Abstract
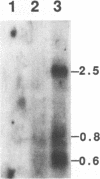
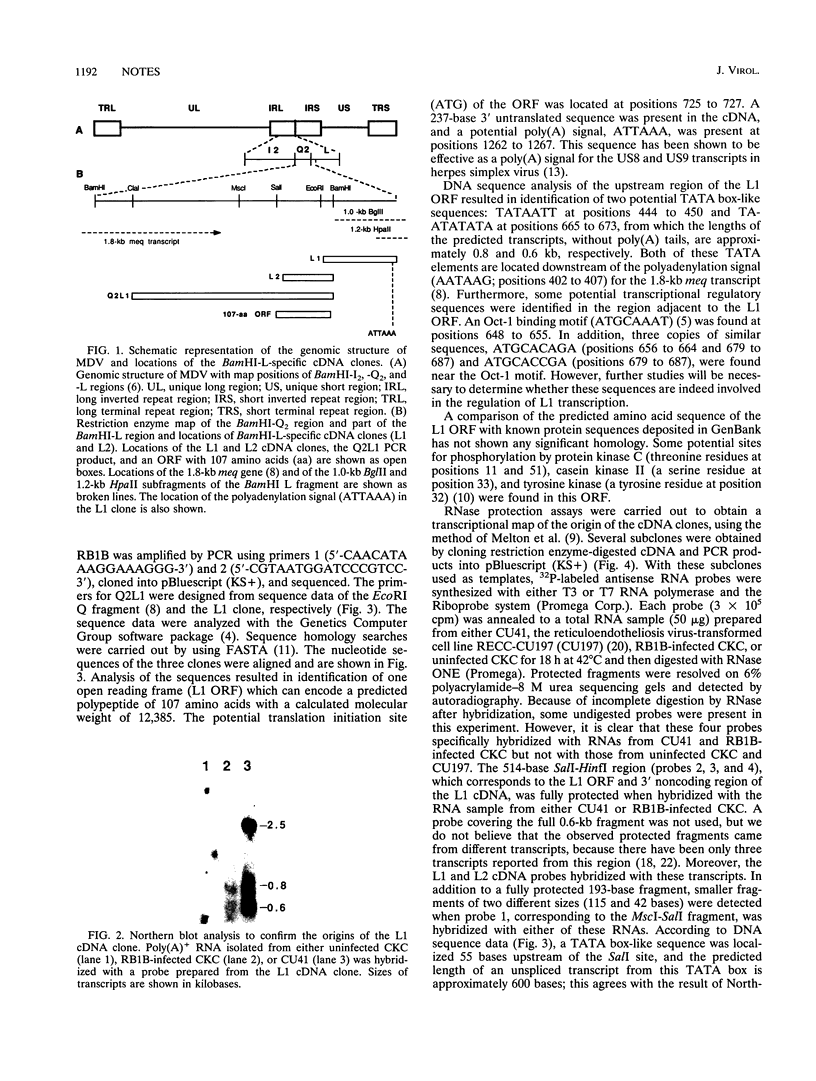
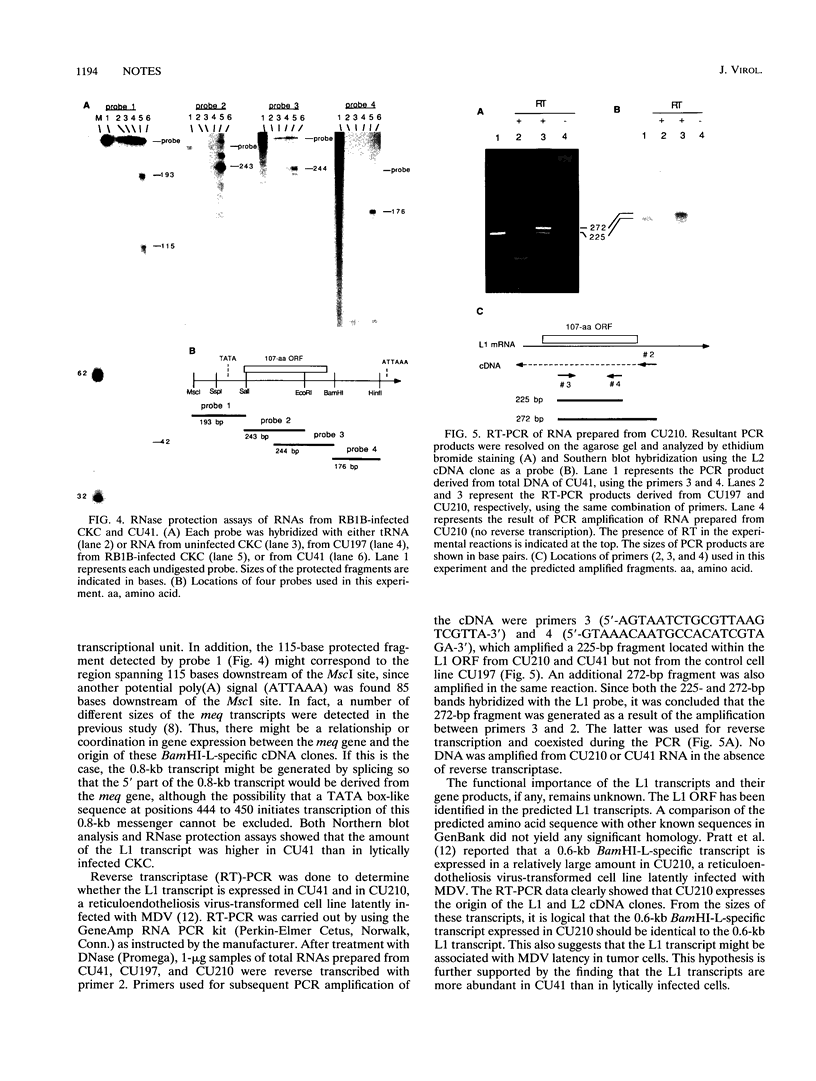
Two Marek's disease (MD) virus BamHI-L-specific cDNA clones were isolated from a cDNA library constructed from poly(A)+ RNA fractions of an MD lymphoblastoid cell line, MDCC-CU41 (CU41). These clones were mapped to the region corresponding to the BamHI-Q2 and L-regions. These clones hybridized with 2.5-, 0.8-, and 0.6-kb transcripts prepared from CU41. The transcriptional unit of the 0.6-kb transcript was determined by RNase protection assays. An open reading frame encoding a 107-amino-acid polypeptide was identified in the 0.6-kb transcript. Reverse transcriptase-PCR demonstrated the presence of this transcript in both CU41 and a reticuloendotheliosis virus-transformed cell line latently infected with MD virus.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson A. S., Francesconi A., Morgan R. W. Complete nucleotide sequence of the Marek's disease virus ICP4 gene. Virology. 1992 Aug;189(2):657–667. doi: 10.1016/0042-6822(92)90589-h. [DOI] [PubMed] [Google Scholar]
- Buckmaster A. E., Scott S. D., Sanderson M. J., Boursnell M. E., Ross N. L., Binns M. M. Gene sequence and mapping data from Marek's disease virus and herpesvirus of turkeys: implications for herpesvirus classification. J Gen Virol. 1988 Aug;69(Pt 8):2033–2042. doi: 10.1099/0022-1317-69-8-2033. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkner F. G., Mocikat R., Zachau H. G. Sequences closely related to an immunoglobulin gene promoter/enhancer element occur also upstream of other eukaryotic and of prokaryotic genes. Nucleic Acids Res. 1986 Nov 25;14(22):8819–8827. doi: 10.1093/nar/14.22.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi K., Sudo M., Lee Y. S., Tanaka A., Nonoyama M. Structure of Marek's disease virus DNA: detailed restriction enzyme map. J Virol. 1984 Jul;51(1):102–109. doi: 10.1128/jvi.51.1.102-109.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D., Lee L., Liu J. L., Kung H. J., Tillotson J. K. Marek disease virus encodes a basic-leucine zipper gene resembling the fos/jun oncogenes that is highly expressed in lymphoblastoid tumors. Proc Natl Acad Sci U S A. 1992 May 1;89(9):4042–4046. doi: 10.1073/pnas.89.9.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt W. D., Morgan R. W., Schat K. A. Characterization of reticuloendotheliosis virus-transformed avian T-lymphoblastoid cell lines infected with Marek's disease virus. J Virol. 1992 Dec;66(12):7239–7244. doi: 10.1128/jvi.66.12.7239-7244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rixon F. J., McGeoch D. J. Detailed analysis of the mRNAs mapping in the short unique region of herpes simplex virus type 1. Nucleic Acids Res. 1985 Feb 11;13(3):953–973. doi: 10.1093/nar/13.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross L. J., Binns M. M. Properties and evolutionary relationships of the Marek's disease virus homologues of protein kinase, glycoprotein D and glycoprotein I of herpes simplex virus. J Gen Virol. 1991 Apr;72(Pt 4):939–947. doi: 10.1099/0022-1317-72-4-939. [DOI] [PubMed] [Google Scholar]
- Sakaguchi M., Urakawa T., Hirayama Y., Miki N., Yamamoto M., Hirai K. Sequence determination and genetic content of an 8.9-kb restriction fragment in the short unique region and the internal inverted repeat of Marek's disease virus type 1 DNA. Virus Genes. 1992 Nov;6(4):365–378. doi: 10.1007/BF01703085. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schat K. A., Buckmaster A., Ross L. J. Partial transcription map of Marek's disease herpesvirus in lytically infected cells and lymphoblastoid cell lines. Int J Cancer. 1989 Jul 15;44(1):101–109. doi: 10.1002/ijc.2910440119. [DOI] [PubMed] [Google Scholar]
- Schat K. A., Pratt W. D., Morgan R., Weinstock D., Calnek B. W. Stable transfection of reticuloendotheliosis virus-transformed lymphoblastoid cell lines. Avian Dis. 1992 Apr-Jun;36(2):432–439. [PubMed] [Google Scholar]
- Silver S., Tanaka A., Nonoyama M. Transcription of the Marek's disease virus genome in a nonproductive chicken lymphoblastoid cell line. Virology. 1979 Feb;93(1):127–133. doi: 10.1016/0042-6822(79)90281-2. [DOI] [PubMed] [Google Scholar]
- Sugaya K., Bradley G., Nonoyama M., Tanaka A. Latent transcripts of Marek's disease virus are clustered in the short and long repeat regions. J Virol. 1990 Dec;64(12):5773–5782. doi: 10.1128/jvi.64.12.5773-5782.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]