Abstract
Plants produce a variety of toxic compounds, which are often used as anticancer drugs. The self-resistance mechanism to these toxic metabolites in the producing plants, however, remains unclear. The plant-derived anticancer alkaloid camptothecin (CPT) induces cell death by targeting DNA topoisomerase I (Top1), the enzyme that catalyzes changes in DNA topology. We found that CPT-producing plants, including Camptotheca acuminata, Ophiorrhiza pumila, and Ophiorrhiza liukiuensis, have Top1s with point mutations that confer resistance to CPT, suggesting the effect of an endogenous toxic metabolite on the evolution of the target cellular component. Three amino acid substitutions that contribute to CPT resistance were identified: Asn421Lys, Leu530Ile, and Asn722Ser (numbered according to human Top1). The substitution at position 722 is identical to that found in CPT-resistant human cancer cells. The other mutations have not been found to date in CPT-resistant human cancer cells; this predicts the possibility of occurrence of these mutations in CPT-resistant human cancer patients in the future. Furthermore, comparative analysis of Top1s of CPT-producing and nonproducing plants suggested that the former were partially primed for CPT resistance before CPT biosynthesis evolved. Our results demonstrate the molecular mechanism of self-resistance to endogenously produced toxic compounds and the possibility of adaptive coevolution between the CPT production system and its target Top1 in the producing plants.
Keywords: evolution, Ophiorrhiza pumila, Camptotheca acuminata
Plants produce a vast array of secondary metabolites to overcome environmental stress and defend themselves against their natural enemies. The broad metabolic diversity represents a process of adaptation that has been subjected to natural selection during evolution (1). Furthermore, humans have benefited from plant-derived compounds that are used as medicines. These compounds, including vincristine, vinblastine, taxol, and camptothecin (CPT), are currently prescribed as anticancer drugs, which function by disrupting basic biological processes in human cells. The fact that toxic metabolite-producing plants are insensitive to the metabolites suggests the presence of a species-specific resistance mechanism to prevent cytotoxicity. How these plants have evolved a mechanism to prevent self-toxicity while simultaneously producing toxic compounds is a long-standing intriguing question that has not been clearly answered to date.
CPT, a monoterpene indole alkaloid, is produced in many distantly related plants, including Ophiorrhiza pumila, Ophiorrhiza liukiuensis, and Camptotheca acuminata. It induces cell death by stabilizing a covalent complex between DNA topoisomerase I (Top1) and the nicked DNA, leading to a DNA lesion (2–4). Although CPT is toxic to most eukaryotic organisms, including higher plants, CPT-producing plants can avoid self-toxicity, thereby suggesting the presence of a species-specific resistance mechanism (5, 6). In contrast, the involvement of either a CPT transporter or a Top1 mutation that prevents CPT binding is known to serve as a CPT-resistance mechanism in CPT-resistant human cancer cells (7, 8). However, no information is available on how CPT-producing plants avoid CPT toxicity (6). Although sequestration of toxic metabolites through vesicles or transporters is generally assumed as a self-resistance mechanism, our previous study (9) indicated that CPT is passively excreted and that no particular sequestration mechanisms operate for CPT, suggesting the functioning of a different mechanism. Here, we report the specific mutations of the target protein Top1 that confer resistance to CPT in CPT-producing plants as a self-resistance mechanism in plants. In addition, we provide evidence regarding adaptive coevolution between the production of CPT and the resistance of Top1.
Results and Discussion
Top1s from both CPT-Producing Plants and Nonproducing Organisms Share a Common Ancestor.
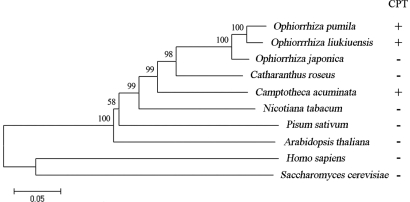
CPT is toxic to most eukaryotic organisms, except CPT-producing plants [supporting information (SI) Fig. S1]. Although most studies on structure and mechanism have focused on human Top1 (hereafter HsTop1) (10, 11), several plant Top1s have been characterized as essential for survival (5, 12). We cloned cDNAs encoding Top1 from three CPT-producing plants; namely, O. pumila, O. liukiuensis, and C. acuminata (hereafter referred to as OpTop1, OlTop1, and CaTop1, respectively). For comparison, Top1 cDNAs were also cloned from CPT-nonproducing plants, including Ophiorrhiza japonica (OjTop1) (closely related to O. pumila and O. liukiuensis) and Catharanthus roseus (CrTop1) (monoterpene indole alkaloid-producing plant). All cDNAs contained the conserved domain of Top1. OpTop1 heterologously expressed in Escherichia coli exhibited topoisomerase activity (Fig. S2). A phylogenetic tree of Top1s is split into two clusters (Fig. 1). HsTop1 and Saccharomyces cerevisiae Top1 form a cluster. The other cluster comprises all plant sequences in which the Top1s of CPT-producing and -nonproducing plants are mixed together. Despite the divergence between HsTop1 and plant Top1s, OpTop1 exhibits considerably high homology (47% identity) with HsTop1. These results suggest the similarity of the overall structure that originated from a common ancestor.
Fig. 1.
Phylogenetic tree of Top1s. Neighbor-joining tree of Top1s from plants, yeast, and humans. The numbers indicate bootstrap values. CPT production is indicated by “+” or “−”.
Amino Acid Substitutions Found Only in Top1s of CPT-Producing Plants.
Because only a few sites in a functional protein are allowed to adapt under natural selection, we first focused only on specific amino acid residues that are involved in CPT binding. The crystal structure of HsTop1 in a covalent complex with duplex DNA containing topotecan, a CPT derivative, suggests that several amino acid residues contribute to its catalytic function or affect CPT binding (13). Comparison of the residues between HsTop1 and plant Top1s shows that only Top1s of CPT-producing plants have 4-aa substitutions in the positions that are proposed to influence CPT binding: Ile-420 to Val, Asn-421 to Lys, Leu-530 to Ile, and Asn-722 to Ser (numbered according to HsTop1) (Fig. 2). No substitution was found in Top1s of CPT-nonproducing plants, including other alkaloid-producing plants such as O. japonica and C. roseus, thereby indicating a functional relationship between these substitutions and CPT resistance (see below). All residues involved in the catalytic function, including the active center Tyr-723, are evolutionarily conserved (Fig. 2). Of particular interest was the substitution at the adjacent position to catalytic Tyr-723, i.e., Asn-722 to Ser, which is found in all CPT-producing plants. Intriguingly, the same change in this position has been reported to confer CPT resistance in human cancer cells isolated from a patient and in yeast (14, 15).
Fig. 2.
Amino acid polymorphism in Top1s of CPT-producing plants and nonproducing organisms. Shown are proposed amino acids that are involved in catalytic function or affect CPT binding. Asp-533 and Ser-722 directly bind to CPT. Other residues that indirectly bind to CPT are important for the proper positioning of Asp-533 and Ser-722. The numbering is based on HsTop1. The red characters indicate the amino acid substitutions in Top1s of CPT-producing plants. Hs, Homo sapiens; At, Arabidopsis thaliana; Cr, Catharanthus roseus; Oj, Ophiorrhiza japonica; Op, Ophiorrhiza pumila; Ol, Ophiorrhiza liukiuensis; Ca, Camptotheca acuminata.
Top1s from CPT-Producing Plants Are Resistant to CPT.
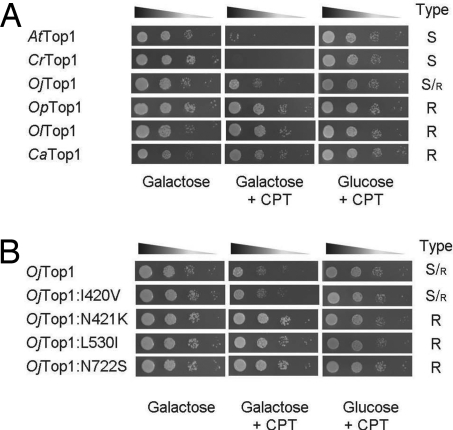
We further investigated whether these plant Top1s are sensitive or resistant to CPT by conducting an in vivo assay using the Top1-deleted S. cerevisiae strain RS190 that has been commonly used to study the interaction of Top1s with antitumor drugs (16–18). In the noninducing condition (glucose-containing medium), CPT did not affect the growth of strain RS190 because Top1, the target of CPT, is absent in this strain. In contrast, overexpression of Top1 by induction with galactose in the presence of CPT caused cell death because CPT blocks the correct reaction of Top1 by stabilizing a covalent complex of the nicked DNA and Top1 leading to a DNA lesion. As shown in Fig. 3A, yeast cells expressing Arabidopsis thaliana Top1 (AtTop1), CrTop1, and OjTop1 exhibited decreased cell viability in the presence of CPT, thereby suggesting that CPT was toxic to these Top1s. In contrast, the expression of Top1s from CPT-producing plants had no significant effect on the cell viability. These results demonstrated that CPT-producing plants possess CPT-resistant Top1s, which are responsible for a self-resistance mechanism in these plants. Moreover, the results implied the effect of a species-specific self-produced toxic metabolite on the evolution of its target molecule in the producing plants.
Fig. 3.
In vivo CPT sensitivity assay of S. cerevisiae expressing Top1. Five microliters of 10-fold serial dilutions (indicated by triangles) of Top1-deleted yeast cells harboring the plasmid encoding plant Top1s (A) or OjTop1 and its mutants (B) were spotted on plates. The expression of exogenous Top1 was induced by galactose and repressed by glucose. The galactose plates were supplemented with 0.25% Me2SO or 5 μg/ml CPT in a final Me2SO concentration of 0.25%. After incubation at 30°C for 2 days, the cell growth was scored as follows: S, CPT-sensitive Top1; S/r, partially CPT-resistant Top1; R, CPT-resistant Top1.
Three Residues of Top1 Involved in CPT Binding Are Important for CPT Resistance.
To evaluate the functional importance of the abovementioned 4-aa substitutions in CPT resistance, we constructed four single OjTop1 mutants: Ile420Val, Asn421Lys, Leu530Ile, and Asn722Ser. We expected these mutants to be CPT-resistant. Three of these four OjTop1 mutants, namely, Asn421Lys, Leu530Ile, and Asn722Ser, were found to be resistant to CPT, indicating that these single-amino-acid substitutions are sufficient to confer resistance (Fig. 3B). In contrast, Ile420Val did not show altered sensitivity (Fig. 3B). In addition, a double mutant of CPT-resistant OpTop1, namely, Ile530Leu and Ser722Asn, became more sensitive to CPT, confirming the importance of these two residues in CPT resistance (Table S1).
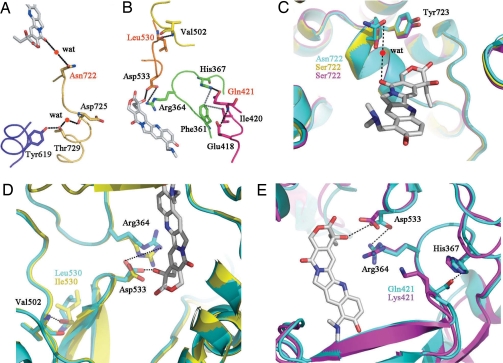
Thus far, several amino acid residues in HsTop1 that affect CPT binding have been characterized (13). Asn-722 forms a water-mediated hydrogen bond with topotecan (Fig. 4A), whereas Asp-533 forms the only direct Top1-topotecan contact (Fig. 4B). The elimination or alteration of other amino acids shown in the figure would negatively affect drug binding (Fig. 4 A and B) (13). To obtain structural information, we constructed structural models of OpTop1 and CaTop1 based on HsTop1 by homology modeling (13). In both OpTop1 and CaTop1, the alteration of Asn-722 to Ser, which has a shorter side chain, leads to the elimination of the water-mediated contact, resulting in the prevention of CPT binding (Fig. 4C) (13). The difference in side chain conformation between Leu and Ile at position 530 could also disrupt CPT binding by shifting the position of Asp-533 that binds to CPT (Fig. 4D). The alteration of Gln-421 (HsTop1) to Lys-421 found in CaTop1 would eliminate the interaction between Gln-421 and His-367, resulting in the disruption of the interaction of Arg-364 and Asp-533 with CPT (Fig. 4E). The substitutions at positions 421 and 530 have thus far not been reported in CPT-resistant Top1s found in human cancer cells. However, because the period for which CPT production has evolved in plants is longer than the period of contact between CPT and human cancer cells, these mutations might occur in the future in human cancer cells resistant to CPT.
Fig. 4.
Schematic representation of the CPT-binding site in Top1. (A and B) Each chain of amino acid residues involved in CPT binding is shown in a different color. Hydrogen bonding or salt bridge interactions are indicated by black dotted lines, and van der Walls interactions are shown by gray dotted lines. Asn-722 forms a hydrogen bond via a water molecule (wat), and Asp-533 forms a direct contact with topotecan (shown in gray). Amino acid substitutions resulting in CPT resistance in CPT-producing plants are represented in red. (C) Superposition of the CPT-binding residue 722 of HsTop1 (blue) in a complex with topotecan and those of OpTop1 (yellow) and CaTop1 (magenta). (D and E) Superposition of the CPT-binding residue Asp-533 of HsTop1 (blue) and OpTop1 (yellow) (D) or CaTop1 (magenta) (E) by substitution Leu530Ile or Gln421Lys. Black dotted lines indicate hydrogen bonding in HsTop1, as indicated in A and B.
CPT Production System Has Coevolved with CPT-Resistant Top1 in the Producing Plants.
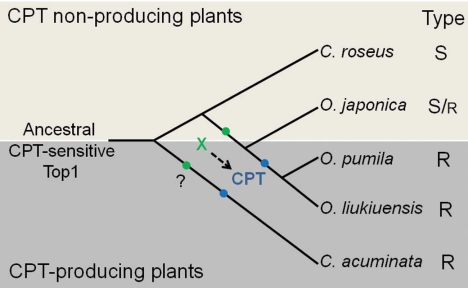
The self-resistance mechanism that develops by target mutation in CPT-producing plants leads us to propose the concept of adaptive coevolution between CPT biosynthesis and Top1. We raised additional questions regarding how CPT-producing plants ensure that they are primed for CPT resistance when the CPT biosynthetic pathway has successfully evolved and how CPT production does not interfere with their survival. The evolutionally primed resistance mechanism exhibited in bacterial antibiotic exporters (19) could also occur in CPT-producing plants, although the resistance mechanism is totally different. The in vivo CPT toxicity assay using the yeast mutants provides additional support to the concept of CPT pre-resistance of Top1. Among the yeast cells containing Top1s from CPT-nonproducing plants, the cells expressing OjTop1 showed better growth in the CPT-containing medium than those expressing either CrTop1 or AtTop1; this finding suggests that OjTop1 is partially resistant to CPT (Fig. 3A and Table S1). These results imply the involvement of additional OjTop1 mutations in conferring partial resistance to CPT. Taken together, we propose that an adaptive coevolution occurred between CPT biosynthesis and Top1 (Fig. 5). Before the speciation of Ophiorrhiza genus, Top1 was pre-adapted to be partially resistant to CPT by the effect of an unidentified compound (X), which might be an intermediate in CPT biosynthesis. This pre-adaptation would have allowed plants belonging to Ophiorrhiza genus, namely, O. pumila, O. liukiuensis, and O. japonica, to ensure a certain degree of CPT resistance before the CPT production was initiated. As soon as the CPT biosynthetic pathway successfully evolved in the species O. pumila and O. liukiuensis, Top1 mutated, i.e., Leu530Ile and Asn722Ser substitutions occurred, due to CPT, thereby rendering the plants completely resistant to CPT. Thus, only those plants that successfully evolved these mutations could maintain the production of CPT. The same scenario is also likely for C. acuminata.
Fig. 5.
Proposed scenario of adaptive coevolution between CPT biosynthesis and Top1. In this model, the pre-adaptation of Top1 for partial resistance occurred in Ophiorrhiza genus before speciation. Subsequently, mutations conferring complete resistance to CPT coevolved with CPT production. The green dots indicate a mutation event caused by an unidentified compound (“X”) that confers partial resistance to CPT. The blue dots indicate another mutation event caused by CPT that confers complete resistance to CPT. The dashed arrow indicates a pathway leading to CPT biosynthesis. The question mark indicates that the Top1 pre-adaptation event may have occurred in C. acuminata. S, CPT-sensitive Top1; S/r, partially CPT-resistant Top1; R, CPT-resistant Top1.
A modified alternative scenario can also be proposed. Small amounts of CPT produced in plants—at concentrations that would not affect the plant survival—might trigger the necessary mutation of Top1 resulting in increased CPT resistance of Top1. This would lead to higher production of CPT in the plants later. This scenario has been actually observed in CPT-resistant human cancer cells. Continuous exposure of cancer cell lines to CPT derivatives resulted in Top1 mutation conferring CPT resistance (20).
Conclusions
In contrast to the self-resistance mechanism in antibiotic-producing bacteria (21–23), self-resistance to toxic metabolites produced in plants has not been studied extensively despite the increasing use of plant-derived medicines. Our study revealed a target mutation-based mechanism of self-resistance to endogenous toxic metabolites in plants, thereby providing insights on a long-standing question in plant biochemistry about the evolution of plant secondary products. This self-resistance mechanism that developed via adaptive coevolution might generally be found in other plants that produce toxic secondary compounds inhibiting basic biological processes, such as antimitotic agents, including paclitaxel in Taxus brevifolia, podophyllotoxin in Podophyllum spp., vinblastine and vincristine in C. roseus, and colchicine in Colchicum autumnale. Although a point mutation of the tubulin-coding gene that confers paclitaxel resistance has been reported in human cancer cells (24, 25), no information on such mutation is available for plants producing paclitaxel. Understanding the self-resistance mechanism can provide useful information not only for developing the strategy of combinatorial biosynthesis and synthetic biology in plant cells but also for elucidating the molecular basis of drug resistance in the clinical field. Comparison of the target sites of the compounds in producing and nonproducing plants may be exploited for obtaining additional information on drug-target interaction and for designing advanced drugs to prevent resistance in cancer cells and combat drug resistance.
Materials and Methods
Plant Materials.
For RNA extraction, we used the hairy roots of O. pumila (26) and O. liukiuensis (27) and young leaves of O. japonica, C. acuminata, and C. roseus, which were cultivated in the Medicinal Plant Gardens of the Graduate School of Pharmaceutical Sciences at Chiba University.
cDNA Cloning.
The cDNA fragments encoding plant Top1s were isolated by degenerate primer PCR with GCIGTIGCIACITAYYTIATHGA as the forward primer and RCACCAIGCIACIGTDATIC as the reverse primer. The obtained sequences were extended in the 5′ and 3′ directions by rapid amplification of cDNA ends (RACE) PCR using the SMART RACE kit (Clontech). Other primers used in RACE are available in Table S2. The cDNA encoding OpTop1 was isolated from the EST library of O. pumila hairy roots.
Phylogenetic Analysis of Top1.
The encoded amino acid sequences were aligned with ClustalW implemented in the MEGA version 4 program (28) by using standard parameters (Fig. S3). A phylogenetic tree was constructed with the same program by using the neighbor-joining method. Bootstrap values were statistically calculated with the default sets of the MEGA program at 500 replicates and seed = 64,238. The protein sequences of other Top1s were retrieved from the National Center for Biotechnology Information. These included Top1s from Nicotiana tabacum (accession no. AAK69776), Pisum sativum (CAA74890), A. thaliana (NP200341), Homo sapiens (AAA61207), and S. cerevisiae (AAA35162).
Expression of Recombinant OpTop1 in E. coli and in Vitro Top1 Assay.
OpTop1s were cloned into pDONR221 by using the Gateway Cloning Technology (Invitrogen), and the integrity of the constructs was verified by DNA sequencing. The Gateway expression vector pDEST17, which has a 6× His N-terminal tag, was used for expression in E. coli. The plasmid was transformed into E. coli BL21-AI (Invitrogen). Cells were grown to a density of ≈0.6 and induced by using l-arabinose to a final concentration of 0.2% for 4 h at 30°C. The recombinant protein was purified by using a HisTrap HP column (Amersham). The plasmid pUC19 was used in the DNA supercoil relaxation activity assay as described in ref. 29.
Yeast Expression and Sensitivity to CPT.
S. cerevisiae strain RS190 (MATa, top1Δ) was purchased from American Type Culture Collection. The genes of plant Top1s were cloned by the Gateway Cloning Technology, and the integrity of the constructs was verified by DNA sequencing. The Gateway expression vector pYES-DEST52 was used for expression in S. cerevisiae. A CPT sensitivity assay was conducted by using yeast cells transformed with various Top1 constructs as described in ref. 14. Individual transformants were grown overnight in a selective medium containing glucose. The cultures were adjusted to an OD600 of 0.3 and serially diluted 10-fold; 5-μl aliquots were spotted onto selective plates supplemented with 2% glucose or galactose and 0 or 5 μg/ml CPT at a final Me2SO concentration of 0.25%.
Construction of Mutant cDNAs.
The mutant cDNAs (OjTop1, I420V; OjTop1, N421K; OjTop1, L530I; and OjTop1, N722S) were prepared by PCR-based mutagenesis (30). PCR was conducted with KOD DNA polymerase (Toyobo). The prepared mutant cDNAs were introduced into pDONR221 (Invitrogen) and sequenced.
Molecular Modeling.
Three-dimensional model structures of OpTop1 and CaTop1 were constructed with MODELLER (31), using the published HsTop1-DNA-topotecan complex (Protein Data Bank entry 1RRJ) as the template (13). Structural alignment figures were prepared with PyMOL (www.pymol.org).
Supplementary Material
Acknowledgments.
We thank T. Hoshino and H. Fuji for help with MODELLER and PyMOL, H. Sudo for the identification of O. japonica, and T. Nagata for providing tobacco BY-2 cells. This work was supported in part by the Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science and by CREST of the Japan Science and Technology Agency. S.S. was the recipient of a postdoctoral fellowship from the Japan Society for the Promotion of Science (hosted by M.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the DNA Data Bank of Japan, www.ddbj.nig.ac.jp. [accession nos. AB372508 (OpTop1), AB372509 (OlTop1), AB372510 (OjTop1), AB372511 (CaTop1), and AB372512 (CrTop1)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0801038105/DCSupplemental.
References
- 1.Benderoth M, et al. Positive selection driving diversification in plant secondary metabolism. Proc Natl Acad Sci USA. 2006;103:9118–9123. doi: 10.1073/pnas.0601738103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsiang YH, Hertzberg R, Hecht S, Liu LF. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 3.Porter SE, Champoux JJ. The basis for camptothecin enhancement of DNA breakage by eukaryotic topoisomerase I. Nucleic Acids Res. 1989;17:8521–8532. doi: 10.1093/nar/17.21.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Staker BL, et al. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc Natl Acad Sci USA. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi T, Matsuhara S, Abe M, Komeda Y. Disruption of a DNA topoisomerase I gene affects morphogenesis in Arabidopsis. Plant Cell. 2002;14:2085–2093. doi: 10.1105/tpc.001925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sirikantaramas S, Yamazaki M, Saito K. Mechanisms of resistance to self-produced toxic secondary metabolites in plants. Phytochem Rev. 2007 Nov 20; doi: 10.1007/s11101-007-9080-2. [DOI] [Google Scholar]
- 7.Rasheed ZA, Rubin EH. Mechanisms of resistance to topoisomerase I-targeting drugs. Oncogene. 2003;22:7296–7304. doi: 10.1038/sj.onc.1206935. [DOI] [PubMed] [Google Scholar]
- 8.Takatani H, et al. Gene mutation analysis and quantitation of DNA topoisomerase I in previously untreated non-small cell lung carcinomas. Jpn J Cancer Res. 1997;88:160–165. doi: 10.1111/j.1349-7006.1997.tb00361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sirikantaramas S, Sudo H, Asano T, Yamazaki M, Saito K. Transport of camptothecin in hairy roots of Ophiorrhiza pumila. Phytochemistry. 2007;68:2881–2886. doi: 10.1016/j.phytochem.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 10.Redinbo MR, Stewart L, Kuhn P, Champoux JJ, Hol WG. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 11.Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 12.Locato V, Balestrazzi A, De Gara L, Carbonera D. Reduced expression of top1beta gene induces programmed cell death and alters ascorbate metabolism in Daucus carota cultured cells. J Exp Bot. 2006;57:1667–1676. doi: 10.1093/jxb/erj194. [DOI] [PubMed] [Google Scholar]
- 13.Chrencik JE, et al. Mechanisms of camptothecin resistance by human topoisomerase I mutations. J Mol Biol. 2004;339:773–784. doi: 10.1016/j.jmb.2004.03.077. [DOI] [PubMed] [Google Scholar]
- 14.Fertala J, Vance JR, Pourquier P, Pommier Y, Bjornsti MA. Substitutions of Asn-726 in the active site of yeast DNA topoisomerase I define novel mechanisms of stabilizing the covalent enzyme-DNA intermediate. J Biol Chem. 2000;275:15246–15253. doi: 10.1074/jbc.275.20.15246. [DOI] [PubMed] [Google Scholar]
- 15.Fujimori A, Harker WG, Kohlhagen G, Hoki Y, Pommier Y. Mutation at the catalytic site of topoisomerase I in CEM/C2, a human leukemia cell line resistant to camptothecin. Cancer Res. 1995;55:1339–1346. [PubMed] [Google Scholar]
- 16.Bjornsti MA, Benedetti P, Viglianti GA, Wang JC. Expression of human DNA topoisomerase I in yeast cells lacking yeast DNA topoisomerase I: Restoration of sensitivity of the cells to the antitumor drug camptothecin. Cancer Res. 1989;49:6318–6323. [PubMed] [Google Scholar]
- 17.Eng WK, Faucette L, Johnson RK, Sternglanz R. Evidence that DNA topoisomerase I is necessary for the cytotoxic effects of camptothecin. Mol Pharmacol. 1988;34:755–760. [PubMed] [Google Scholar]
- 18.Kieber JJ, Tissier AF, Signer ER. Cloning and characterization of an Arabidopsis thaliana topoisomerase I gene. Plant Physiol. 1992;99:1493–1501. doi: 10.1104/pp.99.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tahlan K, et al. Initiation of actinorhodin export in Streptomyces coelicolor. Mol Microbiol. 2007;63:951–961. doi: 10.1111/j.1365-2958.2006.05559.x. [DOI] [PubMed] [Google Scholar]
- 20.Urasaki Y, et al. Characterization of a novel topoisomerase I mutation from a camptothecin-resistant human prostate cancer cell line. Cancer Res. 2001;61:1964–1969. [PubMed] [Google Scholar]
- 21.Cundliffe E. How antibiotic-producing organisms avoid suicide. Annu Rev Microbiol. 1989;43:207–233. doi: 10.1146/annurev.mi.43.100189.001231. [DOI] [PubMed] [Google Scholar]
- 22.Biggins JB, Onwueme KC, Thorson JS. Resistance to enediyne antitumor antibiotics by CalC self-sacrifice. Science. 2003;301:1537–1541. doi: 10.1126/science.1086695. [DOI] [PubMed] [Google Scholar]
- 23.Hopwood DA. How do antibiotic-producing bacteria ensure their self-resistance before antibiotic biosynthesis incapacitates them? Mol Microbiol. 2007;63:937–940. doi: 10.1111/j.1365-2958.2006.05584.x. [DOI] [PubMed] [Google Scholar]
- 24.Giannakakou P, et al. A common pharmacophore for epothilone and taxanes: Molecular basis for drug resistance conferred by tubulin mutations in human cancer cells. Proc Natl Acad Sci USA. 2000;97:2904–2909. doi: 10.1073/pnas.040546297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giannakakou P, et al. Paclitaxel-resistant human ovarian cancer cells have mutant β-tubulins that exhibit impaired paclitaxel-driven polymerization. J Biol Chem. 1997;272:17118–17125. doi: 10.1074/jbc.272.27.17118. [DOI] [PubMed] [Google Scholar]
- 26.Saito K, et al. Feasible production of camptothecin by hairy root culture of Ophiorriza pumila. Plant Cell Rep. 2001;20:267–271. [Google Scholar]
- 27.Asano T, et al. Camptothecin production by in vitro cultures of Ophiorrhiza liukiuensis and O. kuroiwai. Plant Biotechnol. 2004;21:275–281. [Google Scholar]
- 28.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 29.Reddy MK, Nair S, Tewari KK. Cloning, expression and characterization of a gene which encodes a topoisomerase I with positive supercoiling activity in pea. Plant Mol Biol. 1998;37:773–784. doi: 10.1023/a:1006086311875. [DOI] [PubMed] [Google Scholar]
- 30.Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: Study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.