochemistry. 1985;24:2731–2736. doi: 10.1021/bi00332a020. [DOI] [PubMed] [Google Scholar]
- 51.Samuel M, Chowrashi P K, Pepe F A, Chacko S. Biochemistry. 1990;29:7124–7132. doi: 10.1021/bi00482a026. [DOI] [PubMed] [Google Scholar]
- 52.Lowry O H, Rosebrough N J, Farr A L, Randal R J. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 53.Scatchard G. Ann NY Acad Sci. 1949;51:660–672. [Google Scholar]
- 54.Munson P J, Rodbard D. Anal Biochem. 1980;107:220–239. doi: 10.1016/0003-2697(80)90515-1. [DOI] [PubMed] [Google Scholar]
- 55.Chacko S, Eisenberg E. J Biol Chem. 1990;265:2105–2110. [PubMed] [Google Scholar]
- 56.Kron S J, Spudich J A. Proc Natl Acad Sci USA. 1986;83:6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bryan J, Imai M, Lee R, Moore P, Cook R G, Lin W-G. J Biol Chem. 1989;264:13873–13879. [PubMed] [Google Scholar]
- 58.Bryan J. J Muscle Res Cell Motil. 1989;10:95–96. doi: 10.1007/BF01739964. [DOI] [PubMed] [Google Scholar]
- 59.Yamakita Y, Yamashiro S, Matsumura F. J Biol Chem. 1992;267:12022–12029. [PubMed] [Google Scholar]
- 60.Huber P A J, Redwood C S, Avent N D, Tanner M J A, Marston S B. J Muscle Res Cell Motil. 1993;14:385–391. doi: 10.1007/BF00121289. [DOI] [PubMed] [Google Scholar]
- 61.Huber P A, Fraser I D, Marston S B. Biochem J. 1995;312:617–625. doi: 10.1042/bj3120617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trybus K M, Huiatt T W, Lowey S. Proc Natl Acad Sci USA. 1982;79:6151–6155. doi: 10.1073/pnas.79.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Onishi H, wakabayashi T. J Biochem (Tokyo) 1982;92:871–879. doi: 10.1093/oxfordjournals.jbchem.a134001. [DOI] [PubMed] [Google Scholar]
- 64.Hemric M E, Lu F W M, Shrager R, Carey J, Chalovich J M. J Biol Chem. 1993;268:15305–15311. [PMC free article] [PubMed] [Google Scholar]