Abstract
Protein acetylation has been implicated in the regulation of HIV-1 gene transcription. Here, we have exploited the activities of four native histone acetyltransferase (HAT) complexes from yeast to directly test whether acetylation regulates HIV-1 transcription in vitro. HAT activities acetylating either histone H3 (SAGA, Ada, and NuA3) or H4 (NuA4) stimulate HIV-1 transcription from preassembled nucleosomal templates in an acetyl CoA-dependent manner. HIV-1 transcription from histone-free DNA is not affected by the HATs, indicating that these activities function in a chromatin-specific fashion. For Ada and NuA4, we demonstrate that acetylation of only histone proteins mediates enhanced transcription, suggesting that these complexes facilitate transcription at least in part by modifying histones. To address a potential mechanism by which HAT complexes stimulate transcription, we performed a restriction enzyme accessibility analysis. Each of the HATs increases the cutting efficiencies of restriction endonucleases targeting the HIV-1 chromatin templates in a manner not requiring transcription, suggesting that histone acetylation leads to nucleosome remodeling.
Chromatin structures inhibit the binding and function of the numerous proteins that collaborate to produce appropriate levels of eukaryotic transcription (reviewed in ref. 1). It is quite significant, therefore, that recent discoveries have identified multiprotein complexes whose primary function is to help activate gene expression by altering chromatin so that its DNA sequences become more accessible to sequence-specific proteins and the general transcription machinery (reviewed in ref. 2). They provide solid support to the idea that the repressive effects of chromatin on transcription can be counteracted by cellular activities that directly modify nucleosomal structure.
Histone acetyltransferases (HATs) and histone deacetylases comprise a group of chromatin-modifying activities that plays a critical role in gene transcription (for reviews see refs. 3 and 4). Numerous studies have demonstrated a correlation between the acetylation of lysine residues within the amino terminal tails of the core histones and the transcriptional activity of cellular chromatin (reviewed in ref. 5). Hyperacetylated histones appear to accumulate in actively transcribed chromatin (6), whereas hypoacetylated histones are enriched in silent domains (7, 8). Recently, proteins that initially were identified as transcriptional regulators in vivo have been shown to possess HAT or histone deacetylase activity. The initial breakthrough came with the cloning of a nuclear HAT from Tetrahymena, leading to its identification as a homologue of the yeast transcriptional coactivator protein Gcn5 (9). HAT activity since has been demonstrated for several other coactivators, which, as members of protein complexes, are believed to facilitate transcriptional activation by sequence-specific activators. These include p300/CBP (10, 11), pCAF (12), ACTR (13), src-1 (14), and TAFII250 (15). Similarly, histone deacetylases have been shown to associate with corepressor complexes that in turn interact with DNA-bound proteins such as Mad-Max, Ume6, or unliganded receptors (16–22). As a whole, these data provide compelling evidence directly linking pathways of transcriptional regulation with histone acetylation.
Previously, we identified four native HATs from yeast (23). For the complexes termed SAGA and Ada, Gcn5 is the primary, if not only, subunit possessing catalytic HAT activity. In addition, both the 1.8-MDa SAGA complex and 0.8-MDa Ada complex contain Ada2 and Ada3, gene products originally determined to functionally interact with Gcn5 based on genetic screens in yeast for mutants that relieved GAL4-VP16-mediated toxicity (24, 25). SAGA also contains Spt20 (Ada5), Spt3, Spt7, and Spt8, which are members of a family of transcriptional regulators thought to affect TATA box-binding protein function (reviewed in ref. 26), and a subset of TAFII proteins including TAFII90, TAFII68, TAFII60, TAFII25, and TAFII20 (27). SAGA and Ada primarily acetylate nucleosomal histone H3, but also modify H2B to a lesser extent. The additional two HAT activities, termed complex 2 and complex 3, predominantly acetylate histones H4 and H3, respectively. As the subunit composition for these complexes is unknown, they have been named NuA4 (nucleosome acetyltransferase of histone H4) and NuA3 according to their acetylation preferences.
In this study, we attempt to gain a better understanding of the role(s) played by histone acetylation in gene transcription by directly testing whether SAGA, Ada, NuA4, or NuA3 can regulate transcription from nucleosome-assembled templates in vitro. The HIV-1 5′ long terminal repeat was chosen as the experimental promoter, because a clear correlation between acetylation and HIV-1 transcriptional activation has been determined by inducing global hyperacetylation in cells (28), or extracts (29), by treatment with the histone deacetylase inhibitor trichostatin A. We demonstrate that SAGA, Ada, NuA4, and NuA3 stimulate HIV-1 transcription from chromatin templates in an acetyl CoA-dependent fashion. In the absence of transcription, each of the HATs increases access to the nucleosome-reconstituted array for restriction endonucleases. These data suggest a mechanism whereby acetylation leads to remodeling of the HIV-1 nucleosomal array, increasing the access of trans-acting factors, which in turn facilitates transcriptional activation.
MATERIALS AND METHODS
Preparation of the HIV Dinucleosome-5S Array and Nucleosome Reconstitution.
The HIV dinucleosome-5S array was constructed by subcloning an HIV-1 fragment from −225 to +162 into the XhoI sites of pIC-2085S (30), producing a 416-bp insert between the fifth and sixth 5S ribosomal DNA repeats. The resulting plasmid, termed p2085S-HIV(F+), was digested with Asp-718, ClaI, and HhaI, and the array gel was isolated as previously described (30).
For nucleosome reconstitution, 2 μg of the array fragment was incubated with HeLa core histones (1:1 molar ratio of octamers to nucleosomal sites on the DNA), 1 μg of BSA, and 2 M NaCl in a final volume of 10 μl for 15 min at 37°C. The reaction was serially diluted by adding 3.3, 6.7, 5, 3.6, 4.7, 6.7, 10, 30, and 20 μl of 50 mM Hepes (pH 7.5), 1 mM EDTA, 5 mM DTT, and 0.5 mM phenylmethylsulfonyl fluoride (PMSF), with 15-min incubations at 30°C for each dilution step. The reaction was brought to 0.1 M NaCl by adding 100 μl of 10 mM Tris⋅HCl (pH 7.5), 1 mM EDTA, 0.1% Nonidet P-40, 5 mM DTT, 0.5 mM PMSF, 20% glycerol, and 100 μg/ml of BSA, and incubated for 15 min at 30°C. Reconstitutions were stored at 4°C. Structural analyses of array reconstitutions was performed as previously described (30).
HAT Assays and in Vitro Transcription Reactions.
The HIV dinucleosome-5S array template was incubated with purified transcription factors and/or HAT activities for 30 min at 30°C in 20 μl of binding/HAT reaction buffer (10 mM Hepes, pH 7.8/50 mM KCl/5 mM DTT/0.5 mM phenylmethylsulfonyl fluoride/10 mM sodium butyrate/0.25 mg/ml of BSA/5% glycerol). Acetyl CoA was added as indicated in the figure legends. Upon completion of factor binding and histone acetylation, 25 μl of a transcription mix [30 mM Hepes (pH 7.8), 12 mM MgCl2, 60 mM KCl, 10 mM sodium butyrate, 4% polyvinyl alcohol, 12 ng/μl poly{d(I-C)}, 2 ng/μl of E4 control DNA, 36 nM GAL4-AH] was added to each reaction, followed by 5 μl of HeLa nuclear extract (15 μg/μl) (except as indicated in Fig. 4C). E4 DNA and GAL4-AH are included to serve as a control for RNA recovery through subsequent steps. Preinitiation complexes were allowed to form for 20 min at room temperature, and the templates were transcribed for 30 min at 30°C upon the addition of rNTPs to 0.4 mM for each nucleotide. Transcription was terminated by adding 150 μl of stop buffer (267 mM NaCl/20 mM EDTA/1% SDS/33.3 ng/μl of tRNA) to each reaction. Primer extension analysis of the RNA produced during these reactions was performed by using 32P-labeled HIV-1 and E4 primers from positions +50 to +81 and +86 to +110, respectively. Extension products were resolved on 8% polyacrylamide, 8 M urea gels, visualized by autoradiography, and quantitated after PhosphorImager scanning. pG5-E4T was used for the E4 control DNA, which contains 5-GAL4-binding sites upstream of the Ad5 E4 gene sequence from −38 to +250 (31). GAL4-AH and the NF-κB p50 subunit were prepared from bacterial strains as described previously (30).
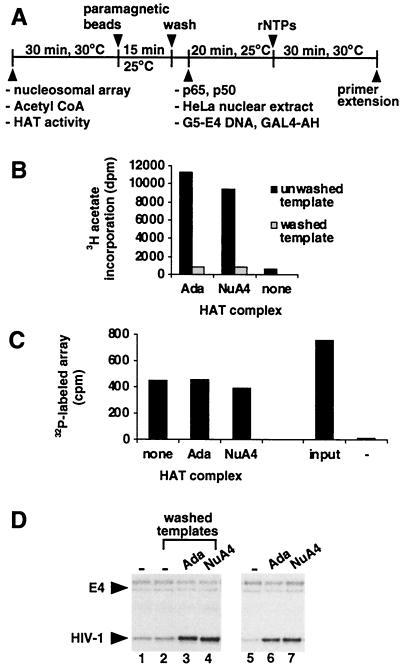
Figure 4.
Acetylation of histones by Ada and NuA4 stimulates HIV-1 transcription. (A) Schematic representation of the transcription protocol using immobilized HIV dinucleosome-5S array templates. (B) Nucleosome-assembled array DNA (10 ng) was treated with the indicated HAT in the presence of acetyl CoA (1 μM) and immobilized onto paramagnetic beads (10 μg). Parallel reactions were either washed or left unwashed, and HAT activity was determined by adding HeLa nucleosomes (1 μg) and (3H)-acetyl CoA (0.25 μCi) and measuring the incorporation of 3H acetate into nucleosomes. (C) 32P-labeled nucleosomal templates were treated with the indicated HAT, immobilized onto beads, and washed as in B. Immobilized templates were detected by scintillation counting. Samples not concentrated on the magnet also are shown. These contain paramagnetic beads with (input) and without (−) the nucleosomal array. (D) Transcription from acetylated and unacetylated immobilized templates was analyzed by adding Ada, NuA4, or no HAT (−). Transcription reactions using washed, immobilized templates are shown in lanes 2–4, and results from unwashed, immobilized templates are shown in lanes 1 and 5–7.
For experiments using immobilized templates, the biotinylated array fragment was attached to Dynabeads Streptavidin (i.e., paramagnetic beads coupled to streptavidin, Dynal). Immobilized templates were washed two times with 40 μl of binding/HAT buffer supplemented to 300 mM KCl, followed by one wash with 40 μl of binding/HAT buffer to remove excess KCl. Washed templates were resuspended in 20 μl of binding/HAT buffer. Neither the wash nor resuspension buffers contained acetyl CoA. For unwashed samples, immobilized templates were concentrated on the magnet, the supernatant was removed, and the beads put back into the original buffer conditions.
Restriction Endonuclease Accessibility Analysis.
Assays were performed similarly to that described for in vitro transcription through the 30-min incubation with nucleotides. After this step, restriction enzymes were added directly to samples and incubated for 15 min at 37°C. Cutting of the 32P-end labeled array was terminated by adding 150 μl of stop buffer to each reaction. Digestion products were resolved by agarose gel electrophoresis. Gels were fixed with 10% acetic acid and 10% methanol, dried, and quantitated after PhosphorImager scanning. Percentage cut was calculated by dividing the value of the digested fragment by the sum of the undigested and digested fragment values for each lane.
Purification of HATs.
HATs were purified as previously described (32). The amounts of Ada, NuA4, NuA3, and SAGA fractions used for the experiments were normalized for HAT activity such that each fraction produced 3,000–6,000 cpm in a standard liquid HAT assay. Liquid HAT assays were performed as described (23).
RESULTS AND DISCUSSION
Establishment of an in Vitro Transcription System with HIV-1 Nucleosomal Templates.
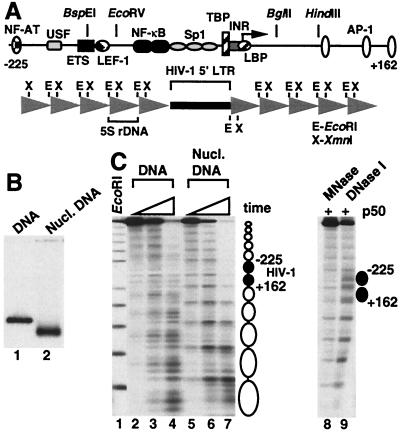
By analyzing cell lines latently infected with HIV-1, Verdin et al. (33) have determined that the HIV-1 genome adopts positioned nucleosomes after integration into the cellular DNA. A MNase-protected region termed nuc-1 resides at −2 to +142, whereas a 250-bp region directly upstream of nuc-1 containing the enhancer and promoter domains is constitutively hypersensitive to DNase I yet resistant to MNase activity. In light of that study, we designed a template containing an HIV-1 fragment from −225 to +162 that can be efficiently assembled from purified components into a nucleosomal array and subsequently used for in vitro transcription experiments. The template contains the HIV-1 fragment flanked on both sides by direct repeats of a 208-bp sequence from a sea urchin 5S rRNA gene (Fig. 1A). This strategy is based on the ability of the 5S rDNA segment to strongly position a histone octamer in vitro such that histone reconstitution of direct repeats of 5S rDNA yields a continuous array of evenly spaced nucleosomes (34). From our previous studies, chromatin assembly of DNA fragments containing target sequences embedded between 5S rDNA repeats has revealed that the 5S nucleosomes promote the positioning of histone octamers on the internal sequences in phase with the neighboring nucleosomes (30, 35).
Figure 1.
Chromatin structure of the HIV dinucleosome-5S array. (A) Schematic representation of the array. A 416-bp insert including HIV-1 sequence from −225 to +162 is flanked on both sides by five direct repeats of a 208-bp 5S ribosomal DNA nucleosome-positioning sequence. (B) Histone-free (DNA) and nucleosome-reconstituted (nucl. DNA) array templates were electrophoresed through a 1.2% agarose gel. (C) 32P-end labeled templates from B were treated with MNase, and the digestion products were resolved by agarose gel electrophoresis (lanes 2–7). For each quantity of MNase assayed, samples were incubated for 0.33-, 1-, and 3-min. EcoRI digestion of the array under limiting conditions is shown in lane 1. Nucleosome positions are represented schematically on the right. In lanes 8 and 9, NF-κB p50 subunit (200 nM) was bound to the array before nuclease digestion.
The transcription template was termed HIV dinucleosome-5S array because the central HIV-1 sequence is large enough to accommodate two histone octamers. This template migrates as a discrete band on agarose gels when reconstituted with histones into nucleosomal DNA, with a mobility distinct from that of histone-free DNA (Fig. 1B). The apparent absence of free DNA in the reconstituted sample indicates that nucleosome assembly occurs efficiently under these conditions. MNase, which preferentially cleaves chromatin in linker DNA, predominantly digests the nucleosome-assembled array at 11 distinct sites to produce a ladder of bands upon gel electrophoresis, whereas MNase digestion of the naked DNA occurs throughout the template (Fig. 1C, lanes 2–7). This finding suggests that 12 positioned nucleosomes occupy the regions between the MNase-cleavage sites, with the central two mapping to the HIV-1 promoter insert. Addition of the p50 subunit of NF-κB to the array does not dramatically alter the MNase digestion pattern, but does increase the level of cutting by DNase I within the nucleosome harboring the NF-κB-binding sites (Fig. 1C, lanes 8 and 9). This observation has been noted in an earlier study (30) and suggests that the binding of NF-κB p50 to the array generates a structure that is reflective of the native chromatin organization for HIV-1 DNA in the region between −225 and +162.
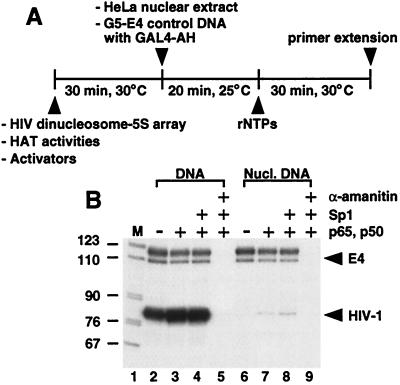
To investigate whether the HIV-1 promoter could direct transcription within the context of the array template, the HIV dinucleosome-5S fragment was incubated with HeLa nuclear extract in the presence and absence of purified activators (Fig. 2). Histone-free array DNA yields high levels of correctly sized HIV-1 transcripts, and, as expected, assembly of the array template into chromatin greatly represses transcription. Compared with naked DNA, the level of transcription directed from the nucleosomal array is reduced approximately 200-fold (Fig. 2B, compare lanes 2 and 6). The NF-κB subunits p65 and p50 stimulate transcription from the nucleosome-reconstituted array 4-fold, whereas addition of Sp1 above that provided by the extract has only a minimal effect. α-Amanitin blocks transcription from both the histone-free and nucleosome-assembled templates, demonstrating that the observed transcripts are properly derived from RNA polymerase II. The data suggest that Sp1 and NF-κB cannot effectively counteract nucleosome-mediated repression when added to preassembled chromatin templates, which is consistent with an earlier study (29). We conclude that these results, combined with the structural studies mentioned above, indicate that the reconstituted array is an appropriate substrate to examine HIV-1 transcription from nucleosomal templates.
Figure 2.
(A) Schematic representation of the in vitro transcription protocol. (B) Activators were incubated with the HIV dinucleosome-5S array (10 ng), present as either histone-free or nucleosomal DNA, and reactions were assayed for transcription. The p65 and p50 subunits of NF-κB each were added to 20 nM. Sp1 was added to 23 nM. Where indicated, α-amanitin was added before nuclear extract to a final concentration of 1 μM. Lane 1 contains DNA size markers.
HAT Complexes Stimulate HIV-1 Transcription in a Chromatin-Specific Manner.
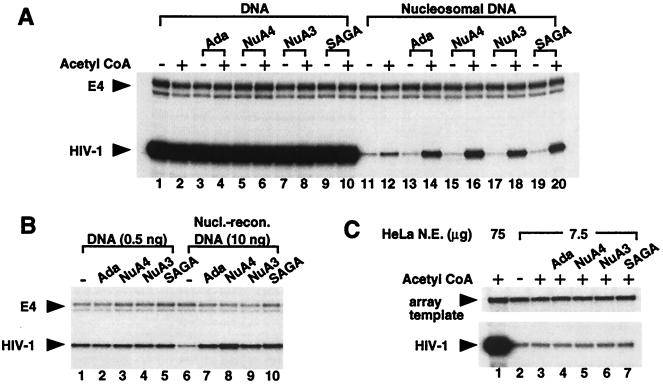
To investigate whether HIV-1 transcription is directly regulated by acetylation, the array template was incubated with either SAGA, Ada, NuA4, or NuA3, and the reactions were passed through the transcription assay. Under these conditions, the HATs acetylate the array in a manner consistent with predetermined histone specificities (data not shown). Fig. 3A reveals that each HAT stimulates the synthesis of HIV-1 RNA in an acetyl CoA-dependent manner from the nucleosome-reconstituted array (lanes 13–20). When comparing adjacent lanes containing the same HAT complex with and without acetyl CoA, the fold increase in transcription ranged from seven for the Ada complex to 12 for NuA4. Although not unexpected, addition of only acetyl CoA to the HeLa nuclear extract induced HIV-1 transcription nearly 2-fold (Fig. 3A, lanes 11 and 12). This effect is likely caused by endogenous acetyltransferases within the extract. No increase in transcription is observed from histone-free DNA in the presence of the HATs (Fig. 3A, lanes 1–10), suggesting that the HAT-mediated transcriptional stimulation is chromatin specific.
Figure 3.
HATs stimulate HIV-1 transcription from chromatin templates. (A) The HIV dinucleosome-5S array fragment (10 ng), existing as either histone-free or nucleosomal DNA, was incubated with and without acetyl CoA (1 μM) and the indicated HATs, and samples were assayed for transcription. NF-κB p65 and p50 each were added to 20 nM to enhance HIV-1 transcription levels. (B) Transcription was performed as described in A, except the amount of HIV-1 template was decreased from 10 ng to 0.5 ng where indicated. Acetyl CoA is present in all lanes. (C) Transcription from histone-free HIV-1 DNA was performed as in A, except the amount of HeLa nuclear extract was decreased from 75 μg to 7.5 μg where indicated. Because 7.5 μg of HeLa extract did not produce detectable RNA signals for the E4 control promoter, the 32P-labeled array template, which migrated high in the gel, was used as an internal control for nucleic acid recovery.
It is possible that HATs only affect conditions yielding low levels of transcription regardless of the chromatin state of the template. For example, maximal stimulation of naked DNA transcription by activators has been shown to occur in nuclear extracts at low template concentrations that yield low basal levels of expression (36). At low template concentrations, it is believed that activators function in part to relieve the repression established by nonspecific DNA-binding proteins present in the nuclear extract. We examined this issue by decreasing the amount of naked DNA 20-fold in the reactions. Consistent with the results above, the HATs did not stimulate transcription from DNA templates in these conditions (Fig. 3B).
It also can be argued that a regulatory protein(s) in the nuclear extract is limiting for transcription from nucleosomal templates, because access to the DNA is inhibited by histones, whereas this factor is not limiting for naked DNA transcription. Furthermore, if the HATs function to enhance the activity of the regulator, their influence on transcription would be observed only on nucleosomal templates. Therefore, to investigate this possibility, we decreased the amount of nuclear extract 10-fold, yet left the amount of histone-free DNA unchanged. HIV-1 transcript levels in these conditions were decreased significantly, indicating that the extract was limiting for transcription on naked DNA templates (Fig. 3C, compare lane 1 with lanes 2 and 3). However, the addition of HATs did not change the level of HIV-1 transcription (lanes 4–7). We conclude from these experiments that the stimulation of HIV-1 transcription by each of the HATs occurs only on chromatin templates.
Histone Acetylation by Ada and NuA4 Facilitates Transcription of the HIV-1 Promoter.
Although Ada, NuA4, NuA3, and SAGA acetylate nucleosomal histones, the data to this point do not demonstrate whether this modification is important to stimulate transcriptional activation. Indeed, the critical step for chromatin-specific regulation by these HATs could involve acetylation of a nonhistone protein(s) present within the HeLa nuclear extract. For example, in addition to histones, p300 will acetylate the tumor suppressor protein, p53, thereby enhancing its DNA-binding activity in vitro (37). Furthermore, components of the general transcription machinery have been demonstrated to be acetylated by both p300 and pCAF (38).
To address this idea, we sought conditions that would enable the removal of HAT activities from acetylated templates before the addition of nuclear extract for transcription. To this end, the HIV dinucleosome-5S array was attached to paramagnetic beads, which allows immobilized templates to be concentrated on a magnet and the supernatant removed (Fig. 4A). After incubation of the nucleosome-assembled array with each of the HATs, attachment of the acetylated array templates to paramagnetic beads, and separation of the beads from the supernatants, we found that all of the bead fractions contained approximately 50% of the input HAT activity (data not shown). Controls demonstrated that the HATs were specifically interacting with the nucleosomal array as opposed to the paramagnetic beads. Therefore, to challenge array-HAT interactions, immobilized templates were washed with buffers containing increasing salt concentrations. As shown in Fig. 4B, a 300 mM KCl wash reduced HAT activity to background levels for Ada and NuA4, indicating that this treatment removes the HATs from acetylated templates. We performed a second control to determine whether the acetylation status of the array affects attachment to the paramagnetic beads, because the array was treated with HATs before bead immobilization to ensure efficient acetylation. Fig. 4C illustrates that approximately 50% of the input array template was immobilized onto the paramagnetic beads under the experimental conditions and that template recovery was not significantly altered after acetylation by Ada and NuA4.
Immobilized templates acetylated by Ada or NuA4, and washed clean of HAT activity, produced RNA levels 2.5-fold higher than that from unacetylated templates when analyzed for transcription (Fig. 4D, compare lanes 3 and 4 with lane 2). Although lower than the fold stimulations reported for templates existing free in solution, these values must be considered within the context of control reactions using unwashed, immobilized templates. In these conditions, where the yeast HATs are present throughout the transcription assay, Ada and NuA4 stimulate transcription 3.5-fold and 4-fold, respectively (Fig. 4D, compare lanes 6 and 7 with lane 5). It appears, therefore, that the ability of Ada and NuA4 to stimulate transcription from immobilized templates is reduced when compared with that for templates existing free in solution. However, the experiment indicates that histone acetylation accounts for the majority of the HAT-mediated stimulation from immobilized templates. Thus, although not ruling out the possibility that there are other nonhistone targets for Ada and NuA4, the data suggest a role for histone acetylation in transcriptional regulation by these HATs.
Information pertaining to SAGA and NuA3 was not obtained from this assay, because these activities confer little or no induction of HIV-1 transcription from immobilized templates under all conditions tested. We can only speculate as to why SAGA and NuA3 stimulate transcription in an acetyl CoA-dependent manner from chromatin templates existing free in solution but not from those attached to paramagnetic beads. For example, in addition to their acetylation activities, SAGA and/or NuA3 may require other functions to facilitate transcription. SAGA has been proposed to coactivate transcription by bridging upstream regulators with the general transcription machinery. If SAGA or any of the other HATs work in this manner, it is conceivable that attachment of the nucleosomal template to paramagnetic beads could inhibit these large protein complexes from interacting with both DNA-bound activators to the basal transcription apparatus. It is also possible that the lysine residues modified by SAGA and NuA3 contribute to transcriptional differences. For example, although Ada and SAGA share the same catalytic subunit, the two native HAT complexes have overlapping, yet distinct, patterns of acetylation (P.A.G., A.E., S.J., and J.L.W., unpublished work). Of course, other possibilities exist. A more definitive determination as to how SAGA and NuA3 function in transcriptional regulation awaits the development of new approaches.
HATs Induce Remodeling of the Nucleosomal Array Structure in the Absence of Transcription.
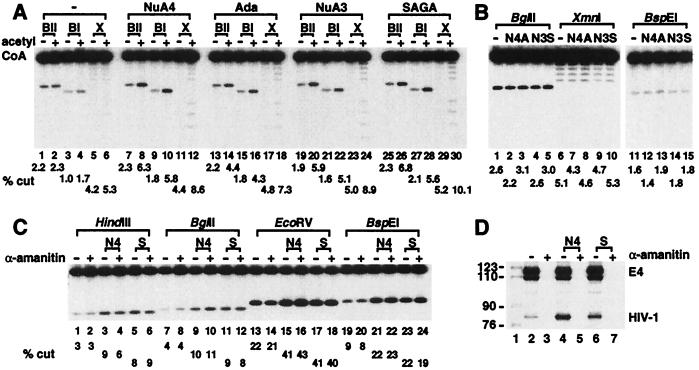
A possible mechanism through which histone acetylation enhances gene transcription involves altering chromatin structure in a manner that increases the accessibility of trans-acting factors for DNA. Indeed, it recently has been demonstrated that nucleosomal templates assembled in vitro with hyperacetylated histones are bound to a greater extent by heat shock factor compared with templates reconstituted with control histones (39). Restriction enzymes often are used to detect changes in chromatin structure because cutting efficiencies for sites buried in nucleosomes usually are reduced compared with those for sites in naked DNA (40). Therefore, to address whether array structure is changed by HAT function, nucleosomal array templates were passed through the transcription conditions. However, rather than subjected to primer extension analysis, samples were treated with restriction endonucleases. As shown in Fig. 5A, each of the HATs increases the cutting efficiencies of restriction endonucleases only in the presence of acetyl CoA. The effect is specific to chromatin because the HATs do not alter digestion of the naked DNA template (Fig. 5B).
Figure 5.
HATs increase access to the nucleosomal array for restriction endonucleases in the absence of transcription. (A) Nucleosome-reconstituted array DNA (10 ng) was incubated with and without acetyl CoA (1 μM) and the indicated HATs. After the addition of nuclear extract and a 30-min incubation with ATP (0.4 mM), samples were treated with BglII (10 units, BII), BspEI (10 units, BI), and XmnI (6 units, X). See Fig. 1A for schematic representation of site locations (B) Histone-free array DNA (10 ng) was treated with NuA4 (N4), Ada (A), NuA3 (N3), and SAGA (S) in the presence of acetyl CoA as in A, however the added amount of each restriction enzyme was reduced 10-fold compared with A. (C) The nucleosomal array was treated with NuA4 (N4) or SAGA (S) in the presence of acetyl CoA, and after the addition of nuclear extract and rNTPs, treated with HindIII (10 units), BglII, EcoRV (20 units), and BspEI. Where indicated, α-amanitin was added before nuclear extract to a final concentration of 1 μM. The sizes of restricted fragments are different from those in A and B because the array was 32P-labeled on the opposite end in this experiment. (D) Parallel reactions from C were passed through primer extension analysis rather than treated with restriction enzymes to verify that transcription was blocked by α-amanitin.
We believe the enhanced accessibility reflects an altered array structure generated in the absence of transcription, because these experiments were performed only with ATP rather than the full complement of rNTPs. In these conditions, we have not been able to detect HIV-1 transcripts from nucleosomal or naked DNA templates (data not shown). To further explore this point, the restriction enzyme accessibility assay was performed in the presence of the RNA polymerase II inhibitor α-amanitin. Fig. 5C illustrates that HAT-mediated increases in restriction enzyme cutting are not affected by α-amanitin, consistent with the idea that acetylation by the HAT complexes facilitates chromatin remodeling of the nucleosomal array independent of transcription.
In vivo a role for acetylation in HIV-1 gene expression has been demonstrated by treating cells latently infected with HIV-1 with histone deacetylase inhibitors. In addition to producing global hyperacetylation of cellular histones, the deacetylase inhibitors cause transcriptional activation of the HIV-1 promoter and reconfiguration of nuc-1 independent of ongoing transcription (28). The in vitro data presented here strengthen these in vivo observations. They provide direct evidence that native protein complexes acetylating nucleosomal histones stimulate HIV-1 transcription from chromatin templates. Moreover, they indicate that acetylation by the HAT complexes induces a change in chromatin structure that renders nucleosomal arrays more accessible to trans-acting factors.
Acknowledgments
We are grateful to Zhaodan Cao of Tularik for providing recombinant p65 prepared from vaccinia virus-infected HeLa cells. We thank members of the Workman lab for many helpful discussions. Support for this work was provided by a grant to J.L.W. from the National Institute of General Medical Sciences. D.J.S. was supported by a postdoctoral fellowship from the Cancer Research Institute. A.E. is a recipient of a postdoctoral fellowship from the Austrian Science Foundation (Fonds zur Förderung der Wissenschaftlichen Forschung). P.A.G. is funded by postdoctoral fellowship PF-98-017-01-GMC from the American Cancer Society. J.L.W. is an Associate Investigator of the Howard Hughes Medical Institute.
ABBREVIATION
- HAT
histone acetyltransferase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1. Owen-Hughes T A, Workman J L. Crit Rev Eukaryotic Gene Expression. 1994;4:403–441. [PubMed] [Google Scholar]
- 2.Workman J L, Kingston R E. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 3.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 4.Wade D A, Pruss D, Wolffe A P. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 5.Turner B M, O’Neill L P. Semin Cell Biol. 1995;6:229–236. doi: 10.1006/scel.1995.0031. [DOI] [PubMed] [Google Scholar]
- 6.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 8.Jeppesen P, Turner B M. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 9.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 10.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 11.Bannister A J, Kouzarides T. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 12.Yang X-J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 13.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 14.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M-J, O’Malley B W. Nature (London) 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 15.Mizzen C A, Yang X-J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J L, Wang L, Berger S L, Kouzarides T, et al. Cell. 1997;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 16.Heinzel T, Lavinsky R, Mullen T-M, Soderstrom M, Laherty C, Torchia W-M, Glass C, Rosenfeld M G. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 17.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Nature (London) 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 18.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 19.Laherty C D, Yang W-M, Sun J-M, Davie J R, Seto E, Eisenman R N. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 21.Kadosh D, Struhl K. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 22.Nagy L, Kao H-Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 23.Grant P A, Duggan L, Côté J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, et al. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 24.Berger S L, Pina B, Silverman N, Marcus G A, Agapite J, Reigier J L, Triezenberg S J, Guarente L. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 25.Piña B, Berger S, Marcus G A, Silverman N, Agapite J, Guarente L. Mol Cell Biol. 1993;13:5981–5989. doi: 10.1128/mcb.13.10.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winston F, Carlson M. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 27.Grant P A, Schieltz D, Pray-Grant M G, Steger D J, Reese J C, Yates J R, Workman J L. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 28.Van Lint C, Emiliani S, Ott M, Verdin E. EMBO J. 1996;15:1112–1120. [PMC free article] [PubMed] [Google Scholar]
- 29.Sheridan P L, Mayall T P, Verdin E, Jones K A. Genes Dev. 1997;11:3327–3340. doi: 10.1101/gad.11.24.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steger D J, Workman J L. EMBO J. 1997;16:2463–2472. doi: 10.1093/emboj/16.9.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin Y S, Carey M F, Ptashne M, Green M R. Cell. 1988;54:659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- 32.Utley R T, Ikeda K, Grant P A, Côté J, Steger D J, Eberharter A, John S, Workman J L. Nature (London) 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 33.Verdin E, Paras P J, Van Lint C. EMBO J. 1993;12:3249–3259. doi: 10.1002/j.1460-2075.1993.tb05994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simpson R T, Thoma F, Brubaker J M. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- 35.Owen-Hughes T, Workman J L. EMBO J. 1996;15:4702–4712. [PMC free article] [PubMed] [Google Scholar]
- 36.Abmayr S M, Workman J L, Roeder R G. Genes Dev. 1988;2:542–553. doi: 10.1101/gad.2.5.542. [DOI] [PubMed] [Google Scholar]
- 37.Gu W, Roeder R G. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 38.Imhof A, Yang X-J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 39.Nightingale K P, Wellinger R E, Sogo J M, Becker P B. EMBO J. 1998;17:2865–2876. doi: 10.1093/emboj/17.10.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gross D S, Garrard W T. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]