Abstract
We examine the occurrence of the ≈300 known protein folds in different groups of organisms. To do this, we characterize a large fraction of the currently known protein sequences (≈140,000) in structural terms, by matching them to known structures via sequence comparison (or by secondary-structure class prediction for those without structural homologues). Overall, we find that an appreciable fraction of the known folds are present in each of the major groups of organisms (e.g., bacteria and eukaryotes share 156 of 275 folds), and most of the common folds are associated with many families of nonhomologous sequences (i.e., >10 sequence families for each common fold). However, different groups of organisms have characteristically distinct distributions of folds. So, for instance, some of the most common folds in vertebrates, such as globins or zinc fingers, are rare or absent in bacteria. Many of these differences in fold usage are biologically reasonable, such as the folds of metabolic enzymes being common in bacteria and those associated with extracellular transport and communication being common in animals. They also have important implications for database-based methods for fold recognition, suggesting that an unknown sequence from a plant is more likely to have a certain fold (e.g., a TIM barrel) than an unknown sequence from an animal.
Keywords: sequence analysis, genome comparison, fold family, databank statistics, protein evolution
There is some evidence that there is a limited number of different protein folds (estimated to be ≈1,000) and that this “molecular parts list” is sufficient for all organisms to get on with life (1, 2). Given that this is true, one is led to ask to what degree the obvious morphological differences among organisms arise from their using different selections from this master parts list. In somewhat extreme terms, are people different from plants because they have distinctly different protein folds? On the opposite extreme, it may be that most folds occur in every organism in the same way that the genetic code and many basic biochemical pathways (such as glycolysis) are almost universally shared. Up to now, it has only been possible to address this question anecdotally in terms of individual examples. Herein we attempt a more comprehensive answer, by structurally characterizing all the known protein sequences in the databanks, i.e., by doing a structural census of the current protein universe (>140,000 sequences). Briefly, we find that the distribution of folds and structural features is different between different groups of organisms (e.g., prokaryote vs. eukaryote), a fact that has strong implications for fold recognition. However, we also find evidence that many folds are shared rather evenly among a wide variety of organisms.
Surveys of the representation of folds in the structure databank (Protein Data Bank, PDB) (3) have been reported and calculations have been done estimating the number and size of sequence families with known folds (1, 4–7). However, there have not been any studies comparing the occurrence of the known fold families among different groups of organisms. This type of comparative work has been done in studies that focused purely on sequences (8–10). For instance, it has been possible to identify sequences, called ancient conserved regions, that have been conserved over long evolutionary time scales between phylogenetically distant species (11, 12). Herein we have similar aims, but endeavor to do the work in a more structural fashion, expecting that the greater conservation of structure (as compared with sequence) and its closer relation to function will reveal more about distant evolutionary relationships (8, 13–15).
Results
Overall Division of the Databank.
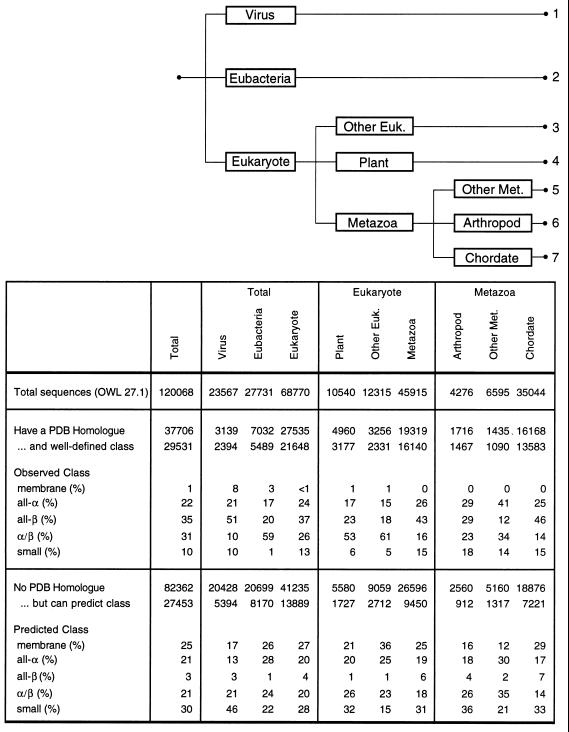
Our approach is straightforward. As shown in Fig. 1, we began with all protein sequences in the publicly accessible databanks [the 142,737 sequences in the OWL composite database (17)]. We then partitioned them in two ways. First, we assigned each sequence to one of the seven groups of organisms shown in Fig. 1 and then divided the sequences into those with and without a homologue in the structure databank. All the taxonometric classification and sequence analysis was done with standard methodology [in particular, the fasta program (22, 23) with conservative thresholds to find sequence homologues]. In the process of partitioning the sequences, we removed from our data set sequence fragments less than 40 residues, sequences that did not fit into our seven taxa (e.g., the few archaean, unclassified, and artificial sequences), and low-complexity sequences. This gave us 120,068 sequences to work with. Most of these (57%) were from eukaryotes with the remainder split between viruses and eubacteria. About 28% of these sequences had a homologue with known structure. Interestingly, eukaryotic sequences (especially chordate ones) were almost twice as likely to have a structural homologue as bacterial or viral sequences (e.g., 46% of chordate sequences had structural homologues versus 25% of bacterial sequences).
Figure 1.
How the total population of sequences is divided into seven taxa in three steps is shown at the top. Other Euk. includes mostly fungi and protists, and Other Met. includes mostly nematodes. Beneath each of the taxa is shown the total number of sequences, the total with and without a structural homologue, and the total with a well-defined structural class. Below this is shown the percentage of the sequences that have each of the five well-defined structural classes.
We analyzed the sequences with structural homologues in detail by using the Structural Classification of Proteins (scop) (19). This classification attempts to comprehensively systematize all known structural resemblances, many of which were originally pointed out on the basis of case-by-case observations of crystal structures [e.g., Rossmann et al. (47) and Harrison (48)]. In total, scop divides the 4,432 structures in the PDB into 8,330 domains, which, in turn, are classified into 318 different fold families. Thus, we were able to associate each sequence matching a structure with a particular scop “fold identifier,” essentially a molecular part number, and then to see how these identifiers were distributed among our seven taxonometric categories. Other classifications of protein structure also divide the structure databank among ≈300 fold families [e.g., CATH, FSSP, Entrez-MMDB, LPFC (5, 13, 49–51)] and should give similar results.
Most of our calculations were done in the most straightforward way based on exhaustive enumeration—counting everything equally. We are fully aware that such an approach tends to give results biased to some degree by the current composition of the sequence databank, i.e., toward proteins that scientists have chosen to study. There are a variety of approaches toward counting sequences (involving differential weighting or polling of selected samples, such as whole genomes) that attempt to address biases in a systematic fashion, and we discuss the application of some of these in detail. However, on a basic level, we do not believe it is possible to remove all traces of “investigator-preference” bias from any sample drawn from the current databanks. Consequently, we believe an approach of straightforward enumeration (just as in governmental censuses) provides the clearest reflection of what we currently know.
Top-10 Folds in Various Taxa.
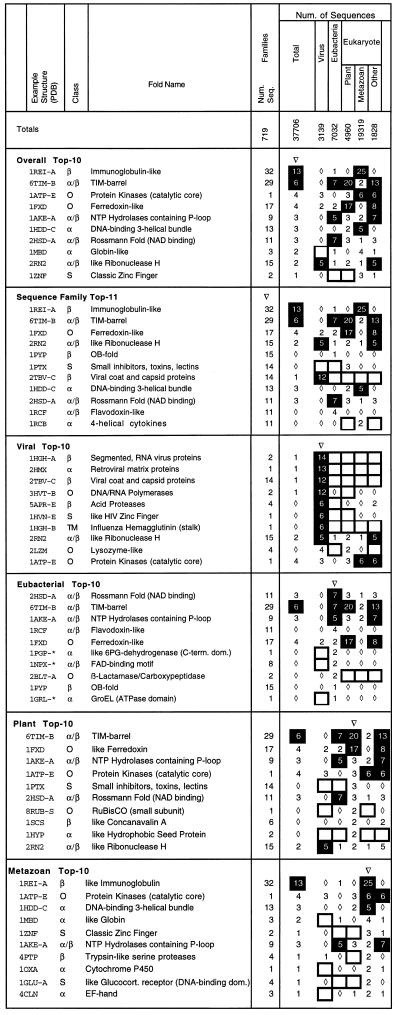
The overall distribution of folds shows that most folds have about ≈130 homologues, but there are a few folds with many more, as shown in the “top-10 lists” in Fig. 2. In particular, the top-7 folds (which include the TIM barrel, the Ig fold, the Rossmann fold, the homeo domain-like three-helix bundle, and the ferrodoxin fold) each have more than 1,000 homologues, and the top-25 folds match almost two-thirds (61%) of the sequences with structural homologues. Some of these folds, such as the nucleotide-binding Rossmann fold, the zinc finger, or the DNA-binding three-helix bundle, perform a single function and tend to be recombined as modules in a variety of proteins. Other folds act as multipurpose parts that can perform a variety of diverse functions within the same structural framework. For instance, the ribonuclease H fold can act either as a structural protein or a nuclease; the ferrodoxin fold appears in ribosomal proteins, transcription factors, and enzymes; and the Ig fold provides a scaffolding for enzymes, transcription factors, and viral envelope proteins, in addition to its well-known role in the immune system.
Figure 2.
Top-10 folds, overall, in terms of number of sequence families and in each of four taxa. In each of the top 10, the number of sequences with a particular fold is shown as a percentage of the total number of sequences in the corresponding taxa that have a structural homologue (this last value is shown as an absolute number at the top). Values: 0%, □; between 0% and 1%, ⋄; greater than 5%, ▪. Also shown in column 1 is a representative structure with that fold. (The syntax is PDB identifier followed by chain. For the three identifiers marked with an ∗, a particular residue selection is also necessary: for 1PGP, residues 177–473; for 1NPX, residues 120–242; for 1GRL, residues 6–136 and 410–523.) In column 2 is the structural class of the fold, derived from scop (S, is small; TM, transmembrane; O, not one of the five well-defined classes—in this case usually an α + β protein). The fold name is in column 3. In column 4, the number of sequence families is shown as an absolute number, not a percentage. This is derived from clustering the domains in the PDB with an e-value threshold of 0.001 (see techniques section). Counting only the number of clusters (i.e., families) effectively represents a particularly stringent form of sequence weighting.
Many folds (125 in all) can be associated with more than one sequence family. That is, the sequences corresponding to each of these folds can be clustered into groups of similar sequences that have no detectable homology between them (see techniques section for more precise definitions). Orengo et al. (5) suggested that protein folds associated with many sequence families, which they dubbed “superfolds,” may represent particularly favorable structural architectures, accommodating to a wide variety of sequences.
We created a new list of superfolds, by ranking the folds considered herein in terms of the number of sequence families they are associated with. [It is not possible to directly compare our most common folds to the superfolds in Orengo et al. (5), because they use slightly different fold definitions and because the databank has grown considerably since their work was published.] Comparing the most common folds in terms of sequence families with the most common overall indicates that most of the common folds are associated with many sequence families and that multifunction folds tend to have more associated families than single-function ones. Specifically, 7 of the top-10 folds have more than nine sequence families (with the exceptions being the globin, protein kinase, and zinc-finger folds, all of which have highly specific functions). However, the converse is not as true: 5 of the folds in the “sequence-family top 11” are not in the overall top 10. These, notably, include the OB fold and four-helical cytokine family, a four-helix bundle.
The most common folds are present in all taxa. However, the degree of their representation varies greatly. This is particularly true for the Ig fold, the most common one. It constitutes 25% of metazoan sequences with structural matches (and 40% of the human sequences), but only ≈1% of the plant, bacterial, and viral sequences. It is, consequently, of interest to look at the most common folds in each of the seven taxa, and this is shown in Fig. 2. Clearly, viruses have the most unique distribution of folds, reflecting their special functional requirements. In fact, four of their top-10 folds do not occur in any of the other six taxa. This is understandable as they are all associated with the viral envelope, which has a highly symmetrical structure unique to viruses. Viruses share with other organisms folds associated with essential viral functions (polymerases, acid proteases, and ribonucleases), but they have few of the folds associated with metabolic enzymes (e.g., TIM barrels, Rossmann folds, or NTP hydrolases). The bacterial top 10, in contrast, shows a great preponderance of folds for metabolic enzymes, in particular glycolytic enzymes. It also contains one fold unique to bacteria (and bacteriophages), that for β-lactamases and d-Ala carboxypeptidases. These enzymes perform functions associated with the unique structure of the bacterial cell wall (i.e., antibiotic resistance and cleavage of d-Ala peptides).
The top-10 folds for multicellular animals (metazoa) are very different from those for bacteria. They contain fewer folds for enzymes and more folds associated with intercellular communication, defense, and transport (e.g., EF hand, Igs, globins, protein kinases, and also within the top 15 are cysteine-knot and four-helical cytokines). There are also three folds of DNA-binding regulatory proteins and one for trypsin-like proteases, which are usually involved in extracellular digestion.
Like the metazoan top 10, the plant top 10 also contains the protein kinase fold, which is involved in signaling. However, it has many more metabolic enzymes, making it in some ways more like the bacterial top 10. It also contains a few folds unique to plants. In particular, the fold of the protein rubisco, which has a crucial role in fixing carbon in photosynthesis, is featured twice in the plant top 10—once for its small subunit, which has a fold unique to plants, and a second time for its large subunit, which contains a ferrodoxin fold.
Top-10 Folds in a Representative Genome.
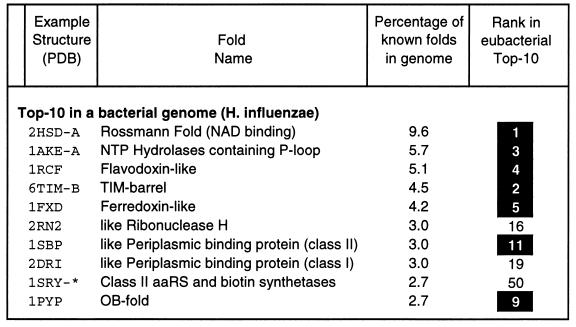
The list of top-10 folds in various taxa, although comprehensive, is to some degree skewed by investigator preference. We can get a sense of this bias by sampling the databanks selectively, specifically, with the sample corresponding to the entire genome of a particular organism. This is done in Fig. 3, which shows a top-10 list derived from the genome of a representative bacterium, Haemophilus influenzae (52). This list is clearly similar to the eubacterial top-10 list in Fig. 2, with 7 of the 10 entries having similar positions. However, it is important to realize that the folds in a genome top 10 are still biased to a degree, by the selection of the representative genome itself and, more importantly, because they depend on the selection of known folds in the structure databank (i.e., in scop and the PDB). With many new genome sequences coming out, it will be possible to perform this common fold analysis, comparatively, on a number of microbial genomes. Some initial analyses show quite revealing differences (53).
Figure 3.
The figure shows the top-10 folds in a representative bacterial genome in a format similar to eubacterial top-10 in Fig. 2. For each of the top-10, column 1 shows a representative structure with that fold. (The syntax is PDB identifier followed by chain. For 1SRY, marked with a “∗”, a particular residue selection is also necessary, A:111–421.) Column 2 gives the fold name, derived from scop. Column 3 shows the number of sequences with a particular fold as a percentage of the 248 sequences in the Haemophilus genome (1,680 sequences in total) that have a structural homologue (using the relatively conservative thresholds described in section on sequence analysis techniques). Column 4 shows the rank of this particular fold in the eubacterial top-10 list in Fig. 2. Folds that appear in roughly the same position in both top-10 lists are shown with black boxes. The data in this table are adapted from the expanded analysis in ref. 53.
A Venn Diagram for Shared Folds.
The variation shown in Figs. 2 and 3 in the common folds among different taxa and selected genomes directly addresses the issue of whether the differences between organisms reflect their having fundamentally different folds. However, it only addresses this issue on a case-by-case basis, in terms of specific folds. In Fig. 4, we attempt a more systematic analysis by asking what fraction of all the known folds (in scop) are present in each of our seven taxa and what fraction of those that are present are shared among different taxa. We performed this analysis by dividing the database into subsets of progressively more related organisms (as shown in Figs. 1 and 4). The major division is between eubacterial and eukaryote sequences with the remainder of the sequences falling into a third miscellaneous division (viral sequences). Next, we divide the largest of these divisions (eukaryotes) into major and minor subsets (plants and metazoa) and a third category that includes the remaining eukaryotic sequences (mostly from protists and fungi). Finally, we again perform a three-way major-minor-miscellaneous division on the largest group of eukaryotic sequences (metazoan ones), partitioning them into chordate, arthropod, and other (mostly round worms).
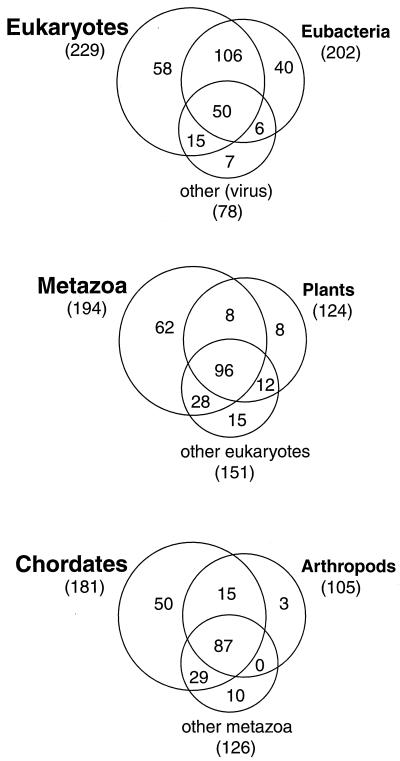
Figure 4.
Venn diagrams showing the number of folds in each group of organisms and how many of these folds are shared between different groups of organisms. Note that in total there are 318 folds in scop. However, we excluded folds associated only with membrane proteins, designed proteins, and model proteins, as well as folds only from archaea and folds not currently in the PDB, giving 282 folds. In the top-most division, these 282 folds are distributed among a major group of sequences (eukaryotes), a minor group (eubacteria), and a miscellaneous group (other and viruses). In the middle division, the major group from the top level (eukaryotic sequences) is subdivided into a major group (metazoa), a minor group (plants), and a miscellaneous group (mostly fungi and protist sequences). This pattern of major, minor, and miscellaneous division is repeated at bottom, where metazoa is subdivided into chordates, arthropods, and other (metazoa).
We find that more related organisms have fewer folds in total but share a larger fraction of them. That is, the “top-level” division, which includes the least related organisms (bacteria, eukaryotes, and others), contains 282 folds in total, but only 18% of these (50 folds) are shared among all three subsets. The next division (plants, metazoa, and others) contains fewer folds (229), but a larger fraction of these are shared (42% or 96 folds). This trend continues in the division containing the most related taxa (chordates, arthropods, and others): these have only 191 folds in total but share 45% of them (87 folds).
If we look only at the two principal subsets at each level of division, we find that they share about half their folds (i.e., eukaryotes and eubacteria share 156 of 275, plants and metazoa share 104 of 214, and chordate and arthropod share 102 of 184). There are only 19 universal folds shared through all divisions. These include the Ig fold, the TIM barrel fold, the Rossmann fold, the ferrodoxin fold, and the ribonuclease H fold.
Characterizing Sequences Without Structural Homologues.
Thus far, we have concentrated on the 37,706 sequences that have a homologue in the structure databank. What can we say about the remaining 82,362 sequences that have no homologue? By using standard methods of secondary structure prediction, we have attempted to place each of these sequences into one of five well-defined structural classes, expanded somewhat from the original class definitions in Levitt and Chothia (45): all-α, all-β, α/β, transmembrane, and small. Most of the sequences with structural homologues can be placed into these five classes by observation. However, because of our fairly strict class definitions and due to a variety of complications (in particular, the difficulty in determining domain definitions in sequences without structural homologues), we could confidently place only about a third of the sequences without a structural homologue into the five categories.
Our results, shown in Fig. 1, indicate that the proportion of proteins in the five well-defined classes varies considerably between taxonomic groups. Most importantly, the results we obtained by looking at sequences that do not have a structural homologue are consistent with the results obtained for sequences that do, suggesting that some of our firmer conclusions about the former can be reliably extrapolated to the later. In particular, we find that the percentage of small proteins is much larger in complex multicellular animals (e.g., arthropods and chordates) than in bacteria and simple eukaryotes (fungi and protists). Also, the percentage of all-β proteins is larger in viruses and eukaryotes than prokaryotes, and within eukaryotes the percentage increases as one moves from the simpler organisms to the more complex metazoans (e.g., arthropods and chordates). This may be understandable for the sequences that have a structural homologue in terms of the large number of all-β Igs in vertebrates. However, note that it is also true for the sequences that do not have structural homologues (and consequently probably do not have Ig folds).
Continuing our focus on structural class, we repeated the Venn-diagram analysis in Fig. 4, this time looking at the number of distinct folds in each taxa individually for each of the five structural classes. Except for small proteins, we found that each structural class gave essentially similar results to the overall results shown in Fig. 4. However, eukaryotes, especially chordates, had a far greater proportion of the small folds. In particular, of the 35 small protein folds, 30 occur in eukaryotes but only 8 occur in bacteria, and of the 30 in eukaryotes, 27 occur in metazoa and 23 occur in chordates. Thus, there is prevalence of small proteins in eukaryotes, both in terms of relative numbers of sequences (with and without structural homologues) and in terms of number of folds. In some sense, this is counterintuitive as one might expect simpler smaller organisms, such as bacteria, to have more small folds. However, it is explained to some degree by the number of small protein folds involved in intercellular communication and regulation in vertebrates (e.g., insulin, kringle domains, fibronectin, or zinc fingers).
Implications, Especially for Fold Recognition
We have conducted a census of the current population of proteins. It is in a sense skewed and incomplete because we do not have all possible proteins for a given taxa. However, because the number of protein sequences is growing at a tremendous rate [more than doubling every 2 years (17)], we would expect this situation to improve to some degree in the future and we would hope that our conclusions give one a clear taste of what is to come. Furthermore, having a clear grasp of the current state of the databanks provides an important yardstick for measuring the results of future sequencing projects and for assessing the hidden biases in database-based methods for structure prediction and fold recognition.
Specifically, we have found that although there are large numbers of folds shared between organisms, different organisms have a markedly different distribution of folds. In addition to its obvious evolutionary implications, this finding is very important for approaches to fold recognition, the matching of a query sequence to a target structure, where there is no detectable homology between the sequence and the structure (29, 30). That is, our census indicates that knowing the species of an unknown sequence gives one clear clues as to its fold. For instance, there are 282 folds in the current protein universe (Fig. 4). However, a priori it is reasonable to rule out 80 of them (282–202) for an unknown bacterial sequence. In particular, we would not expect this unknown sequence to have many of the common eukaryotic folds associated with transcription or signaling, such as zinc fingers or EF hands. Likewise, knowing that an unknown sequence is from a plant means that it would be much more likely to have a TIM-barrel fold than if it were from an animal, in which case it would be much more likely to have an Ig fold.
Sequence Analysis Techniques Employed
A Relational Database of Folds, Sequences, and Taxa.
Our census was greatly expedited by use of simple relational database implemented by using dbm and “object-oriented” perl (version 5) (16). Relational tables linked the 142,737 sequence identifiers, the 37,706 structure matches, the 282 fold identifiers, and the 7 taxonometric ranks. We will make available over the Internet a number of these tables at the following URL: http://bioinfo.mbb.yale.edu/census.
Sequences were taken from the OWL composite database (April 1996) (17, 18) and the Haemophilus genome project website (http://www.tigr.org), structures were from the PDB, and fold defintions were from scop 1.32 (May 1996) (19, 20). We assigned specific taxonometric ranks to sequences based on the classification scheme associated with GenBank (21). All archaean, artificial, and unclassified sequences were excluded.
Sequence Comparison and Clustering into Families.
All sequence matching was done with the fasta program (version 2.0) (22–24) with k-tup 1 and an e-value cutoff of 0.001. This is a very conservative threshold, and empirical tests have shown that it should give one error every 1,000 comparisons (25, 26). Low complexity sequences were filtered out first by using the seg program (27, 28).
There are more sensitive methods of comparing sequences to structures than the fasta program, e.g., profiles, Hidden–Markov models, and threading (29–32). These methods would be expected to find more homologues for certain folds. However, the sensitivity improvement would not be uniform over all folds. The more sensitive methods tend to do better for large fold families (with many associated sequences) or for fold families with clearly defined sequence motifs. Thus, using these methods would bias the results even more toward highly populated and well-characterized folds. This is not advantageous because for a large-scale census, where uniform sampling and treatment of the data are more important than sensitivity (as one is more concerned with relative rather than absolute numbers).
The number of sequence families for each fold is derived from clustering all the domains in the PDB (using scop domain definitions). fasta is used for the sequence comparisons; a pair of domains matching with an e-value of 0.001 or less is taken as connected (significantly related). Each cluster consists of the domains connected to one another by at least one linkage. This is a similar approach to that taken in Hobohm et al. (33) but with a somewhat different method of sequence comparison.
Sequence Weighting and Databank Sampling.
We did not attempt to use explicit sequence weighting in our census [see Altschul et al. (34), Sander and Schneider (6), Vingron and Sibbald (35), Gerstein et al. (36), and Miyazawa and Jernigan (37)]. Thus, our conclusions to some degree directly reflect the biases inherent in the databanks. We feel that completely removing these biases is impossible and that assessing them is to a large degree a subjective issue [e.g., see Altschul et al. (34)]. Furthermore, insofar as our conclusions about the current state of the databanks reveal bias, we feel they are useful for assessing hidden biases in database-based structure-prediction methods.
Note, however, that aspects of our census did involve four forms of implicit (and reasonable) sequence weighting. (i) In compiling the OWL composite databank, all mutant and identical sequences were removed. This is, in effect, a very simple type of sequence weighting that removes one of the major problems in doing calculations on the PDB, the problem of compensating for the many structures (e.g., T4 lysozyme) solved in different liganded states or as mutants. (ii) The enumeration of sequence families shown in Fig. 2 is a specific and particularly stringent form of sequence weighting (see above for the method). It greatly down weights highly homologous sequences, giving all the sequences in a family, even a large one, an aggregate weight of 1.0. (iii) Many of the conclusions in Fig. 2 and all the conclusions in Fig. 4 are completely independent of sequence weighting because they are only concerned with membership, whether or not a given fold is present in a particular taxa. (iv) Finally, the genome top-10 list in Fig. 3 is constructed from a complete genome sequence that is not biased by the preferences of investigators to sequence proteins of functional importance. However, it is still skewed by the selection of the known structures in the PDB matched against the genome.
For the numbers reported herein that are the result of exhaustive enumeration—counting everything—we found that we could achieve essentially the same results through randomly sampling (i.e., polling) small subsets, the same approach that has been argued to be effective in the American governmental census (38).
Class Prediction.
For the class predictions, we used a standard “off-the-shelf” approach. We first divided all sequences on the basis of length. Those with less than 40 residues were excluded, and those with between 40 and 80 residues were classed as small. For sequence with more than 80 residues, we applied the following protocol: Based on the annotations [from Swiss-Prot (39)], we decided whether or not a sequence was transmembrane. By doing this, we found that only about 10% of the sequences without structural homologues are transmembrane. This is probably an underestimate, and we tried to assess its magnitude by testing each sequence with the Kyte–Doolittle and GES hydropathy scales (40–42). By using a strict threshold, we found 5% of the sequences to be transmembrane but by using the lax one, we found that 30% were (but with many documented false positives).
After removing small and transmembrane proteins, we ran the gor program for secondary-structure prediction (43). We used commonly accepted thresholds (44) for placing a protein in the various classes: all-α has α > 40% and β < 5%, all-β has β > 40% and α < 5%, and α/β has α > 30% and β > 20%. Sequences that did not fit in any of the previous classes were considered not to have a “well-defined” class. Note this includes (i) sequences with a structural homologue where the structure has the α + β class (45), (ii) sequences without a structural homologue that code for single-domain proteins naturally falling into the α + β class, and (iii) sequences without a structural homologue that code for multidomain proteins where each domain has a well-defined class (e.g., all-α and all-β) but where the protein is considered as whole for class prediction. We tested our class predictions against the observed classes in scop and found about 80% agreement. We also tested our structural class predictions by comparing a sample of them with the results of running the PHD server (46) and found substantial agreement.
Acknowledgments
We thank L. Stryer, A. Murzin, S. Brenner, C. Chothia, T. Johnson, and E. Brodkin for helpful conversations or reading the manuscript. M.G. acknowledges the Office of Naval Research for support (Young Investigator Grant N00014–97-1–0725) and M.L. acknowledges the Department of Energy (DOE DE-FG03-95ER62135).
ABBREVIATIONS
- PDB
Protein Data Bank
- scop
Structural Classification of Proteins
References
- 1.Chothia C. Nature (London) 1992;357:543–544. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- 2.Dorit R L, Schoenbach L, Gilbert W. Science. 1990;250:1377–1382. doi: 10.1126/science.2255907. [DOI] [PubMed] [Google Scholar]
- 3.Bernsteinm F C, Koetzle T F, Williams G J B, Meyer E F, Jr, Brice M D, Rodgers J R, Kennard O, Shimanouchi T, Tasumi M. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 4.Holm L, Sander C. Science. 1996;273:595–602. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 5.Orengo C A, Jones D T, Thornton J M. Nature (London) 1994;372:631–634. doi: 10.1038/372631a0. [DOI] [PubMed] [Google Scholar]
- 6.Sander C, Schneider R. Proteins Struct Funct Genet. 1991;9:56–68. doi: 10.1002/prot.340090107. [DOI] [PubMed] [Google Scholar]
- 7.Pascarella S, Argos P. Protein Eng. 1992;5:121–137. doi: 10.1093/protein/5.2.121. [DOI] [PubMed] [Google Scholar]
- 8.Doolittle R F. Annu Rev Biochem. 1995;64:287–314. doi: 10.1146/annurev.bi.64.070195.001443. [DOI] [PubMed] [Google Scholar]
- 9.Koonin E V, Tatusov R L, Rudd K E. Proc Natl Acad Sci USA. 1995;92:11921–11925. doi: 10.1073/pnas.92.25.11921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ouzounis C, Kyrpides N. FEBS Lett. 1996;390:119–123. doi: 10.1016/0014-5793(96)00631-x. [DOI] [PubMed] [Google Scholar]
- 11.Green P, Lipman D, Hillier L, Waterston R, States D, Claverie J M. Science. 1993;259:1711–1716. doi: 10.1126/science.8456298. [DOI] [PubMed] [Google Scholar]
- 12.Green P. Curr Opin Struct Biol. 1994;4:404–412. [Google Scholar]
- 13.Gibrat J F, Madej T, Bryant S H. Curr Opin Struct Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 14.Chothia C, Lesk A M. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chothia C, Gerstein M. Nature (London) 1997;385:579–581. doi: 10.1038/385579a0. [DOI] [PubMed] [Google Scholar]
- 16.Wall L, Christiansen D, Schwartz R. Programming Perl. Sebastapol, CA: O’Reilly and Associates; 1996. [Google Scholar]
- 17.Bleasby A J, Akrigg D, Attwood T K. Nucleic Acids Res. 1994;22:3574–3577. [PMC free article] [PubMed] [Google Scholar]
- 18.Bleasby A J, Wootton J C. Protein Eng. 1990;3:153–159. doi: 10.1093/protein/3.3.153. [DOI] [PubMed] [Google Scholar]
- 19.Murzin A, Brenner S E, Hubbard T, Chothia C. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 20.Brenner S, Chothia C, Hubbard T J P, Murzin A G. Methods Enzymol. 1996;266:635–642. doi: 10.1016/s0076-6879(96)66039-x. [DOI] [PubMed] [Google Scholar]
- 21.Benson D A, Boguski M, Lipman D J, Ostell J. Nucleic Acids Res. 1996;24:1–5. doi: 10.1093/nar/24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipman D J, Pearson W R. Science. 1985;227:1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- 23.Pearson W R, Lipman D J. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearson W R. Methods Enzymol. 1996;266:227–259. doi: 10.1016/s0076-6879(96)66017-0. [DOI] [PubMed] [Google Scholar]
- 25.Brenner S, Hubbard T, Murzin A, Chothia C. Nature (London) 1995;378:140. doi: 10.1038/378140a0. [DOI] [PubMed] [Google Scholar]
- 26.Brenner, S., Chothia, C. & Hubbard, T. (1997) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 27.Wootton J C, Federhen S. Computers and Chemistry. 1993;17:149–163. [Google Scholar]
- 28.Wootton J C, Federhen S. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 29.Jones D T, Thornton J M. Curr Opin Struct Biol. 1996;6:210–216. doi: 10.1016/s0959-440x(96)80076-5. [DOI] [PubMed] [Google Scholar]
- 30.Bowie J U, Eisenberg D. Curr Opin Struct Biol. 1993;3:437–444. [Google Scholar]
- 31.Eddy S R. Curr Opin Struct Biol. 1996;6:361–365. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- 32.Gribskov M, Lüthy R, Eisenberg D. Methods Enzymol. 1990;183:146–159. doi: 10.1016/0076-6879(90)83011-w. [DOI] [PubMed] [Google Scholar]
- 33.Hobohm W, Scharf M, Schneider R, Sander C. Protein Sci. 1992;1:409–417. doi: 10.1002/pro.5560010313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul S F, Carroll R J, Lipman D J. J Mol Biol. 1989;207:647–653. doi: 10.1016/0022-2836(89)90234-9. [DOI] [PubMed] [Google Scholar]
- 35.Vingron M, Sibbald P R. Proc Natl Acad Sci USA. 1993;90:8777–8781. doi: 10.1073/pnas.90.19.8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerstein M, Sonnhammer E, Chothia C. J Mol Biol. 1994;236:1067–1078. doi: 10.1016/0022-2836(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 37.Miyazawa S, Jernigan R L. J Mol Biol. 1996;256:623–644. doi: 10.1006/jmbi.1996.0114. [DOI] [PubMed] [Google Scholar]
- 38.Ladd, E. C. Tempest in a Census. Wall Street Journal, 30 July 1997, A14.
- 39.Bairoch A, Boeckmann B. Nucleic Acids Res. 1992;20:2019–2022. doi: 10.1093/nar/20.suppl.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 41.Engelman D M, Steitz T A, Goldman A. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 42.Jähnig F. Trends Biochem Sci. 1990;15:93–95. doi: 10.1016/0968-0004(90)90188-h. [DOI] [PubMed] [Google Scholar]
- 43.Garnier J, Gibrat J F, Robson B. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 44.Rost B. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 45.Levitt M, Chothia C. Nature (London) 1976;261:552–558. doi: 10.1038/261552a0. [DOI] [PubMed] [Google Scholar]
- 46.Rost B, Sander C. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 47.Rossmann M G, Liljas A, Branden C I, Banaszak L J. Enzymes. 1975;11:61–102. [Google Scholar]
- 48.Harrison S C. Nature (London) 1991;353:715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- 49.Holm L, Sander C. Nucleic Acids Res. 1994;22:3600–3609. [PMC free article] [PubMed] [Google Scholar]
- 50.Orengo C A, Flores T P, Taylor W R, Thornton J M. Protein Eng. 1993;6:485–500. doi: 10.1093/protein/6.5.485. [DOI] [PubMed] [Google Scholar]
- 51.Schmidt R, Gerstein M, Altman R. Protein Sci. 1997;6:246–248. doi: 10.1002/pro.5560060127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 53.Gerstein, M. (1997) J. Mol. Biol., in press.