Abstract
In cross-sectional or cohort studies with binary outcomes, it is biologically interpretable and of interest to estimate the relative risk or prevalence ratio, especially when the response rates are not rare. Several methods have been used to estimate the relative risk, among which the log-binomial models yield the maximum likelihood estimate (MLE) of the parameters. Because of restrictions on the parameter space, the log-binomial models often run into convergence problems. Some remedies, e.g., the Poisson and Cox regressions, have been proposed. However, these methods may give out-of-bound predicted response probabilities. In this paper, a new computation method using the SAS Nonlinear Programming (NLP) procedure is proposed to find the MLEs. The proposed NLP method was compared to the COPY method, a modified method to fit the log-binomial model. Issues in the implementation are discussed. For illustration, both methods were applied to data on the prevalence of microalbuminuria (micro-protein leakage into urine) for kidney disease patients from the Diabetes Control and Complications Trial. The sample SAS macro for calculating relative risk is provided in the appendix.
Keywords: Prevalence ratio, PROC NLP, relative risk, risk difference
1 Introduction
Recently, there has been much discussion and interest in the literature concerning the appropriateness of estimating relative risk (RR) versus odds ratio (OR) in cross-sectional and cohort studies, for example, Schouten et al. [1], Axelson et al. [2], McNutt et al. [3, 4], Skov et al. [5] and Zhang and Yu [6], among others. In case-control studies, the OR may be preferable because of the different sampling fractions in the case and control groups. However, the RR is often more interpretable than the OR [2], especially in cohort studies. When the outcome is rare, the RR can be approximated by the OR, but the approximation is questionable if the outcome is common.
Relative risk can be calculated from a binomial model with a log link function [7], referred to as the log-binomial model (LBM). The LBM was implemented in SAS PROC GENMOD [5]. Alternatively, Lee [8] and Lee and Chia [9] used the Cox proportional hazards model to estimate the RR in cross-sectional studies. Zou [10] proposed an information sandwich estimator to obtain a robust variance estimate from the Poisson regression; Carter et al. [11] proposed a quasi-likelihood estimator from a Poisson regression.
In the ideal situation when the calculations converge, all methods should produce consistent parameter estimates. However, none of the available methods based on LBMs solve the convergence problem completely. In addition, estimated probabilities from the Poisson/Cox regressions could be out-of-bound. In this article we propose a new computation method using SAS PROC NLP. The new method nearly always converges, and it guarantees that the predicted probabilities are between 0 and 1.
The rest of the paper is organized as follows: In the next section, we review the available estimation methods and describe the NLP method. We then evaluate and compare the performance of the proposed method and the COPY method [12] in Section 3 and discuss several important issues of implementation in Section 4. As an example, both methods are applied to data on the prevalence of microalbuminuria for kidney disease patients from the Diabetes Control and Complications Trial. The conclusion includes a discussion of tips for implementing the methods.
2 The estimation methods
Let Y = 0 (1) denote the absence (presence) of the event. For a subject with covariates X = (x1,…, xk), the response probability in a log-binomial model is defined as
| (1) |
where β = (β1, …, βk) is the vector of parameters. The loglikelihood function is given by
| (2) |
The RR of xj increasing by one unit is calculated as P (Y = 1|xj + 1)/P (Y = 1|xj), j = 1, …, k. The popular methods of estimating the RR include the LBM [7] and its modification called the COPY method [12], the Cox/Poisson regression with robust variance estimates [10, 11] and the method of adjusting OR [6].
Since P(Y = 1|X) ∈ (0, 1), the estimate β̂ should satisfy the constraint β̂ X < 0 for all possible X values in the data. Wacholder [7] constrains the likelihood to prevent the response probability from approaching 0. It requires an extra step that determines if exp(β̂Xi) ≤ P*, where P* is a predetermined maximum possible value such as 0.99. When the estimates are on or near the boundaries of the valid parameter space, the estimation algorithm will not converge. The convergence problem is most likely to happen when the model contains a continuous or polychotomous covariate, or the response prevalence is high [13, 14]. The simulation studies by Carter et al. [11] show that the estimates have poor properties when the success probability approaches 1. Deddens and Petersen [12] provide a remedy called the COPY method. The COPY method consists of expanding the original data set to include a large number of copies of the original data set together with one copy of the original data set with cases and controls reversed. Estimates from the COPY method are a good approximation of the MLEs. Deddens et al. [15] suggest using the COPY method with 1000 copies of the original data set, as been used in the present paper.
Lee [8] and Lee and Chia [9] recommend to use the Cox proportional hazards model with the hazard function
where h0(t) is the baseline hazard. When the follow-up time is equal for all individuals, the hazard ratio estimated by Cox regression equals the RR in cross-sectional studies [8, 16]. Correspondingly the time t is set to a constant in Lee’s method. The Poisson regression is used when the outcome is the number of events m over the time at risk t and the model formulation is
In both the Cox and Poisson regressions, the underlying distribution of the response is Poisson. As the actual response follows a binomial distribution, the variance of the coefficient β̂k tends to be overestimated [13]. Barros and Hirakata [13] and Zou [10] propose a robust variance estimator in the Poisson/Cox regression to adjust for over dispersion.
Zhang and Yu [6] propose a method to convert the OR estimated by logistic regression to the RR:
where P0 is the event rate in the unexposed group. However, this method produces a biased estimate when confounding is present [4] and it is not recommended [15].
Barros and Hirakata [13] compare the Cox/Poisson regression with robust variance estimator and the log-binomial model to the Mantel-Haenszel estimator of RR. They find that the Cox/Poisson regression with robust variance estimates and the log-binomial regression performed equally well. The Poisson/Cox regression could produce an estimated prevalence greater than 1, however. Based on several simulation studies by Skov et al. [5] and McNutt et al. [4], the LBM is preferred since the parameter estimates are asymptotically unbiased, and the prevalence estimates are between 0 and 1. Recently, Blizzard and Hosmer [19] compare the LBM, the logistic regression method by Schouten et al. [1] and the Poisson regression approach. They find that the failure rates (non-convergence, out-of-bounds predicted probabilities) are rather high for all three methods. In a summary paper, Lumley et al. [17] review a number of estimation algorithms, compared the relative merits of different estimators, and give some guidelines for implementation in popular software. They urge that RR regression commands be implemented in standard software.
Given that LBMs have a high non-convergence rate and the Poisson regressions could give predicted probabilities greater than 1, it is desirable to find a computation method that produces estimates with small biases and predicted probabilities between 0 and 1, and more importantly, that have a high or even 100% convergence rate. Here, we propose using the SAS Nonlinear Programming (NLP) procedure [18]. The NLP procedure offers a set of optimization techniques for minimizing or maximizing a continuous nonlinear function with parameters β = (β1, …, βk) with lower and upper bounds, linear and nonlinear equations, and equality and inequality constraints [18]. For calculating RR, this amounts to maximizing the loglikelihood function ℓ(β) in Equation (2) for βi ∈ ℛ,i = 1, …, k, subject to linear inequality constraints
where Xi = (xi1, …, xik) is the vector of covariates for the i-th subject. PROC NLP provides a variety of optimization techniques, e.g., the Newton-Raphson method with line search or ridging, the Quasi-Newton methods, etc., and the estimates explicitly satisfy the constraints. Hence, the NLP procedure is appropriate for estimating the RR. The NLP method can also be extended to other scales, for example, risk difference. To estimate the RR, the response probabilities should satisfy that 0 < P(Y = 1|X) = βX′ < 1.
3 Simulation
Blizzard and Hosmer [19] used simulations to compare the relative bias, mean square error (MSE) of the estimates and the coverage rates of the 95% confidence intervals (CIs) from the LBM, Schouten’s expanded logistic regression and the Poisson regression. We used the same simulation settings and compared the results from the COPY method and those from the proposed NLP method. The parameters used in the simulation are shown in Table 1.
Table 1.
The parameters used in the simulations
| Simulation I: | ||||
|---|---|---|---|---|
| Case | β0 | β1 | a | |
| 1 | −2.30259 | 0.38376 | 6.0 | |
| 2 | −2.30259 | 0.38376 | 4.0 | |
| 3 | −1.20397 | 0.56687 | 2.0 | |
| 4 | −1.20397 | 0.56687 | 1.0 | |
| 5 | −0.69315 | 0.65200 | 1.0 | |
| 6 | −0.69315 | 0.65200 | 0.0 | |
| 7 | −0.35667 | 0.70808 | 0.5 | |
| 8 | −0.35667 | 0.70808 | −0.5 | |
|
| ||||
| Simulation II: | ||||
|
| ||||
| Case | β0 | βD | βU | p |
|
| ||||
| 9 | log(0.3) | log(1.5) | 0.18 | 0.2 |
| 10 | log(0.3) | log(1.5) | 0.18 | 0.5 |
| 11 | log(0.3) | log(2.0) | 0.10 | 0.2 |
| 12 | log(0.3) | log(2.0) | 0.10 | 0.5 |
|
| ||||
| Simulation III: | ||||
|
| ||||
| Case | β0 | βD | βU | p |
|
| ||||
| 13 | log(0.15) | log(1.5) | 0.36 | 0.2 |
| 14 | log(0.15) | log(1.5) | 0.36 | 0.5 |
| 15 | log(0.15) | log(2.0) | 0.20 | 0.2 |
| 16 | log(0.15) | log(2.0) | 0.20 | 0.5 |
In Simulation I, the response probability P(Y = 1|x) = exp(β0 +β1x) and x ~ U(−6, a), a uniform random variable with upper bound a. Simulation II consisted of both dichotomous and continuous covariates and P(Y = 1|xU, xD) = exp(β0 + βDxD + βU xU ). The dichotomous variable xD ~ B(p) and the continuous variable xU were generated from xD by a uniform distribution xU |xD ~ U(−6 + 2xD, 2 + 2xD). In Simulation II, the coefficients for the continuous variable, βU, took moderate values 0.18 and 0.10. To test the convergence of the estimation methods in a slightly more extreme situation, we doubled the coefficients in Simulation III so that there was a larger difference between the minimum and maximum response probabilities. We also generated 1000 data sets with sample size 500. The measures for comparison were the average percent relative , 100 times the average , where θ̂r was the estimate for the r-th data replication and θ was the true parameter value.
Table 2 shows the convergence rate and summary measures, i.e, bias and MSE, of the parameter estimates for Simulation I with sample size 500. Columns 2–4 are convergence rates from the three methods, i.e., PROC GENMOD using LBM (GM), the COPY1000 method (COPY) and PROC NLP (NLP). Convergence rates for PROC GENMOD were much lower, below 50% for Case 1 and 7. Both the COPY and the NLP method converged 100%. The bias and MSE of the parameters were calculated using the converged estimates. The MSEs from both methods were very close. The relative biases from the NLP method were smaller for Case 1, 3, 5, 7, and the COPY method was better for the other cases. Note that we used the average percent relative bias. Since the true parameter values were rather small, the absolute biases from both methods were very small, also. When PROC GENMOD failed to converge, both methods had a negative bias because the estimates were bounded on one side. The COPY method appeared to be more affected by this than the PROC NLP method.
Table 2.
Summary of convergence rates and parameter estimates for Simulation I
| Case | Convergence rate (%) | COPY | NLP | ||||
|---|---|---|---|---|---|---|---|
| GM | COPY | NLP | Bias | MSE | Bias | MSE | |
| 1 | 48 | 100 | 100 | −1.57 | 0.17 | −0.99 | 0.17 |
| 2 | 90 | 100 | 100 | −0.06 | 0.60 | 1.19 | 0.67 |
| 3 | 79 | 100 | 100 | −1.56 | 0.48 | −0.88 | 0.50 |
| 4 | 88 | 100 | 100 | −0.71 | 1.09 | 0.41 | 1.21 |
| 5 | 69 | 100 | 100 | −1.24 | 0.58 | −0.60 | 0.59 |
| 6 | 89 | 100 | 100 | 0.02 | 1.56 | 1.20 | 1.75 |
| 7 | 49 | 100 | 100 | −1.53 | 0.58 | −0.95 | 0.59 |
| 8 | 90 | 100 | 100 | 0.03 | 1.89 | 1.23 | 2.11 |
As the simulation scenarios were not exactly the same, e.g., random number generator and seed, we could not replicate the simulation by Blizzard and Hosmer [19] exactly. It is useful to compare the above Table 2 with their simulation results, however. Blizzard and Hosmer [19] showed that, if converged, the LBM (PROC GENMOD) yielded a smallert bias and MSE for the parameter estimates than either the Poisson regression or the expanded data logistic model [1]. Among all five methods, i.e., two in Table 2 and three from Blizzard and Hosmer [19], the estimates from the COPY method and the NLP method had very small biases and they both had MSE very similar to the LBM.
Table 3 shows the convergence rate and parameter estimates for simulations II and III. Again we see that both the COPY and NLP methods converged 100%, and the bias and the MSEs from both methods were comparable.
Table 3.
Summary of convergence rates and parameter estimates for Simulations II and III.
|
βD |
βU |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Convergence rate (%) | COPY | NLP | COPY | NLP | ||||||
| GM | COPY | NLP | Bias | MSE | Bias | MSE | Bias | MSE | Bias | MSE | |
| 9 | 100 | 100 | 100 | −37.18 | 7.79 | −37.03 | 7.79 | −18.43 | 0.31 | −18.12 | 0.30 |
| 10 | 100 | 100 | 100 | −2.36 | 4.16 | −2.03 | 4.17 | −13.37 | 0.19 | −13.11 | 0.19 |
| 11 | 100 | 100 | 100 | −27.71 | 8.19 | −27.50 | 8.15 | −42.67 | 0.33 | −42.49 | 0.33 |
| 12 | 100 | 100 | 100 | 7.81 | 3.45 | 8.11 | 3.49 | −15.45 | 0.08 | −15.31 | 0.08 |
|
| |||||||||||
| 13 | 73 | 100 | 100 | 8.32 | 13.00 | 8.58 | 13.17 | −19.33 | 1.07 | −18.61 | 1.05 |
| 14 | 99 | 100 | 100 | 15.27 | 10.39 | 16.02 | 10.56 | −13.82 | 0.63 | −13.31 | 0.61 |
| 15 | 99 | 100 | 100 | −3.36 | 10.65 | −2.87 | 10.74 | −6.15 | 0.44 | −5.47 | 0.44 |
| 16 | 100 | 100 | 100 | −3.77 | 8.86 | −3.20 | 8.93 | −9.68 | 0.31 | −9.25 | 0.30 |
Based on the simulations, we found that the estimates from both the COPY and NLP methods were very close to the true values. This confirmed that both methods performed well. The NLP method explicitly imposes the constraints of [0, 1] to the estimated prevalences and can be used as an alternative to the COPY method.
4 Implementation
There are several important issues related to the implementation of the COPY and the NLP methods. Blizzard and Hosmer [19] reported a convergence problem with the COPY method. This problem appeared to be due to poor starting values. Deddens et al. [15] suggested using the start value option intercept = −4 in the COPY method. In Simulation I, the convergence rate of the COPY method was reduced to 90% if the option was omitted. This indicates the importance of appropriate starting values.
The original macro by Deddens et al. [15] made physical copies of the data, rather than using weights. We found results from both approaches to have been the same. Using weights will save memory and computation time when the number of copies is very large. We suggest using weights instead of making physical copies of the data. Deddens et al. [15] suggested a two-step process. Step 1 was to use the results of PROC GENMOD if it converged on the original data set, and Step 2 was to use the results of PROC GENMOD on the modified data set if PROC GENMOD did not converge on the original data set. We did the second step in both cases. When PROC GENMOD converges, the PROC GENMOD and PROC NLP results are essentially the same.
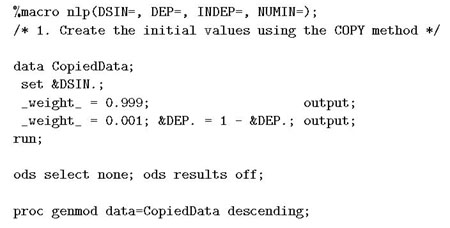
In the implementation of the NLP method, we also used a two-step approach (see appendix): first we used the COPY method to generate initial values for PROC NLP; then used PROC NLP to find the MLE based on the starting value. The SAS macro provided in the appendix can be used can be used as a working engine for RR estimation.
5 Example
Since the discovery of insulin in 1921, the medical community debated the hypothesis that the marked elevation of blood glucose (hyperglycemia) associated with diabetes mellitus was responsible for the development and progression of the microvascular complications of type 1 or insulin-dependent diabetes: retinopathy leading to blindness, nephropathy leading to end-stage kidney disease, and neuropathy leading to loss of sensation, ulceration and amputation [21]. The earliest sign of kidney disease is the leakage of small amounts of protein (albumin) into urine, which can be measured by the albumin excretion rate (AER) expressed as mg/24h of albumin excreted into the urine. For healthy people, the AER should be less than 40mg/24h and some would say no greater than 20 or 30mg/24h. The earliest sign of possible diabetic nephropathy is microalbuminuria (MA), defined as an AER > 40 mg/24h (but < 300mg/24h).
The Diabetes Control and Complications Trial (DCCT) was launched by the National Institute of Diabetes, Digestive and Kidney Diseases in 1981 in order to definitively answer whether a program of intensive therapy aimed at near normal levels of glycemia, as compared to conventional therapy aimed at maintenance of clinical well being, would affect onset and progression of microvascular complications (The Diabetes Control and Complications Research Group, 1995). The DCCT involved 1441 patients enrolled in 29 clinical centers in the US and Canada, and followed for an average of 6.5 years (4–9) years. Of these, 726 patients comprising the primary prevention cohort were free of any microvascular complications (AER≤40 mg/24h and no retinopathy, among other features) and 715 patients comprising the secondary intervention cohort had minimal pre-existing levels of albuminuria (AER < 200 mg/24h) and mild retinopathy. Patients were randomly assigned to receive either intensive or conventional treatment. Intensive treatment employed all available means (self-monitoring four or more times a day with three or more multiple daily injections or a pump, in conjunction with diet and exercise) to obtain levels of blood glucose as close as possible to the normal range while attempting to avoid hypoglycemia (blood glucose below a physiologically safe level). Conventional treatment, on the other hand, consisted of one or two daily injections of insulin and less frequent self-monitoring with the goal of maintaining clinical well-being, but without any specific glucose targets.
Although intensive therapy was associated with excess weight gain and greater risk of hypoglycemia, risks of microvascular complications over the average of 6.5 years of follow-up were significantly reduced with intensive versus conventional therapy [21]. As the earliest stage of nephropathy, MA is highly predictive of progression to overt albuminuria, which is associated with glomerular destruction. Hence, it is also of interest to assess the effect of intensive therapy on the prevalence of MA. Lachin [21] presented the analysis of factors related to the prevalence of MA in those people evaluated at the sixth year, the mean follow-up time. He considered a subset of 172 patients in the secondary intervention cohort with 15 ≤ AER≤ 40 mg/24h. In addition to the intensive versus conventional treatment group (intensive = 0 or 1), the analysis adjusted for the percent of HbA1c at baseline (HbA1c), prior duration of diabetes in years (years), systolic blood pressure in mmHg (sbp), and gender (female) (1 if female and 0 if male).
HbA1c is a measure of the average level of blood glucose control in the 4–6 weeks prior to the trial. The underlying hypothesis is that HbA1c level and duration of diabetes together determine the risk of further disease progression. Progression of nephropathy may also be associated with increased levels of blood pressure leading to kidney damage. These effects may also differ for men and women. Thus the objective was to obtain an adjusted assessment of the effect of intensive versus conventional treatment and also to explore the association between these baseline factors and the risk of MA [21].
Our analysis was based on the subset of 172 patients, and the outcome was the presence of MA, defined as an AER > 40 mg/24h. Table 4 shows percentages for the binary variables and ranges for continuous variables. The prevalence of MA is moderate (24.4%). However, the values for HbA1c, diabetes duration and sbp had wide ranges. For a subject with sbp = 148, it was likely that the linear predictor βX′ was close to the boundary 0 and the predicted probability was so close to 1 that it causes non-convergence. When the LBM was fitted to data using PROC GENMOD, then calculation did not converge. The estimates of OR and RR of MA for the treatment and major baseline risk factors are presented in Table 4. The OR estimates are calculated from SAS PROC LOGISTIC and the RR estimates are calculated from the COPY and NLP methods. We see that the RR estimates from both methods are almost identical to two decimal places. This confirms that the RR estimates are consistent and reliable.
Table 4.
Estimates of odds ratio and relative risk of MA
| Odds ratio | Relative risk (NLP) | Relative risk (COPY) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variable | Percent/Range | OR | 95% CI | p | RR | 95% CI | p | RR | 95% CI | p |
| intensive | 52% | 0.21 | (0.09, 0.47) | <.001 | 0.35 | (0.19, 0.64) | <.001 | 0.35 | (0.19, 0.64) | <.001 |
| female | 45% | 0.41 | (0.17, 0.99) | 0.046 | 0.48 | (0.26, 0.89) | 0.021 | 0.48 | (0.26, 0.89) | 0.020 |
| HbA1c | (6.7, 14.4) | 1.76 | (1.33, 2.34) | <.001 | 1.31 | (1.15, 1.48) | <.001 | 1.30 | (1.15, 1.48) | <.001 |
| years | (1.3, 15.0) | 1.01 | (0.89, 1.14) | 0.880 | 0.99 | (0.92, 1.05) | 0.681 | 0.99 | (0.92, 1.05) | 0.681 |
| sbp | (90, 148) | 1.02 | (0.98, 1.07) | 0.262 | 1.01 | (0.98, 1.03) | 0.677 | 1.01 | (0.98, 1.03) | 0.676 |
As an example, the estimates for intensive therapy from both methods were RR = 0.350. If we were to use OR (0.205) as the measure, we would report a 1.00-0.205 = 79.5% reduction of odds. Since RR = 0.350, the relative reduction of risk rate would be 1.00-0.35 = 65%, which is more relevant for health practitioners. The difference between the OR and RR for HbA1c is more pronounced. As we see from Table 4, the difference between RR and OR may change the interpretation and impression of the treatment effect. For epidemiologic studies where the outcome is common, we would expect that such difference to be more noticeable.
6 Discussion
The OR always overestimates the magnitude of association as compared to the RR when their logs are both greater than 0, i.e., 0 ≤ log(RR) ≤ log(OR). The magnitude of the difference between the estimates of OR and RR could be noticeable when the prevalence of outcome is common. However, in practice, OR are seemingly always interpreted as RR, regardless of the prevalence of the outcome.
In this paper, we propose a new method of calculating the RR. This method uses the SAS NLP procedure and is widely available. As PROC NLP explicitly imposes the constraints, the corresponding estimates are guaranteed to be within the bounds. Different methods should produce similar estimates if they converge. In practice, we suggest examining the consistency of the estimates from different methods to ensure that the results are correct.
The NLP procedure can also be used to estimate the risk difference, where the log link function is replaced by the identity link and the constraints are 0 < βXi < 1, i = 1, …, n. It is also useful to extend the estimation technique to multivariate correlated binary data. Once the log-binomial model reaches convergence, the goodness of fit of the model can be assessed using the method described by Blizzard and Hosmer [19].
Acknowledgments
The research of the first author was supported in part by the Intramural Research Program of the National Institute on Aging. The authors wish to thank two anonymous reviewers for their helpful suggestions and Ms. Caroline Phillips for proof reading the paper.
Appendix. Sample code


Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schouten EG, Dekker JM, Kok FJ, le Cessie S, van Houl-welingen HC, Pool J. Risk ratio and rate ratio estimation in case cohort designs: hypertension and cardiovascular mortality. Statistics in Medicine. 1993;12:17331745. doi: 10.1002/sim.4780121808. [DOI] [PubMed] [Google Scholar]
- 2.Axelson O, Fredriksson M, Ekberg K. Use of the prevalence ratio versus the prevalence odds ratio as a measure of risk in cross sectional studies (Correspondence) Occupational Environmental Medicine. 1994;51:574. doi: 10.1136/oem.51.8.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McNutt L, Hafner J, Xue X. Correcting the odds ratio in cohort studies of common outcomes (Letter) Journal of the American Medical Association. 1999;282:529. doi: 10.1001/jama.282.6.529. [DOI] [PubMed] [Google Scholar]
- 4.McNutt L, Wu C, Xue X, Hafner JP. Estimating the relative risk in cohort studies and clinical trials of common outcomes. American Journal of Epidemiology. 2003;157:940–943. doi: 10.1093/aje/kwg074. [DOI] [PubMed] [Google Scholar]
- 5.Skov T, Deddens J, Petersen M, Endahl L. Prevalence proportion ratios: estimation and hypothesis testing. International Journal of Epidemiology. 1998;27:91–95. doi: 10.1093/ije/27.1.91. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, Yu K. What’s relative risk? A method of correcting the odds ratio in cohort studies of common outcomes. Journal of the American Medical Accociation. 1998;280:1690–1691. doi: 10.1001/jama.280.19.1690. [DOI] [PubMed] [Google Scholar]
- 7.Wacholder S. Binomial regression in GLIM: estimating risk ratios and risk differences. American Journal of Epidemiology. 1986;123:174–184. doi: 10.1093/oxfordjournals.aje.a114212. [DOI] [PubMed] [Google Scholar]
- 8.Lee J. Odds ratio or relative risk for cross-sectional data? International Journal of Epidemiology. 1994;23:201–203. doi: 10.1093/ije/23.1.201. [DOI] [PubMed] [Google Scholar]
- 9.Lee J, Chia KS. Prevalence odds ratio vs. prevalence ratio - a response. Occupational Environmental Medicine. 1995;52:781–782. doi: 10.1136/oem.52.11.781-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou G. A modified Poisson regression approach to prospective studies with binary data. American Journal of Epidemiology. 2004;159:702–706. doi: 10.1093/aje/kwh090. [DOI] [PubMed] [Google Scholar]
- 11.Carter RE, Lipstiz SR, Tilley BC. Quasi-likelihood estimation for relative risk regression models. Biostatistics. 2005;6:39–44. doi: 10.1093/biostatistics/kxh016. [DOI] [PubMed] [Google Scholar]
- 12.Deddens JA, Petersen MR, Lei X. Estimation of prevalence ratios when PROC GENMOD does not converge. Proceeding of the 28th Annual SAS Users Group International Conference; Cary NC, SAS Institute Inc. 2003. pp. 270–278. http://www2.sas.com/proceedings/sugi28270-28.pdf. [Google Scholar]
- 13.Barros AJD, Hirakata VN. Alternatives for logistic regression in cross-sectional studies: an emprirical comaprison of models that directly estimate the prevalence ratio. BMC Medical Research Methodology. 2003;3:21. doi: 10.1186/1471-2288-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J. Estimation of prevalence rate ratios from cross sectional data: a reply. International Journal of Epidemiology. 1995;24:1066–1067. doi: 10.1093/ije/24.5.1064. [DOI] [PubMed] [Google Scholar]
- 15.Deddens JA, Petersen MR. Re: “Estimating the relative risk in cohort studies and clinical trials of common outcomes” (Letter to the editor) American Journal of Epidemiology. 2004;159:213–214. doi: 10.1093/aje/kwh022. [DOI] [PubMed] [Google Scholar]
- 16.Breslow N. Covariance analysis of censored survival data. Biometrics. 1974;30:89–99. [PubMed] [Google Scholar]
- 17.Lumley T, Kronmal R, Ma S. Relative risk regression in medical research: models, contrasts, estimators, and algorithms. University of Washington Biostatistics Working Paper Series. 2006 Working Paper 293. [Google Scholar]
- 18.SAS Institute Inc. SAS/OR® 9.1.3 Users Guide: Mathematical Programming 3.2. Cary, NC: SAS Institute Inc; 2007. pp. 1–4. [Google Scholar]
- 19.Blizzard L, Hosmer DW. Parameter estimation and goodness-of-fit in log binomial regression. Biometrical Journal. 2006;48:5–22. doi: 10.1002/bimj.200410165. [DOI] [PubMed] [Google Scholar]
- 20.The Diabetes Control and Complications Research Group. Effect of intensive therapy on the development and progression of diabetic nephropathy in the Diabetes Control and Complications Trial. Kidney International. 1995;47:1703–1720. doi: 10.1038/ki.1995.236. [DOI] [PubMed] [Google Scholar]
- 21.Lachin JL. Biostatistical Methods: The Assessment of Relative Risks. John Wiley and Sons; New York: 2000. [Google Scholar]


