Abstract
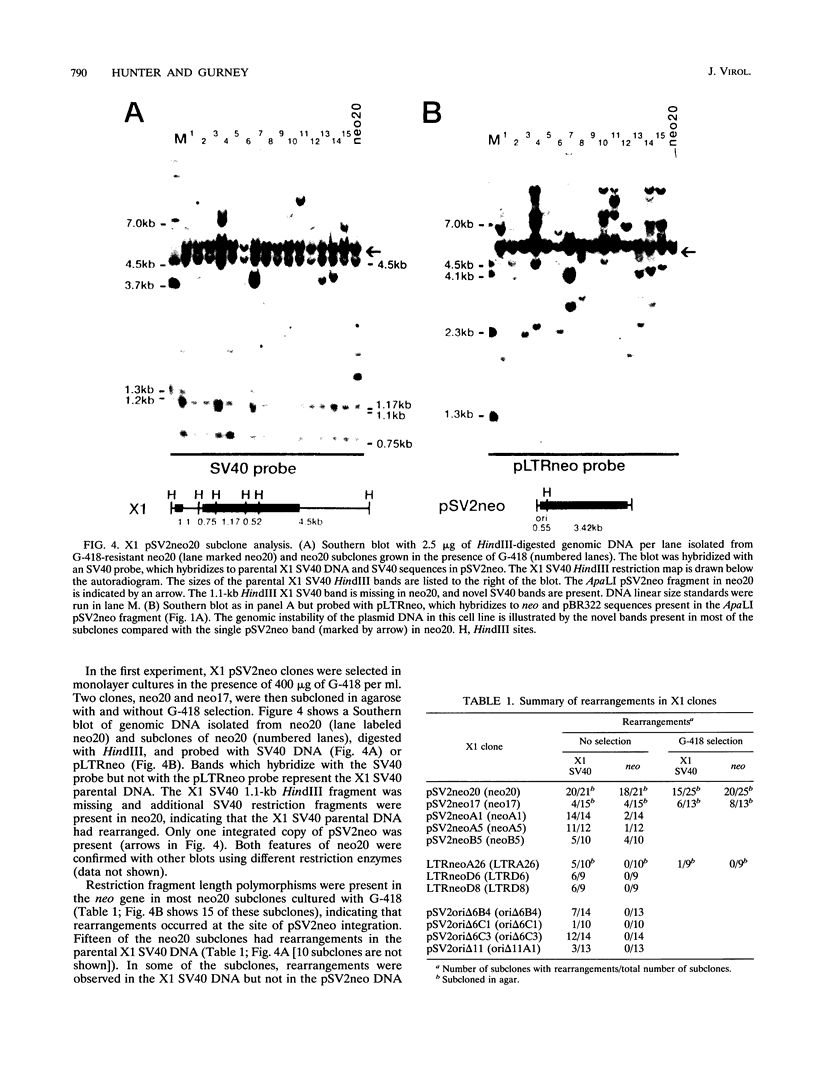
DNA rearrangements in the form of deletions and duplications are found within and near integrated simian virus 40 (SV40) DNA in nonpermissive cell lines. We have found that rearrangements also occur frequently with integrated pSV2neo plasmid DNA. pSV2neo contains the entire SV40 control region, including the origin of replication, both promoters, and the enhancer sequences. Linearized plasmid DNA was electroporated into X1, an SV40-transformed mouse cell line that expresses SV40 large T antigen (T Ag) and shows very frequent rearrangements at the SV40 locus, and into LMtk-, a spontaneously transformed mouse cell line that contains no SV40 DNA. Stability was analyzed by subcloning G-418-resistant clones and examining specific DNA fragments for alterations in size. Five independent X1 clones containing pSV2neo DNA were unstable at both the neo locus and the T Ag locus. By contrast, four X1 clones containing mutants of pSV2neo with small deletions in the SV40 core origin and three X1 clones containing a different neo plasmid lacking SV40 sequences were stable at the neo locus, although they were still unstable at the T Ag locus. Surprisingly, five independent LMtk- clones containing pSV2neo DNA were unstable at the neo locus. LMtk- clones containing origin deletion mutants were more stable but were not as stable as the X1 clones containing the same plasmid DNA. We conclude that the SV40 origin of replication and early control region are sufficient viral components for the genomic instability at sites of SV40 integration and that SV40 T Ag is not required.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baran N., Lapidot A., Manor H. Formation of DNA triplexes accounts for arrests of DNA synthesis at d(TC)n and d(GA)n tracts. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):507–511. doi: 10.1073/pnas.88.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender M. A., Brockman W. W. Rearrangement of integrated viral DNA sequences in mouse cells transformed by simian virus 40. J Virol. 1981 Jun;38(3):872–879. doi: 10.1128/jvi.38.3.872-879.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsma D. J., Olive D. M., Hartzell S. W., Subramanian K. N. Territorial limits and functional anatomy of the simian virus 40 replication origin. Proc Natl Acad Sci U S A. 1982 Jan;79(2):381–385. doi: 10.1073/pnas.79.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan M., Topp W., Sambrook J. Studies on simian virus 40 excision from cellular chromosomes. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):709–719. doi: 10.1101/sqb.1979.043.01.079. [DOI] [PubMed] [Google Scholar]
- Breitman M. L., Tsui L. C., Buchwald M., Siminovitch L. Introduction and recovery of a selectable bacterial gene from the genome of mammalian cells. Mol Cell Biol. 1982 Aug;2(8):966–976. doi: 10.1128/mcb.2.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock P., Miller J., Botchan M. Effects of poly[d(pGpT).d(pApC)] and poly[d(pCpG).d(pCpG)] repeats on homologous recombination in somatic cells. Mol Cell Biol. 1986 Nov;6(11):3948–3953. doi: 10.1128/mcb.6.11.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butner K. A., Lo C. W. High frequency DNA rearrangements associated with mouse centromeric satellite DNA. J Mol Biol. 1986 Feb 20;187(4):547–556. doi: 10.1016/0022-2836(86)90333-5. [DOI] [PubMed] [Google Scholar]
- Caddle M. S., Lussier R. H., Heintz N. H. Intramolecular DNA triplexes, bent DNA and DNA unwinding elements in the initiation region of an amplified dihydrofolate reductase replicon. J Mol Biol. 1990 Jan 5;211(1):19–33. doi: 10.1016/0022-2836(90)90008-A. [DOI] [PubMed] [Google Scholar]
- Carroll S. M., Gaudray P., De Rose M. L., Emery J. F., Meinkoth J. L., Nakkim E., Subler M., Von Hoff D. D., Wahl G. M. Characterization of an episome produced in hamster cells that amplify a transfected CAD gene at high frequency: functional evidence for a mammalian replication origin. Mol Cell Biol. 1987 May;7(5):1740–1750. doi: 10.1128/mcb.7.5.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Grass D. S., Blanck G., Hoganson N., Manley J. L., Pollack R. E. A functional simian virus 40 origin of replication is required for the generation of a super T antigen with a molecular weight of 100,000 in transformed mouse cells. J Virol. 1983 Nov;48(2):492–502. doi: 10.1128/jvi.48.2.492-502.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daya-Grosjean L., Monier R. Presence of free viral DNA in simian virus 40-transformed nonproducer cells. J Virol. 1978 Aug;27(2):307–312. doi: 10.1128/jvi.27.2.307-312.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornreiter I., Erdile L. F., Gilbert I. U., von Winkler D., Kelly T. J., Fanning E. Interaction of DNA polymerase alpha-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992 Feb;11(2):769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornreiter I., Höss A., Arthur A. K., Fanning E. SV40 T antigen binds directly to the large subunit of purified DNA polymerase alpha. EMBO J. 1990 Oct;9(10):3329–3336. doi: 10.1002/j.1460-2075.1990.tb07533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F., Ragsdale C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988 Jul 11;16(13):6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folger K. R., Wong E. A., Wahl G., Capecchi M. R. Patterns of integration of DNA microinjected into cultured mammalian cells: evidence for homologous recombination between injected plasmid DNA molecules. Mol Cell Biol. 1982 Nov;2(11):1372–1387. doi: 10.1128/mcb.2.11.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard R. D., Montelone B. A., Walter C. F., Innis J. W., Scott W. A. Role of specific simian virus 40 sequences in the nuclease-sensitive structure in viral chromatin. Mol Cell Biol. 1985 Jan;5(1):52–58. doi: 10.1128/mcb.5.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluzman Y., Frisque R. J., Sambrook J. Origin-defective mutants of SV40. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):293–300. doi: 10.1101/sqb.1980.044.01.033. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Gurney T., Jr, Gurney E. G. Spontaneous rearrangement of integrated simian virus 40 DNA in nine transformed rodent cell lines. J Virol. 1989 Jan;63(1):165–174. doi: 10.1128/jvi.63.1.165-174.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Lane D., Lipsich L., Wigler M., Botchan M. Characteristics of an SV40-plasmid recombinant and its movement into and out of the genome of a murine cell. Cell. 1980 Aug;21(1):127–139. doi: 10.1016/0092-8674(80)90120-8. [DOI] [PubMed] [Google Scholar]
- Heartlein M. W., Knoll J. H., Latt S. A. Chromosome instability associated with human alphoid DNA transfected into the Chinese hamster genome. Mol Cell Biol. 1988 Sep;8(9):3611–3618. doi: 10.1128/mcb.8.9.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J. B., Murphy D., Defendi V. Instability of integrated viral DNA in mouse cells transformed by simian virus 40. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1736–1740. doi: 10.1073/pnas.78.3.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J., Murphy D., Defendi V. Amplification and rearrangement of integrated SV40 DNA sequences accompany the selection of anchorage-independent transformed mouse cells. Cell. 1980 Nov;22(2 Pt 2):535–543. doi: 10.1016/0092-8674(80)90363-3. [DOI] [PubMed] [Google Scholar]
- Hudziak R. M., Laski F. A., RajBhandary U. L., Sharp P. A., Capecchi M. R. Establishment of mammalian cell lines containing multiple nonsense mutations and functional suppressor tRNA genes. Cell. 1982 Nov;31(1):137–146. doi: 10.1016/0092-8674(82)90413-5. [DOI] [PubMed] [Google Scholar]
- Hyrien O., Debatisse M., Buttin G., de Saint Vincent B. R. A hotspot for novel amplification joints in a mosaic of Alu-like repeats and palindromic A + T-rich DNA. EMBO J. 1987 Aug;6(8):2401–2408. doi: 10.1002/j.1460-2075.1987.tb02518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis J. W., Scott W. A. DNA replication and chromatin structure of simian virus 40 insertion mutants. Mol Cell Biol. 1984 Aug;4(8):1499–1507. doi: 10.1128/mcb.4.8.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongstra J., Reudelhuber T. L., Oudet P., Benoist C., Chae C. B., Jeltsch J. M., Mathis D. J., Chambon P. Induction of altered chromatin structures by simian virus 40 enhancer and promoter elements. Nature. 1984 Feb 23;307(5953):708–714. doi: 10.1038/307708a0. [DOI] [PubMed] [Google Scholar]
- KIT S., DUBBS D. R., PIEKARSKI L. J., HSU T. C. DELETION OF THYMIDINE KINASE ACTIVITY FROM L CELLS RESISTANT TO BROMODEOXYURIDINE. Exp Cell Res. 1963 Aug;31:297–312. doi: 10.1016/0014-4827(63)90007-7. [DOI] [PubMed] [Google Scholar]
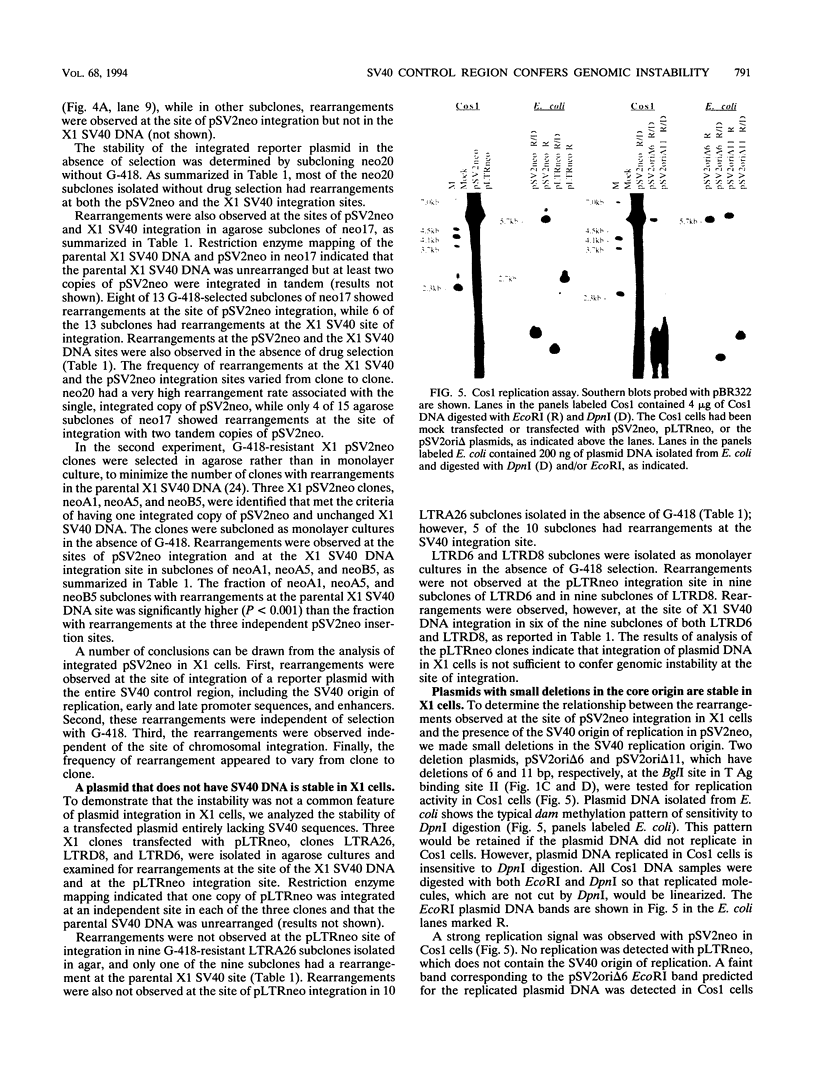
- Kaufman R. J., Sharp P. A. Amplification and expression of sequences cotransfected with a modular dihydrofolate reductase complementary dna gene. J Mol Biol. 1982 Aug 25;159(4):601–621. doi: 10.1016/0022-2836(82)90103-6. [DOI] [PubMed] [Google Scholar]
- Leach D. R., Stahl F. W. Viability of lambda phages carrying a perfect palindrome in the absence of recombination nucleases. 1983 Sep 29-Oct 5Nature. 305(5933):448–451. doi: 10.1038/305448a0. [DOI] [PubMed] [Google Scholar]
- Leu T. H., Hamlin J. L. Activation of a mammalian origin of replication by chromosomal rearrangement. Mol Cell Biol. 1992 Jun;12(6):2804–2812. doi: 10.1128/mcb.12.6.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo C. W. Transformation by iontophoretic microinjection of DNA: multiple integrations without tandem insertions. Mol Cell Biol. 1983 Oct;3(10):1803–1814. doi: 10.1128/mcb.3.10.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur J. G., Beitel L. K., Chamberlain J. W., Stanners C. P. Elements which stimulate gene amplification in mammalian cells: role of recombinogenic sequences/structures and transcriptional activation. Nucleic Acids Res. 1991 May 11;19(9):2477–2484. doi: 10.1093/nar/19.9.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur J. G., Stanners C. P. A genetic element that increases the frequency of gene amplification. J Biol Chem. 1991 Mar 25;266(9):6000–6005. [PubMed] [Google Scholar]
- Mink M., Basak A. N., Küntzel H. Restoration of the yeast LEU2 gene by transcriptionally controlled recombination between tandem repeats. Mol Gen Genet. 1990 Aug;223(1):107–113. doi: 10.1007/BF00315802. [DOI] [PubMed] [Google Scholar]
- Mougneau E., Birg F., Rassoulzadegan M., Cuzin F. Integration sites and sequence arrangement of SV40 DNA in a homogeneous series of transformed rat fibroblast lines. Cell. 1980 Dec;22(3):917–927. doi: 10.1016/0092-8674(80)90569-3. [DOI] [PubMed] [Google Scholar]
- Mounts P., Kelly T. J., Jr Rearrangements of host and viral DNA in mouse cells transformed by simian virus 40. J Mol Biol. 1984 Aug 15;177(3):431–460. doi: 10.1016/0022-2836(84)90294-8. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Wobbe C. R., Weissbach L., Dean F. B., Hurwitz J. Role of DNA polymerase alpha and DNA primase in simian virus 40 DNA replication in vitro. Proc Natl Acad Sci U S A. 1986 May;83(9):2869–2873. doi: 10.1073/pnas.83.9.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnane J. P. Inducible gene expression by DNA rearrangements in human cells. Mol Cell Biol. 1986 Feb;6(2):549–558. doi: 10.1128/mcb.6.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnane J. P. Influence of cellular sequences on instability of plasmid integration sites in human cells. Somat Cell Mol Genet. 1990 May;16(3):195–209. doi: 10.1007/BF01233356. [DOI] [PubMed] [Google Scholar]
- Murnane J. P., Yezzi M. J. Association of high rate of recombination with amplification of dominant selectable gene in human cells. Somat Cell Mol Genet. 1988 May;14(3):273–286. doi: 10.1007/BF01534588. [DOI] [PubMed] [Google Scholar]
- Murnane J. P., Yezzi M. J., Young B. R. Recombination events during integration of transfected DNA into normal human cells. Nucleic Acids Res. 1990 May 11;18(9):2733–2738. doi: 10.1093/nar/18.9.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnane J. P., Young B. R. Nucleotide sequence analysis of novel junctions near an unstable integrated plasmid in human cells. Gene. 1989 Dec 7;84(1):201–205. doi: 10.1016/0378-1119(89)90157-1. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Rich A. Negatively supercoiled simian virus 40 DNA contains Z-DNA segments within transcriptional enhancer sequences. Nature. 1983 Jun 23;303(5919):674–679. doi: 10.1038/303674a0. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Passananti C., Davies B., Ford M., Fried M. Structure of an inverted duplication formed as a first step in a gene amplification event: implications for a model of gene amplification. EMBO J. 1987 Jun;6(6):1697–1703. doi: 10.1002/j.1460-2075.1987.tb02420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden K. W., Pipas J. M., Pearson-White S., Nathans D. Isolation of mutants of an animal virus in bacteria. Science. 1980 Sep 19;209(4463):1392–1396. doi: 10.1126/science.6251547. [DOI] [PubMed] [Google Scholar]
- Perry M. E., Commane M., Stark G. R. Simian virus 40 large tumor antigen alone or two cooperating oncogenes convert REF52 cells to a state permissive for gene amplification. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8112–8116. doi: 10.1073/pnas.89.17.8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli A. S., Smith G. R. Chromosomal context dependence of a eukaryotic recombinational hot spot. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):227–231. doi: 10.1073/pnas.89.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray F. A., Peabody D. S., Cooper J. L., Cram L. S., Kraemer P. M. SV40 T antigen alone drives karyotype instability that precedes neoplastic transformation of human diploid fibroblasts. J Cell Biochem. 1990 Jan;42(1):13–31. doi: 10.1002/jcb.240420103. [DOI] [PubMed] [Google Scholar]
- Sager R., Anisowicz A., Howell N. Genomic rearrangements in a mouse cell line containing integrated SV40 DNA. Cell. 1981 Jan;23(1):41–50. doi: 10.1016/0092-8674(81)90268-3. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Botchan M., Gallimore P., Ozanne B., Pettersson U., Williams J., Sharp P. A. Viral DNA sequences in cells transformed by simian virus 40, adenovirus type 2 and adenovirus type 5. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):615–632. doi: 10.1101/sqb.1974.039.01.075. [DOI] [PubMed] [Google Scholar]
- Sanes J. R., Rubenstein J. L., Nicolas J. F. Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 1986 Dec 1;5(12):3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimke R. T., Sherwood S. W., Hill A. B., Johnston R. N. Overreplication and recombination of DNA in higher eukaryotes: potential consequences and biological implications. Proc Natl Acad Sci U S A. 1986 Apr;83(7):2157–2161. doi: 10.1073/pnas.83.7.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurman R., Keulen W. Modified protocol for DNA sequence analysis using Sequenase 2.0. Biotechniques. 1991 Feb;10(2):185–185. [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- St-Onge L., Bouchard L., Bastin M. High-frequency recombination mediated by polyomavirus large T antigen defective in replication. J Virol. 1993 Apr;67(4):1788–1795. doi: 10.1128/jvi.67.4.1788-1795.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg B., Pollack R., Topp W., Botchan M. Isolation and characterization of T antigen-negative revertants from a line of transformed rat cells containing one copy of the SV40 genome. Cell. 1978 Jan;13(1):19–32. doi: 10.1016/0092-8674(78)90134-4. [DOI] [PubMed] [Google Scholar]
- Stewart N., Bacchetti S. Expression of SV40 large T antigen, but not small t antigen, is required for the induction of chromosomal aberrations in transformed human cells. Virology. 1991 Jan;180(1):49–57. doi: 10.1016/0042-6822(91)90008-y. [DOI] [PubMed] [Google Scholar]
- Tenen D. G., Haines L. L., Livingston D. M. Binding of an analog of the simian virus 40 T antigen to wild-type and mutant viral replication origins. J Mol Biol. 1982 May 25;157(3):473–492. doi: 10.1016/0022-2836(82)90472-7. [DOI] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989 Feb 24;56(4):619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Thomas K. R., Folger K. R., Capecchi M. R. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986 Feb 14;44(3):419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- Treco D., Arnheim N. The evolutionarily conserved repetitive sequence d(TG.AC)n promotes reciprocal exchange and generates unusual recombinant tetrads during yeast meiosis. Mol Cell Biol. 1986 Nov;6(11):3934–3947. doi: 10.1128/mcb.6.11.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treco D., Thomas B., Arnheim N. Recombination hot spot in the human beta-globin gene cluster: meiotic recombination of human DNA fragments in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Aug;5(8):2029–2038. doi: 10.1128/mcb.5.8.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Robert de Saint Vincent B., DeRose M. L. Effect of chromosomal position on amplification of transfected genes in animal cells. Nature. 1984 Feb 9;307(5951):516–520. doi: 10.1038/307516a0. [DOI] [PubMed] [Google Scholar]
- Wahls W. P., Moore P. D. Homologous recombination enhancement conferred by the Z-DNA motif d(TG)30 is abrogated by simian virus 40 T antigen binding to adjacent DNA sequences. Mol Cell Biol. 1990 Feb;10(2):794–800. doi: 10.1128/mcb.10.2.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls W. P., Wallace L. J., Moore P. D. Hypervariable minisatellite DNA is a hotspot for homologous recombination in human cells. Cell. 1990 Jan 12;60(1):95–103. doi: 10.1016/0092-8674(90)90719-u. [DOI] [PubMed] [Google Scholar]
- Ward P., Berns K. I. In vitro rescue of an integrated hybrid adeno-associated virus/simian virus 40 genome. J Mol Biol. 1991 Apr 20;218(4):791–804. doi: 10.1016/0022-2836(91)90267-a. [DOI] [PubMed] [Google Scholar]
- Wegner M., Schwender S., Dinkl E., Grummt F. An amplification-promoting sequence from mouse genomic DNA: interaction with a trans-acting factor that also affects gene expression. DNA Cell Biol. 1990 Jun;9(5):311–321. doi: 10.1089/dna.1990.9.311. [DOI] [PubMed] [Google Scholar]
- Wegner M., Schwender S., Dinkl E., Grummt F. Interaction of a protein with a palindromic sequence from murine rDNA increases the occurrence of amplification-dependent transformation in mouse cells. J Biol Chem. 1990 Aug 15;265(23):13925–13932. [PubMed] [Google Scholar]
- Wegner M., Zastrow G., Klavinius A., Schwender S., Müller F., Luksza H., Hoppe J., Wienberg J., Grummt F. Cis-acting sequences from mouse rDNA promote plasmid DNA amplification and persistence in mouse cells: implication of HMG-I in their function. Nucleic Acids Res. 1989 Dec 11;17(23):9909–9932. doi: 10.1093/nar/17.23.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. C., Körner A. Cleavage of cruciform DNA structures by an activity from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6445–6449. doi: 10.1073/pnas.82.19.6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannis-Hadjopoulos M., Frappier L., Khoury M., Price G. B. Effect of anti-cruciform DNA monoclonal antibodies on DNA replication. EMBO J. 1988 Jun;7(6):1837–1844. doi: 10.1002/j.1460-2075.1988.tb03016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Gralla J. D. In situ nucleoprotein structure at the SV40 major late promoter: melted and wrapped DNA flank the start site. Genes Dev. 1989 Nov;3(11):1814–1822. doi: 10.1101/gad.3.11.1814. [DOI] [PubMed] [Google Scholar]
- Zhang L., Gralla J. D. In situ nucleoprotein structure involving origin-proximal SV40 DNA control elements. Nucleic Acids Res. 1990 Apr 11;18(7):1797–1803. doi: 10.1093/nar/18.7.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Ambrosio E., Furano A. V. DNA synthesis arrest sites at the right terminus of rat long interspersed repeated (LINE or L1Rn) DNA family members. Nucleic Acids Res. 1987 Apr 10;15(7):3155–3175. doi: 10.1093/nar/15.7.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]