Summary
Stabilization of actin filaments is critical for supporting actomyosin-based contractility and for maintaining stable cellular structures. Tropomyosin is a well-characterized ubiquitous actin stabilizer that inhibits ADF/cofilin-dependent actin depolymerization. Here, we show that UNC-87, a calponin-related Caenorhabditis elegans protein with seven calponin-like repeats, competes with ADF/cofilin for binding to actin filaments and inhibits ADF/cofilin-dependent filament severing and depolymerization in vitro. Mutations in the unc-87 gene suppress the disorganized actin phenotype in an ADF/cofilin mutant in the C. elegans body wall muscle, supporting their antagonistic roles in regulating actin stability in vivo. UNC-87 and tropomyosin exhibit synergistic effects in stabilizing actin filaments against ADF/cofilin, and direct comparison reveals that UNC-87 effectively stabilizes actin filaments at much lower concentrations than tropomyosin. However, the in vivo functions of UNC-87 and tropomyosin appear different, suggesting their distinct roles in the regulation of actomyosin assembly and cellular contractility. Our results demonstrate that actin binding via calponin-like repeats competes with ADF/cofilin-driven cytoskeletal turnover, and is critical for providing the spatiotemporal regulation of actin filament stability.
Keywords: Actin dynamics, ADF/cofilin, Calponin, Tropomyosin
Introduction
Actin filaments are intrinsically dynamic cytoskeletal structures. At the cellular level, this dynamic behavior is enhanced or suppressed by a number of actin regulatory proteins (Pollard and Borisy, 2003). Actin assembly is accelerated by actin nucleation factors, such as the Arp2/3 complex (Goley and Welch, 2006) and members of the formin family (Higgs, 2005), whereas disassembly is enhanced by actin severing and/or depolymerizing proteins, such as gelsolin and actin depolymerizing factor (ADF)/cofilin (Ono, 2007). On the one hand, coordinated regulation of actin assembly and disassembly produces persistent actin treadmilling that drives actin-based motility (Pollard and Borisy, 2003). On the other hand, the actin cytoskeleton must be stabilized when actin supports myosin-based contractility or when actin maintains stable cellular structures. F-actin binding proteins can stabilize actin filaments by preventing depolymerization or by cross-linking filaments to provide mechanical strength (McGough, 1998). How actin stability is spatially and temporally regulated in the cell is, however, is not completely understood.
ADF/cofilin enhances actin filament turnover by accelerating the rate of depolymerization at the pointed ends and by severing filaments (Ono, 2007). The filament severing activity of ADF/cofilin has been directly observed (Ichetovkin et al., 2000; Maciver et al., 1991b; Ono et al., 2004; Pavlov et al., 2007). However, the depolymerizing activity at the pointed ends has been demonstrated indirectly by spectroscopic methods (Carlier et al., 1997; Maciver et al., 1998), and a recent study has shown that ADF/cofilin does not significantly increase the rate of depolymerization at the pointed ends (Andrianantoandro and Pollard, 2006). In cultured cells, ADF/cofilin enhances depolymerization of actin filaments and increases actin monomer concentrations (Hotulainen et al., 2005; Kiuchi et al., 2007). It also promotes actin polymerization by nucleating filament assembly (Andrianantoandro and Pollard, 2006; Chen et al., 2004; Kudryashov et al., 2006) or by severing filaments, with a concomitant increase in the number of exposed barbed ends (Ghosh et al., 2004). Thus, ADF/cofilin promotes overall actin filament dynamics, but the precise mechanism by which ADF/cofilin regulates actin dynamics is complex and not completely understood.
ADF/cofilin is required for rapid actin turnover in vivo, and inhibition of ADF/cofilin activity stabilizes the actin cytoskeleton (Lappalainen and Drubin, 1997). ADF/cofilin can be inhibited by phosphorylation or by binding to phospholipids, and spatial and temporal control of these regulatory mechanisms is important for coordinated cytoskeletal function during directional cell migration (Mouneimne et al., 2006; Nishita et al., 2005). In addition, tropomyosin, a well-characterized F-actin binding protein, stabilizes actin filaments (Cooper, 2002) by preventing ADF/cofilin-dependent actin filament disassembly (Bernstein and Bamburg, 1982; Ono and Ono, 2002). ADF/cofilin and tropomyosin often localize to different compartments of the cell and contribute to spatial separation of dynamic and stable populations of actin filaments (DesMarais et al., 2002; Gupton et al., 2005; Iwasa and Mullins, 2007). However, there are a number of other less well-characterized F-actin-binding proteins that also localize to different cytoskeletal compartments. The mechanisms underlying their functional interactions with ADF/cofilin-dependent actin dynamics remain elusive.
The body wall muscle of the nematode Caenorhabditis elegans has been used as a model to study assembly and maintenance of striated muscle (Moerman and Fire, 1997). We previously demonstrated that actin filament disassembly activities of UNC-60B (ADF/cofilin) (Ono et al., 2003; Ono et al., 1999) and actin-interacting protein 1 (UNC-78) (Mohri et al., 2006; Ono, 2001) are required for organized assembly of actin filaments in C. elegans muscle, and that tropomyosin stabilizes actin filaments antagonistically (Ono and Ono, 2002; Yu and Ono, 2006). Tropomyosin is a major component of isolated nematode thin filaments. Although removal of tropomyosin by high salt increased ADF/cofilin binding to thin filaments in vitro, the binding still appeared partially inhibited. Therefore, additional factor(s) that prevent ADF/cofilin from binding to actin must exist (Ono and Ono, 2002). We hypothesized that UNC-87 is one of these additional actin stabilizers. The unc-87 gene is implicated in protecting myofibrils from mechanical damage during contraction (Goetinck and Waterston, 1994b). UNC-87 is a calponin-related protein with seven calponin-like (CLIK) repeats, but it lacks a calponin-homology (CH) domain, which is present in the calponin and SM22/transgelin families of actin binding proteins (Gimona et al., 2002). UNC-87 localizes to the thin filaments (Goetinck and Waterston, 1994a) and directly binds to actin filaments in vitro (Kranewitter et al., 2001). However, the precise mode of actin binding and the nature of the actin stabilizing function of UNC-87 are not clearly understood.
The CLIK repeat is an actin-binding motif that is unique to calponin-related proteins from yeast to vertebrates. CLIK repeats are present in the C-terminal halves of calponins and SM22/transgelins and mediate the binding to F-actin, whereas the N-terminal CH domains of calponin and SM22/transgelin are dispensable for actin binding (Gimona and Mital, 1998; Goodman et al., 2003). Ectopic expression of CLIK repeats from calponin and UNC-87 in cultured mammalian cells inhibits dynamic reorganization of actin filaments (Gimona et al., 2003; Lener et al., 2004), suggesting that CLIK repeats negatively regulate actin cytoskeleton turnover. In this study, we show that UNC-87 inhibits severing and depolymerization activities of ADF/cofilin in vitro and in vivo, and that this effect is more potent and stable than that of tropomyosin. From our results, we propose that actin-binding via CLIK repeats antagonizes ADF/cofilin-driven actin cytoskeleton remodeling at specialized cellular regions.
Results
UNC-87 and ADF/cofilin bind to F-actin in a mutually exclusive manner
To test our initial hypothesis that UNC-87 stabilizes actin filaments, we first examined whether UNC-87 inhibits ADF/cofilin-mediated actin turnover in vitro, and whether UNC-87 and ADF/cofilin compete for binding to actin filaments, as was previously shown for tropomyosin and ADF/cofilin (Bernstein and Bamburg, 1982; Ono and Ono, 2002). To correlate in vitro and in vivo studies in C. elegans, we used actin, UNC-87 and UNC-60B (C. elegans muscle-specific ADF/cofilin) from C. elegans for most of the biochemical assays. In an actin co-pelleting assay at high speed, UNC-60B co-sedimented with actin filaments with a saturation at an approx. 1:1 molar ratio (Fig. 1Aa,b). However, when actin was preincubated with 10 μM UNC-87, the amount of UNC-60B in the pellets was decreased, indicating inhibition of actin binding (Fig. 1A, compare a with b). The inhibitory effect of UNC-87 was concentration dependent, and a 30–50% reduction in actin-UNC-60B binding was detected at 2.5 μM UNC-87 (actin:UNC-87 = 4:1) (Fig. 1Ac). This inhibition was, however, not complete (Fig. 1Ac), because UNC-87 was dissociated from actin and increased in the supernatants as the concentrations of UNC-60B were increased (Fig. 1Ab). These results indicate that UNC-87 and UNC-60B compete for binding to F-actin.
Fig. 1.
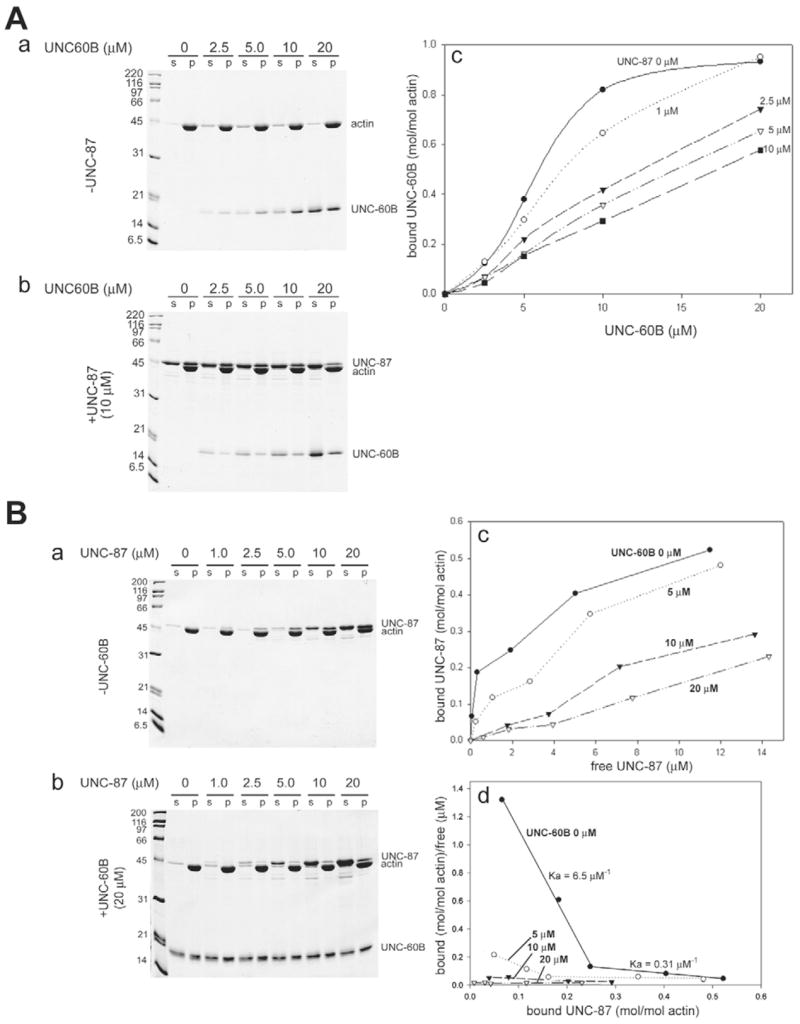
UNC-87 or UNC-60B interacts with Ce-actin in a mutually exclusive manner. (A) 10 μM Ce-actin was pre-incubated with 0–10 μM UNC-87 for 30 minutes, and then, various concentrations of UNC-60B (0–20 μM) were added to the mixtures. After 30 minutes, the samples were centrifuged at 285,000 g for 20 minutes, and the supernatant (s) and pellets (p) were analyzed by SDS-PAGE (12% acrylamide gel). Representative gels from experiments in the absence (a) or in the presence of 10 μM UNC-87 (b) are shown. Quantitative analysis of the results by densitometry is shown in c, in which the amounts of actin-bound UNC-60B (mol/mol actin) were plotted as a function of total UNC-60B concentrations (μM). (B) 10 μM Ce-actin was pre-incubated with 0–20 μM UNC-60B for 30 minutes, and then, various concentrations of UNC-87 (0–20 μM) were added to the mixtures. After 30 minutes, the samples were centrifuged at 285,000 g for 20 minutes, and the supernatant (s) and pellets (p) were analyzed by SDS-PAGE. Representative gels of experiments in the absence (a) and in the presence of 20 μM UNC-60B (b) are shown. Quantitative analysis of the results by densitometry is shown in c, in which the amounts of actin-bound UNC-87 (mol/mol actin) were plotted as a function of free UNC-87 concentrations (μM). (d) The quantitative results were further analyzed using a Scatchard plot.
The monomeric UNC-87 protein is capable of bundling actin filaments (Kranewitter et al., 2001), suggesting that multiple actin-binding sites are present in a single molecule. When we tested the binding of 1.0–20 μM UNC-87 to 10 μM F-actin by the co-sedimentation assay, binding did not reach saturation within this concentration range (Fig. 1Ba,c, closed circles). Similarly, binding did not reach saturation using 2 μM F-actin (our unpublished data). Higher concentrations of UNC-87 were technically difficult to analyze quantitatively because of the close electrophoretic mobilities of UNC-87 and actin on SDS-PAGE (Fig. 1Ba). Nonetheless, Scatchard analysis of the data strongly suggested the presence of a strong actin-binding site (Ka = 6.5 ± 0.32 μM−1) and a weak actin-binding site (Ka = 0.31 ± 0.0067 μM−1) (Fig. 1Bd, closed circles), supporting the presence of two separate regions of interaction with F-actin. Preincubation of F-actin with UNC-60B (note the reverse order of incubation as compared with Fig. 1A) inhibited binding of UNC-87 to F-actin (Fig. 1Bb) in a concentration-dependent manner (Fig. 1Bc). Scatchard analyses of these data indicated that the strong actin-binding site of UNC-87 was effectively inhibited by low concentrations of UNC-60B (Fig. 1Bd, see 5 μM shown by open circles).
As a consequence of the above, we detected that UNC-60B inhibited the actin-bundling activity of UNC-87 (Fig. 2). In a low-speed sedimentation assay, UNC-87 induced the formation of actin bundles that sedimented under these conditions (Fig. 2Aa), whereas, when actin was pre-incubated with UNC-60B, actin-bundling and hence actin filament pelleting at low g forces was efficiently inhibited (Fig. 2Ab,c). When UNC-87 and UNC-60B were simultaneously added to F-actin, the inhibitory effect on bundling was apparently reduced, most probably due to their competitive binding to the side of actin filaments (our unpublished data). We further confirmed these effects on actin bundling by direct observation of fluorescently labeled actin filaments (Fig. 2B). In the presence of UNC-87, thick actin bundles were formed (Fig. 2Bb, arrows), whereas UNC-60B induced the fragmentation of actin filaments without bundling (Fig. 2Bc). When UNC-87 was incubated with actin that had been pre-treated with UNC-60B, only short individual filaments were present, but actin bundles were not detected (Fig. 2Bd). Gelsolin-severed short actin filaments (up to 1:50 molar ratios of gelsolin and actin) were efficiently bundled by UNC-87 (our unpublished data), suggesting that the inhibitory effect of UNC-60B is not simply due to shortening of the filaments. This antagonistic effect contrasts the previously reported synergistic effects on actin bundling between ADF/cofilin and μ-actinin (Maciver et al., 1991a) or elongation factor 1μ (Gungabissoon et al., 2001).
Fig. 2.
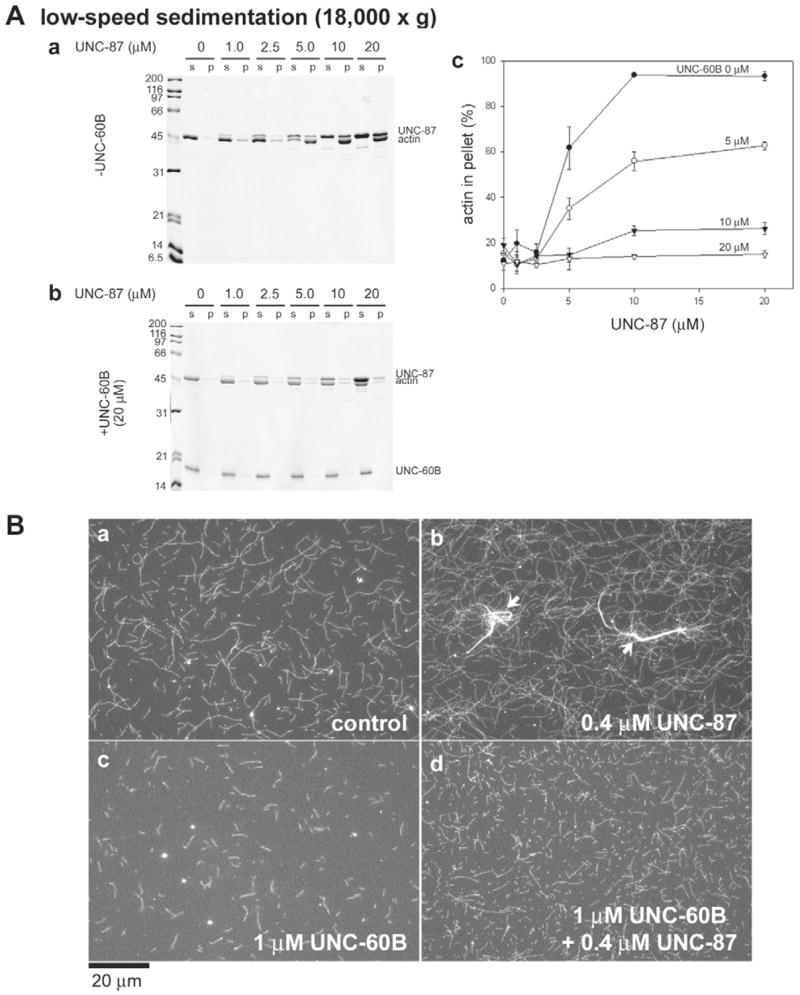
UNC-60B inhibits actin-bundling activity of UNC-87. (A) 10 μM rabbit F-actin was pre-incubated with 0–20 μM UNC-60B for 30 minutes, and varied concentrations of UNC-87 (0–20 μM) were added to the mixtures. After 30 minutes, the samples were centrifuged at 18,000 g for 10 minutes, and the supernatant (s) and pellets (p) were analyzed by SDS-PAGE. Representative gels of the experiments in the absence (a) and presence of 20 μM UNC-60B (b) are shown. Quantitative analysis of the results by densitometry is shown in (c), in which the percentages of sedimented (bundled) actin were plotted as a function of total UNC-87 concentrations (μM). Data are means ± s.d. of three experiments. (B) 1 μM Alexa Fluor 488-biotin-labeled rabbit F-actin was incubated with buffer only (a), 0.4 μM UNC-87 (b), 1 μM UNC-60B (c), or 1 μM UNC-60B and 0.4 μM UNC-87, and the filaments were observed by fluorescence microscopy. UNC-87 induced actin bundling (b, arrows), but the bundling was inhibited by the presence of UNC-60B (d). Bar, 20 μm.
UNC-87 Is a potent inhibitor of ADF/cofilin-mediated actin turnover
The competitive binding of UNC-87 and UNC-60B to F-actin suggests that UNC-87 influences the rate of actin turnover that is regulated by UNC-60B. Our preliminary experiments indicated that UNC-87 alone inhibited latrunculin A-induced actin depolymerization in vitro (our unpublished data). Furthermore, using fluorescence microscopy, we found that UNC-87 has a strong protective effect against filament severing by UNC-60B (Fig. 3). Fluorescently labeled actin filaments were immobilized to a glass surface in a perfusion chamber, exposed to these proteins in two sequential perfusion steps, and examined for their stability. In control experiments, actin filaments at 3 minutes after the first perfusion with buffer only (Fig. 3a) and at 3 minutes after the second perfusion with buffer only (Fig. 3b) showed only minimal alterations in filament morphology. Addition of 2 μM UNC-60B in the second perfusion step caused the severing of filaments (Fig. 3d) that had been pre-treated with buffer only (Fig. 3c). However, incubation of filaments with 2 μM UNC-87 in the first perfusion step (Fig. 3e) efficiently protected actin filaments from severing by UNC-60B (Fig. 3f). A tenfold lower concentration (0.2 μM UNC-87) also had a strong protective effect against severing by UNC-60B (Fig. 3h), and only at 0.02 μM was the effect of UNC-87 was reduced (Fig. 3i, see arrows for severed filaments).
Fig. 3.
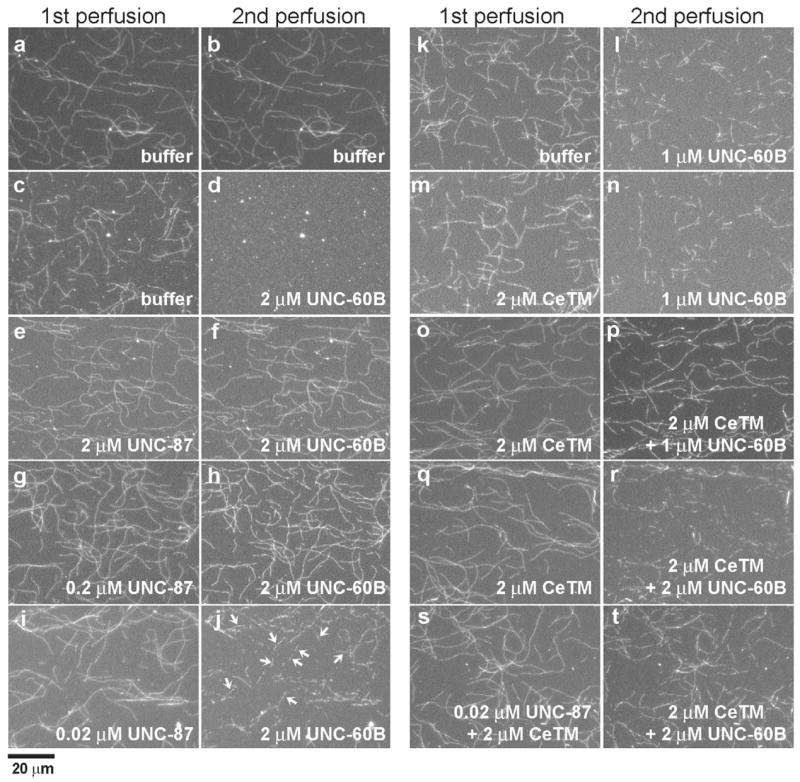
UNC-87 and tropomyosin inhibit actin filament severing by UNC-60B. Alexa Fluor 488-biotin-labeled rabbit actin filaments in perfusion chambers were treated in two sequential perfusion steps with a buffer containing proteins as indicated on the figure. Arrows in j indicate severed filaments. Micrographs of the same fields were taken after both perfusion steps. Bar, 20 μm.
Tropomyosin is well-known to stabilize actin filaments against depolymerization by ADF/cofilin in vitro and in vivo (Cooper, 2002). The C. elegans lev-11 gene (Y105E8B.1) is the single tropomyosin gene that encodes multiple splice isoforms (Kagawa et al., 1995). C. elegans tropomyosin (CeTM) that was purified from wild-type worms consists primarily of high molecular mass isoforms, CeTMI and or CeTMII (Ono and Ono, 2002). A direct comparison of the effects of UNC-87 and CeTM on actin severing revealed that UNC-87 is a more potent stabilizer of actin filaments than tropomyosin. Pre-incubation of actin filaments with 2 μM CeTM had only a minimal protective effect against 1 μM UNC-60B (Fig. 3, compare l with n). The presence of 2 μM CeTM in both the first and second perfusion steps, however, elicited a significant protective effect against 1 μM UNC-60B (Fig. 3o,p), suggesting that CeTM is dissociated from the filaments in the absence of free CeTM. However, when UNC-60B was increased to 2 μM, the protective effect of 2 μM CeTM was minimal (Fig. 3q,r), indicating that high concentrations of tropomyosin are required to protect actin filaments from severing by ADF/cofilin, whereas UNC-87 displays a robust and pronounced effect at significantly lower protein concentrations.
Recombinant UNC-87 and turkey smooth muscle tropomyosin have been shown to simultaneously bind to F-actin in vitro (Kranewitter et al., 2001), and we confirmed that UNC-87 and CeTM bind to C. elegans actin (Ce-actin) in a similar manner (Fig. 4A). When Ce-actin (10 μM) was pre-incubated with 0–10 μM UNC-87 and subsequently with 2 μM CeTM, nearly the same amounts of CeTM co-sedimented with actin (Fig. 4Aa, compare lanes p for different UNC-87 concentrations). Similarly, when Ce-actin (10 μM) was pre-incubated with 0–5 μM CeTM and subsequently with 5 μM UNC-87, nearly the same amounts of UNC-87 co-sedimented with actin (Fig. 4Ab, compare lanes p for different CeTM concentrations). In addition, we detected that UNC-87 and CeTM showed cooperative stabilization of actin filaments. UNC-87 at 0.02 μM (Fig. 3i,j) or CeTM (2 μM) alone (Fig. 3q,r) had only weak protective effects against UNC-60B, but, when both proteins were combined at these concentrations, the stabilizing effect was significantly enhanced (Fig. 3s,t).
Fig. 4.
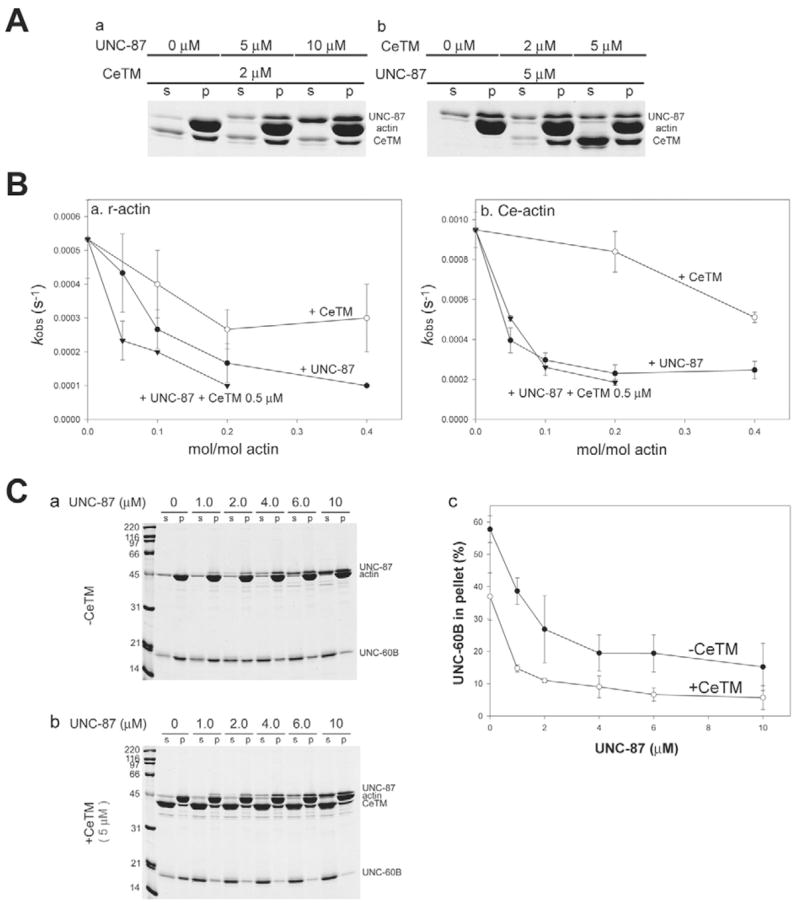
UNC-87 and tropomyosin inhibit UNC-60B-mediated actin turnover. (A) Simultaneous binding of UNC-87 and CeTM to Ce-actin. (a) Ce-actin (10 μM) was pre-incubated with 0–10 μM UNC-87 for 30 minutes and subsequently incubated with 2 μM CeTM for 30 minutes. The reactions were analyzed by pelleting assays (s, supernatants; p, pellets). (b) Ce-actin (10 μM) was pre-incubated with 0–5 μM CeTM for 30 minutes and subsequently incubated with 5 μM UNC-87 for 30 minutes. The reactions were analyzed by pelleting assays as shown in a. (B) Effects of UNC-87 or CeTM on UNC-60B-accelerated actin turnover were measured by nucleotide exchange. F-actin (5 μM) from rabbit muscle (a) or C. elegans (b) was mixed with UNC-60B (2.5 μM) and UNC-87 (0, 0.25, 0.5, 1, or 2 μM; black circles), UNC-60B (2.5 μM) and CeTM (0, 0.5, 1, or 2 μM; white circles), or UNC-60B (2.5 μM), CeTM (0.5 μM) and UNC-87 (0, 0.5, 1, or 2 μM; black triangles) in the presence of 40 μM etheno-ATP, and the fluorescence of etheno-ATP was monitored over time. The data were fitted to exponential curves and the rate of increase in the fluorescence (kobs: 1/second) were calculated and plotted as a function of concentration of UNC-87 or CeTM. Values are mean ± s.d. of three experiments. (C) Co-pelleting assay of 10 μM Ce-actin with 10 μM UNC-60B after pre-incubation with various concentrations of UNC-87 in the absence (a; black circles in c) or presence of 5 μM CeTM (b; white circles in c). Relative amounts (%) of actin-bound UNC-60B in the pellets were plotted as a function of total UNC-87 concentrations (c). Values are means ± s.d. of three experiments.
Next, we quantitatively compared the F-actin stabilizing effects of UNC-87 and CeTM by determining the exchange rate of actin-bound nucleotides as a measure for the rate of actin depolymerization (Mohri and Ono, 2003) (Fig. 4A). In the absence of UNC-60B, F-actin is stable and the rate of nucleotide exchange was below 0.0001/second for both rabbit and Ce-actin (data not shown) (Mohri and Ono, 2003). UNC-60B enhanced the rate to ~0.0005 and ~0.0009/second for rabbit actin and Ce-actin, respectively (Fig. 4B). By contrast, UNC-87 inhibited the nucleotide exchange rate of rabbit and Ce-actin, and did so more potently than CeTM (Fig. 4B). The difference in their inhibitory effects was more pronounced for Ce-actin than for rabbit actin (Fig. 4B). Mixtures of UNC-87 and CeTM had stronger effects on rabbit actin than UNC-87 or CeTM alone (Fig. 4Ba), supporting the observation of their cooperative effects on the inhibition of actin severing from microscopy. The cooperation between UNC-87 and CeTM on Ce-actin was not significant (Fig. 4Bb), presumably because UNC-87 already exhibited the maximal inhibitory effects under these conditions. In co-sedimentation assays, the pre-incubation of actin with both UNC-87 and CeTM inhibited co-sedimentation of UNC-60B and actin more strongly than pre-incubation with UNC-87 alone (Fig. 4C). Taken together, these biochemical results identify cooperation between UNC-87 and CeTM in stabilizing actin filaments against UNC-60B-mediated severing and depolymerization.
AlthoughUNC-87 co-purifies with C. elegans thin filaments, the molar ratio of UNC-87 to actin in the thin filament fraction was only ~1:15 as determined by quantitative western blot analysis (Table 1). This substoichiometric value is in sharp contrast to the stoichiometric association of tropomyosin with actin in the thin filaments (Table 1). The amount of thin filament-associated UNC-60B was very low (Table 1), because a major portion of UNC-60B was recovered in the cytosolic fraction (Ono and Ono, 2002). Nonetheless, our in vitro studies using pure proteins (Figs 1–4) suggest that such low amounts of UNC-87 can be sufficient to inhibit ADF/cofilin-dependent actin severing and depolymerization in vivo.
Table 1.
Quantification of actin and UNC-87 in the thin filament fraction
| Amounts in 10 μg of isolated thin filaments
|
|||
|---|---|---|---|
| μg | nmole | mol/mol | |
| Actin | 4.22 ± 0.11* | 100.4 ± 2.6 | 1 |
| UNC-87 | 0.28 ± 0.08 | 6.79 ± 2.1 | 0.068 (~1/15) |
| CeTM | – | – | 0.141*† (~1/7) |
| UNC-60B | – | – | 9.44μ10−7† (~1/1.1μ106) |
Data are means ± s.d. of three separate experiments.
Data from Ono and Ono (2002).
In vivo functional interactions among UNC-87, tropomyosin and UNC-60B (ADF/cofilin) in muscle actin filament organization
In a previous study, Goetinck and Waterston reported that UNC-87 localizes to the thin filaments in the C. elegans body wall muscle (Goetinck and Waterston, 1994a). Detailed immunolocalization studies revealed a preferential localization of UNC-87 to the center of the I-band region, whereas it was found to be absent from the distal edges of the thin filaments (Fig. 5). Double staining of UNC-87 and actin showed a similar striated localization patterns for both proteins with no detectable signal at the dense bodies in the center of the striations (Fig. 5a–c). However, the band of UNC-87 was slightly narrower than that of actin (Fig. 5, compare a with b), indicating that UNC-87 is absent from the distal portions of the thin filaments (near the pointed ends of the actin filaments). By contrast, CeTM has been shown previously to localize to the distal portion of the thin filaments (Ono et al., 2006; Ono and Ono, 2002), and, double staining of CeTM and UNC-87 indeed identified CeTM at the distal portion and UNC-87 at the central portion of the actin bands, albeit with some overlap (Fig. 5g–i). UNC-60B localized to the center of the actin bands between dense bodies (Fig. 5e), together with UNC-87 (Fig. 5d–f). However, with the exception of this partial overlap, UNC-87 and UNC-60B mostly localized to different regions of the thin filaments. These distinct localization patterns of UNC-87, CeTM and UNC-60B along the thin filaments (Fig. 5j) further support that these molecules regulate actin by different mechanisms.
Fig. 5.
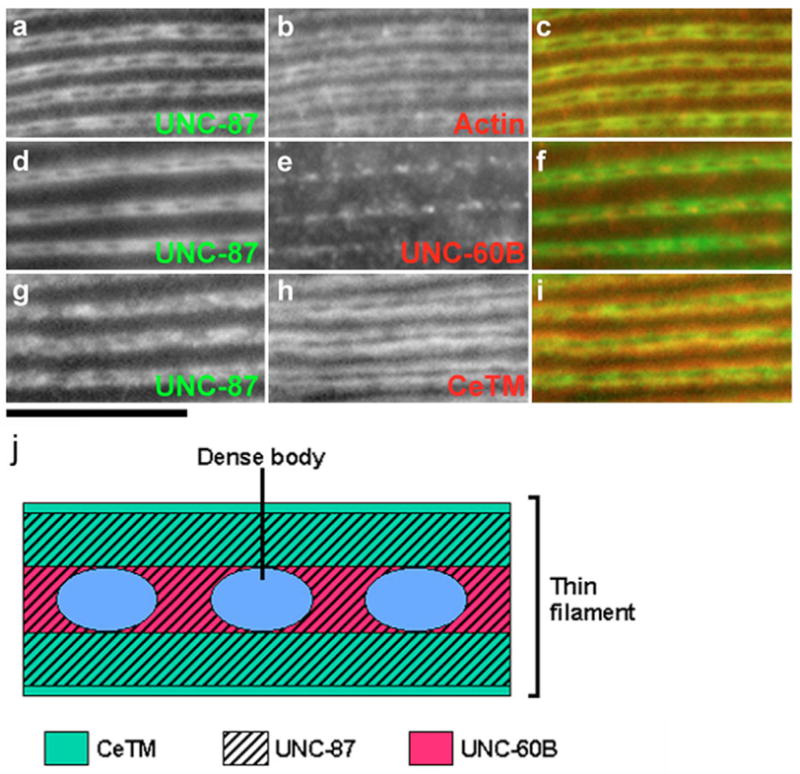
Localization of UNC-87, CeTM and UNC-60B in adult body wall muscle. Wild-type adult nematodes were double-stained with anti-UNC-87 (a,d,g) and anti-actin (b) or anti-UNC-60 (e) or anti-CeTM (h) antibodies. Merged images are shown in c,f,i. Bars, 10 μm. (j) Schematic representation of locations of UNC-87, CeTM and UNC-60B in the thin filaments.
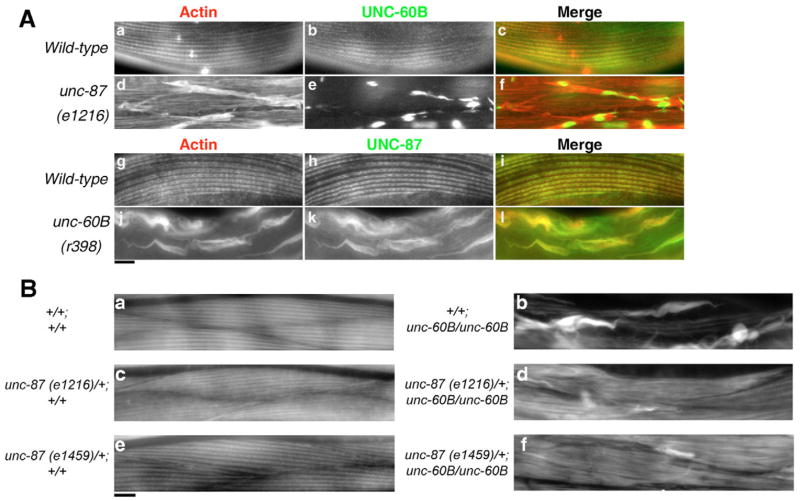
Mutations in the unc-87 gene cause disorganization of actin filaments and the formation of actin aggregates at the periphery of the muscle cells (Goetinck and Waterston, 1994a; Goetinck and Waterston, 1994b). In this study, we used two loss-of-function unc-87 alleles, unc-87(e1216) and unc-87(e1459). Homozygous unc-87(e1459) animals show stronger muscle phenotypes than homozygous unc-87(e1216) animals (Goetinck and Waterston, 1994a). In unc-87(e1216) mutant animals, UNC-60B was found in these actin aggregates (Fig. 6Ad–f), and with a particular concentration in the core (Fig. 6Ae,f). Mutations in unc-60B also cause the formation of actin aggregates in the body wall muscle (Ono et al., 1999). In unc-60B(r398) mutant animals, mislocalization of UNC-87 into these aggregates was apparent, and the pattern was nearly identical to that of actin (Fig. 6Aj–l). These changes in localization pattern of UNC-87 and UNC-60B suggest a functional interaction involving the regulation of actin dynamics.
Fig. 6.
Genetic interaction between UNC-87 and UNC-60B in adult body wall muscle. (A) Adult body wall muscle of wild-type (a–c, g–i), unc-87(e1216) (d–f), or unc-60B(r398) (j–l) animals was double stained with anti-actin (a,d,g,j) and anti-UNC-60B (b,e) or anti-UNC-87 (h,k) antibodies. Merged images are shown in c,f,i,l (red: actin, green: UNC-60B, or UNC-87). (B) Actin filament organization in adult body wall muscle was visualized by staining with tetramethylrhodamine-phalloidin. Micrographs of wild-type (a), +/+; unc-60B(r398) (b), unc-87(e1216)/+; +/+ (c), unc-87(e1216)/+; unc-60B(r398) (d), unc-87(e1459)/+; +/+ (e), and unc-87(e1459)/+; unc-60B(r398) (f) muscle. Bars, 10 μm.
To further determine the potential functional relationship between UNC-87 and UNC-60B, we next tested the effects of their double mutations on actin organization. Our biochemical studies indicate an antagonistic regulation of actin stability and turnover by UNC-87 and UNC-60B. Thus, a double mutation of unc-87 and unc-60B was expected to improve the actin phenotype, as we have previously demonstrated for CeTM and UNC-60B (Ono and Ono, 2002; Yu and Ono, 2006). Unexpectedly, unc-87(e1216 or e1459);unc-60B(r398) double homozygotes were embryonic lethal, whereas each of the single mutants was homozygous viable. The unc-87(e1216 or e1459);unc-60B(r398) double homozygotes developed to threefold stage embryos, but were severely paralyzed and subsequently failed to hatch (our unpublished data). The body wall muscles of the double mutant embryos were disorganized with the formation of actin aggregates, reminiscent of unc-60B(r398) single mutant embryos (our unpublished data). The cause for the observed lethality in the double mutants is currently unclear, but the severe phenotype indicates that both UNC-87 and UNC-60B are critical for the regulation of actin dynamics during embryonic development. Notably, double mutants, in which unc-87(e1216 or e1459) was heterozygous and unc-60B was homozygous (unc-87(e1216 or e1459)/+;unc-60B/unc-60B), grew into adults and showed only reduced formation of actin aggregates (Fig. 6Bd,f), compared with unc-60B (Fig. 6Bb) or unc-87(e1216) single mutants (Fig. 6Ad). Finally, heterozygous unc-87 mutation alone (unc-87(e1216 or e1459)/+) did not cause alterations in the organization of the actin cytoskeleton (Fig. 6Bc,e) compared with wild-type animals (Fig. 6Ba). Two unc-87 alleles, e1216 (moderate loss-of-function) and e1459 (strong loss-of-function), showed highly penetrant genetic interaction with unc-60B (Fig. 6Bd,f), as 74% (n=46) of unc-87(e1216)/+;unc-60B and 87% (n=39) of unc-87(e1459)/+;unc-60B worms exhibited reduction in actin aggregates. These genetic interactions between unc-87 and unc-60B in the adult muscle further support their antagonistic roles in the regulation of actin stability.
Both UNC-87 and CeTM stabilize actin filaments in vitro by inhibiting depolymerization and severing by UNC-60B. If both UNC-87 and CeTM also play similar roles in actin stabilization in vivo, knockdown of both UNC-87 and CeTM is expected to enhance actin disorganization, as a result of the increased accessibility of actin filament interaction sites for UNC-60B and a concomitantly increased ability to sever and depolymerize the filaments. However, if UNC-87 and CeTM have different in vivo functions, double knockdown of UNC-87 and CeTM may cause other phenotypes such as suppression of actin disorganization. In unc-87 mutants, CeTM and actin co-localized in the aggregates in a nearly identical manner (Fig. 7Ad–f). In a wild-type background, RNA interference (RNAi) of two long CeTM isoforms (CeTMI and CeTMII) or two long and two short CeTM isoforms (CeTMIII and CeTMIV) induced a wavy appearance of the actin filaments (Fig. 7Bb,c) (Ono and Ono, 2002). By contrast, in the unc-87(e1216 or e1459) mutant background, RNAi of CeTM suppressed the formation of actin aggregates (Fig. 7Be,f,h,i), and the characteristic wavy actin bundles were diminished (Fig. 7Be,f,h,i). However, the defined striated pattern of actin filaments was not completely restored in both unc-87 mutants (Fig. 7Be,f,h,i). RNAi of four CeTM isoforms caused a stronger phenotypic suppression than RNAi of CeTMI and CeTMII, and unc-87(e1217) (a moderate loss-of-function allele) was more strongly suppressed than unc-87(e1459) (a strong loss-of-function allele) (Table 2). By contrast, RNAi of CeTM reduced worm motility of unc-87 mutants instead of suppressing the phenotype (our unpublished data), suggesting that reduced contractility contributed to suppressing the actin disorganization in unc-87 mutants (see Discussion). These results suggest that UNC-87 and CeTM do not simply function in a similar manner, and that an intricate balance of the activities of UNC-87, CeTM and UNC-60B is important for actin filament organization and proper localization of these proteins.
Fig. 7.
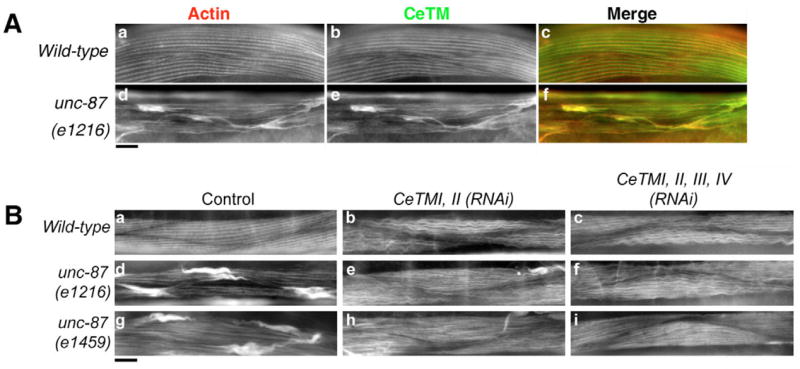
Genetic interaction between UNC-87 and CeTM in body wall muscle. (A) Adult body wall muscle of wild-type (a–c) or unc-87(e1216) (d–f) animals was double stained with anti-actin (a and d) and anti-CeTM (b,e) antibodies. Merged images are shown in c and f (red: actin, green: CeTM). (B) Actin filament organization in adult body wall muscle was visualized by staining with tetramethylrhodamine-phalloidin. Wild-type (a–c), unc-87(e1216) homozygous (d–f), or unc-87(e1459) homozygous (g–i) animals were treated with control RNAi (a,d,g), CeTMI,II (RNAi) (b,e,h), or CeTMI,II,III,IV (RNAi) (c,f,i). Bars, 10 μm.
Table 2.
Effects of CeTM knockdown on actin aggregate formation
| Percentages of adult worms with actin aggregates (%)
|
|||
|---|---|---|---|
| Strain | Control RNAi | CeTMI,II(RNAi) | CeTMI,II,III,IV(RNAi) |
| Wild type | 0 (0/107) | 0 (0/102) | 0 (0/101) |
| unc-87(e1216) | 100 (115/115) | 34 (34/100) | 5.9 (4/68) |
| unc-87(e1459) | 100 (106/106) | 73 (58/80) | 26 (18/70) |
Discussion
In this study, we demonstrate that the calponin-repeat protein UNC-87 is an effective stabilizer of the actin cytoskeleton and protects actin filaments against ADF/cofilin-dependent actin filament depolymerization and severing. In vitro, the actin-stabilizing effect of UNC-87 is more pronounced and persistent than that of tropomyosin. In adult C. elegans muscle, the functions of UNC-87 and ADF/cofilin (UNC-60B) appear antagonistic with respect to regulating actin organization, which supports their opposing functions on actin filament stability. However, the genetic interaction between UNC-87 and tropomyosin (CeTM) reveal their distinct functions in the regulation of actin organization.
UNC-87 stabilizes actin filaments by competitively inhibiting the binding of ADF/cofilin to the filaments. Although UNC-87 bundles actin filaments in vitro (Kranewitter et al., 2001), this bundling activity is not required for the actin-stabilizing effect. Immobilized actin filaments, which are decorated but not bundled by UNC-87, are still protected from severing by ADF/cofilin (Fig. 3). Although tropomyosin has a similar stabilizing function (Bernstein and Bamburg, 1982; Ono and Ono, 2002) our direct comparison of the effects of UNC-87 and tropomyosin demonstrates that UNC-87 is more efficient than tropomyosin in this respect, suggesting that UNC-87 and tropomyosin bind to F-actin in different ways. The simultaneous binding of tropomyosin and UNC-87 to actin filaments in vitro (Kranewitter et al., 2001) supports their association with non-overlapping binding sites on the actin filament. Tropomyosin interacts along the long pitch groove of the actin filaments. Low and high molecular mass tropomyosin isoforms contain periodic amino acid repeats that correspond to 6 and 7 actin binding sites, respectively, and multiple tropomyosin isoforms regulate the actin cytoskeleton in both muscle and non-muscle cells (Gunning et al., 2005). Although the actin-stabilizing function against ADF/cofilin appears a common feature for many (if not all) tropomyosin isoforms (Cooper, 2002), the structural basis for the interaction of most non-muscle tropomyosins with actin is not completely understood. ADF/cofilin alters the twist of the actin filament (Galkin et al., 2001; McGough et al., 1997), and an altered actin conformation may be favorable for the cooperative binding of ADF/cofilin, but unfavorable for the interaction of tropomyosin and other actin binding proteins. At present, the exact mode of interaction between UNC-87 and F-actin remains speculative. A recent structural analysis of the calponin-F-actin interaction has demonstrated that the C-terminal region of smooth muscle h1 calponin, which contains all three CLIK repeats, binds to actin subdomains 1 and 2 in the filament (Galkin et al., 2006). This region overlaps with the known interaction sites of ADF/cofilin (McGough et al., 1997; Ono et al., 2001). Therefore, ADF/cofilin and CLIK repeats of UNC-87 perhaps compete for the same binding site on F-actin.
We have observed slight differences in the localization patterns of UNC-87 and CeTM. The localization of CeTM results from its competition with Ce-kettin, a large immunoglobulin-like repeat protein, near the dense bodies (Ono et al., 2006). UNC-87 appears excluded from the distal region of the thin filaments near the pointed ends of F-actin, and ectopically expressed GFP-tagged UNC-87, as well as overexpressed and endogenous h1 calponin are likewise excluded from peripheral actin structures in cultured mammalian cells (Gimona et al., 2003). However, the precise mechanisms that regulate the exclusion of UNC-87 from the pointed ends of F-actin remain speculative. Tropomodulin that concentrates at the actin pointed ends may inhibit association of UNC-87 with actin. Alternatively, constant ADF/cofilin-driven turnover of actin subunits and spontaneous elongation at the pointed ends may inhibit stable association of UNC-87. ADF/cofilin preferentially binds to ADP-actin that is generally enriched towards the pointed ends (Maciver and Weeds, 1994). In cultured striated muscle, exchange of actin subunits at the pointed ends controls the length of the thin filaments (Littlefield et al., 2001). Therefore, persistent exclusion of UNC-87 from the actin pointed ends may be required to allow ADF/cofilin-dependent actin turnover to maintain the myofibril structure, whereas UNC-87 may be required for prolonged stabilization of F-actin at more proximal regions. This hypothesis is consistent with the phenotypes of the unc-87 mutants alone or in combination with the unc-60B mutation (Fig. 6). In the unc-87 mutants, actin filament stability is reduced and the filaments are more prone to rapid disassembly by active UNC-60B, which may cause the disorganization of actin filaments. When the activity of UNC-60B is low, actin disassembly is less frequent, and disorganization of the filaments can be suppressed.
Although the adult muscle phenotypes of the unc-87 single and unc-87;unc-60B double mutants are consistent with the proposed antagonistic roles of UNC-87 and UNC-60B on actin remodeling, the embryonic phenotypes were not. The unc-87;unc-60B double homozygous mutants were lethal at late embryonic stages. By contrast, unc-87 or unc-60B single mutants are homozygous viable (Goetinck and Waterston, 1994b; Ono et al., 1999). Such enhancement in the phenotype cannot be explained by a simple antagonistic mechanism, but rather suggests additional cooperative functions between UNC-87 and UNC-60B. Indeed, under certain conditions, ADF/cofilin itself displays opposing functions: it nucleates actin assembly and promotes polymerization (Andrianantoandro and Pollard, 2006; Chen et al., 2004; Kudryashov et al., 2006). It is conceivable that, during embryonic stages when the actin cytoskeleton is reorganized to form myofibrils, both UNC-60B and UNC-87 act to increase the amount of F-actin. Actin filaments in the unc-87;unc-60B double mutant embryos were similarly disorganized to those in the unc-60B mutants, and comparison of the actin phenotypes did not provide further insight into the functional relationship between UNC-60B and UNC-87. Nonetheless, our results demonstrate that UNC-60B and UNC-87 play different yet profound roles in actin organization, and further dissection of the functions of UNC-60B (severing/depolymerization and nucleation) and UNC-87 at different developmental stages is required to clarify this issue.
The cooperative stabilization of F-actin by UNC-87 and tropomyosin antagonizes the activities of ADF/cofilin in vitro, albeit our results on the genetic interaction between UNC-87 and CeTM also demonstrate distinct functions in vivo. Combination of unc-87 mutation and CeTM RNAi suppresses the disorganization of actin filaments in body wall muscle rather than enhancing the phenotype. In part, this can be due to the known effects of muscle contractility on actin organization, since active muscle contraction can damage myofibrils (Friden and Lieber, 1992). Indeed, disorganized actin filaments in unc-87 mutants is suppressed by a myosin mutation that reduces contraction frequency and magnitude (Goetinck and Waterston, 1994b), further suggesting that, in addition to the action of UNC-60B, actomyosin contractility also contributes to the actin phenotype in unc-87 mutants. Knockdown of CeTM reduces the contractile activity of body wall muscle (Ono and Ono, 2002; Yu and Ono, 2006), thus suppressing the phenotype of unc-87 mutants. However, the actin phenotype in unc-60B mutants is only weakly suppressed by reduced myosin activity (Yu and Ono, 2006), suggesting that the mechanisms of actin disorganization in unc-87 and unc-60B mutants are different. By contrast, proper actomyosin interaction is required for the organized myofibril assembly (De Deyne, 2000; Kagawa et al., 2006). Currently, the role of myosin in actin dynamics is not completely understood, but recent studies in dividing cells suggest that myosin II enhances actin dynamics (Guha et al., 2005; Murthy and Wadsworth, 2005). To better understand the function of UNC-87, we are currently investigating the potential role of UNC-87 in actomyosin contraction. Preliminary results indicate an inhibitory effect of UNC-87 on actomyosin contractility (our unpublished data), suggesting a significant involvement in the regulation of actin organization. Notably, a similar function is attributed to calponin family members in mammalian muscle and non-muscle cells, and this function requires the interaction of calponin with the actin filaments via the CLIK repeats. The three CLIK repeats in h1 calponin reduce Arp2/3-mediated actin polymerization during podosome formation (Gimona et al., 2003), inhibit cell motility and increase resistance to pharmacological inhibitors of actin polymerization (Danninger and Gimona, 2000).
Taken together, our study provides evidence for the inhibition of ADF/cofilin-dependent actin dynamics by CLIK repeats, and provides a first mechanistic insight into a possibly conserved function of CLIK repeats in the stabilization of actin filaments that has been observed in yeast (Goodman et al., 2003) and cultured mammalian cells (Gimona et al., 2003). Genetic analysis in budding yeast revealed that mutation of the calponin homolog scp1 induces cooperative effects with the sac6 (fimbrin) mutation, but not with cofilin or tropomyosin mutation (Goodman et al., 2003). Notably, UNC-87 is a unique protein that contains seven CLIK repeats, whereas, in yeast SCP1 and mammalian SM22/transgelin, only one repeat is present. A deletion analysis of UNC-87 has demonstrated that multiple CLIK repeats are required for strong F-actin binding and bundling (Kranewitter et al., 2001), and that the effects on actin turnover directly correlate with the number of CLIK repeats (Lener et al., 2004). CLIK repeats are intrinsically unfolded protein domains that adopt a stable conformation only upon contact with actin filaments. Here, we describe the formation of two independent actin binding sites of different affinities by the seven CLIK repeats of UNC-87. Actin filament stabilization appears independent of the bundling activity, and UNC-87 may use both filament interaction sites to bind the same filament with different affinities and potentially also to two different regions. It should be noted that also the prominent actin cross-linking protein α-actinin was shown recently to employ both actin-binding sites for lateral binding to actin filament (Hampton et al., 2007).
Although, at present, no calponin-like protein has been identified in vertebrate striated muscle, several actin-stabilizing proteins including nebulin (McElhinny et al., 2005), Xin (Pacholsky et al., 2004) and myotilin (Salmikangas et al., 2003) also have repetitive actin-binding motifs. By contrast, C. elegans does not have nebulin or Xin, but Ce-kettin (Ono et al., 2006) has immunoglobulin-like repeats and shows some similarity to myotilin. Thus, UNC-87 might be functionally related to these actin stabilizing proteins with repetitive actin-binding motifs. Importantly, mutations in nebulin and myotilin are associated with nemaline myopathy (Clarkson et al., 2004) and limb girdle muscular dystrophy (Olive et al., 2005), respectively, suggesting that impaired actin stabilizing activity may cause these inherited muscle diseases. Nebulin deficiency causes muscle defects in mice (Bang et al., 2006; Witt et al., 2006), whereas myotilin-deficient mice show no abnormal muscle phenotype (Moza et al., 2007). Thus, these actin-stabilizing proteins may have different roles in specialized vertebrate muscles. Future studies should provide further insight into the spatial regulation of actin dynamics and stability in vivo employing the C. elegans model as a useful system to study the functions of actin-stabilizing proteins in striated muscle.
Materials and Methods
Nematode strains
Wild-type C. elegans strain N2 was obtained from the Caenorhabditis Genetics Center (Minneapolis, MN). unc-87(e1216) and unc-87(e1459) (Goetinck and Waterston, 1994a) were provided by Pamela Hoppe (Western Michigan University, Kalamazoo, MI) and Robert Waterston (University of Washington, Seattle, WA). unc-60B(r398) (McKim et al., 1988) was originally provided by David Baillie (Simon Fraser University, Burnaby, Canada), and a further outcrossed strain (Yu and Ono, 2006) was used. Double mutant strains, unc-87(e1216); unc-60B(r398) and unc-87(e1459); unc-60B(r398) were generated by standard crosses. However, the double mutants were homozygous lethal. Therefore, the strains were maintained as unc-87/+ I; unc-60B/unc-60B V, where wild-type chromosome I was marked by dpy-5, and heterozygosity and homozygosity of the mutant alleles was confirmed by sequencing genomic DNA.
Proteins
C. elegans actin (Ce-actin) (Ono, 1999) and C. elegans tropomycin (CeTM) (Ono and Ono, 2002) were purified from wild-type strain N2 as described. Rabbit muscle actin was purified as described previously (Pardee and Spudich, 1982). Recombinant UNC-60B (Ono and Benian, 1998) and UNC-87 (Kranewitter et al., 2001) were expressed in E. coli and purified as described.
Assays for F-actin binding and bundling by pelleting
Co-pelleting assays of rabbit muscle actin or Ce-actin with UNC-60B, UNC-87, and CeTM were performed as described previously in a buffer containing 0.1 M KCl, 2 mM MgCl2, 20 mM Hepes-NaOH, 1 mM DTT, pH 7.5 (Ono and Ono, 2002). Ultracentrifugation was performed in a Beckman TLA100 rotor at 80,000 rpm (285,000 g) for 20 minutes at 20°C. To examine actin-bundling, the mixtures were assembled in the same manner as the co-pelleting assays, and centrifuged at 18,000 g for 10 minutes. The amounts of proteins that were contained in the supernatants and pellets were analyzed by SDS-PAGE and densitometry as described previously. Scatchard analysis was performed with SigmaPlot 2000 (Systat Software), and dissociation constants were calculated by linear regression of the data.
Direct observation of actin filament severing and bundling
Observation of actin filament severing by fluorescence microscopy was performed as described previously (Ono et al., 2004) with slight modifications. Briefly, Alexa Fluor 488 and biotin-labeled actin were attached to a perfusion chamber using anti-biotin antibody (Molecular Probes). After washing twice with wash buffer (50 mM KCl, 5 mM EGTA, 2 mM MgCl2, 1 mM ATP, 1 mM DTT, 0.5 mg/ml bovine serum albumin and 20 mM Hepes-NaOH, pH 7.5), UNC-87 and/or CeTM in wash buffer were perfused and incubated for 5 minutes (first perfusion). After washing twice with anti-bleaching buffer (wash buffer plus 100 mM DTT, 0.036 mg/ml catalase, 0.2 mg/ml glucose oxidase, 6 mg/ml glucose), UNC-60B in anti-bleaching buffer was perfused into the chamber and incubated for 3 minutes (second perfusion). Micrographs of the fluorescent actin filaments were taken after the first and second perfusions in the same field.
To examine actin bundling, 1 μM Alexa Fluor 488 and biotin-labeled actin was incubated with or without 1 μM UNC-60B in wash buffer for 3 minutes, and then incubated with or without 0.4 μM UNC-87 in wash buffer for 3 minutes at room temperature. The mixtures were diluted tenfold with wash buffer and the filaments were attached to perfusion chambers using anti-biotin antibody. After washing twice with anti-bleaching buffer, micrographs were taken.
Monitoring kinetics of actin turnover by nucleotide exchange
The kinetics of actin turnover was monitored as described previously (Mohri and Ono, 2003) by measuring exchange rate of actin-bound nucleotide.
Quantitative analysis of the thin filament proteins by western blot
Nematode thin filaments were isolated as described previously (Ono and Ono, 2002). Amounts of actin and UNC-87 were quantified by western blot with mouse monoclonal anti-actin (C4) and guinea pig anti-UNC-87 antibodies using purified actin and UNC-87 as standards, respectively. Densitometric quantification was performed in ranges where band intensity of the standard proteins was linearly correlated with their amounts.
Fluorescence microscopy
Imunofluorescent staining of adult nematodes was performed as described previously (Finney and Ruvkun, 1990). Primary antibodies used were mouse anti-actin monoclonal (C4, MP Biomedicals), rabbit anti-UNC-60B (Ono et al., 1999), guinea pig anti-CeTM (Ono and Ono, 2002), rabbit anti-UNC-87 (Goetinck and Waterston, 1994a) (provided by P. Hoppe and R. Waterston), and guinea pig anti-UNC-87 antibodies. Guinea pig anti-UNC-87 was prepared using purified UNC-87 as immunogen at Quality Controlled Biochemicals (Hopkinton, MA). Secondary antibodies were Alexa Fluor 488-goat anti-guinea pig IgG (Invitrogen), Cy3-donkey anti-rabbit IgG and anti-mouse (Jackson ImmunoResearch). Staining of adult worms using tetramethylrhodamine-phalloidin was performed as described elsewhere (Ono, 2001).
Samples were observed by epifluorescence using a Nikon Eclipse TE2000 inverted microscope with a CFI Plan Fluor ELWD 40× (dry, NA 0.60) or a CFI Plan Apo 60× (oil, NA 1.40) objective. Images were captured by a SPOT RT Monochrome CCD camera (Diagnostic Instruments) and processed by the IPLab imaging software (BD Biosciences) and Adobe Photoshop 6.0.
RNA interference experiments
Nematodes were treated with RNAi for CeTM by feeding them with E. coli expressing double stranded RNA as described previously (Ono and Ono, 2002). L4 larvae were treated with RNAi, and phenotypes were characterized in their F1 generation.
Acknowledgments
We thank P. Hoppe and R. Waterston for antibody and worm strains, D. Baillie for worm strains, and J. H. Woo for technical support. Some worm strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH NCRR. This work was supported by a Marie Curie Excellence Grant (MEXT-CT-2003–002573) of the European Union to M.G. and an NIH grant (R01 AR48615) to S.O.
References
- Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Bang ML, Li X, Littlefield R, Bremner S, Thor A, Knowlton KU, Lieber RL, Chen J. Nebulin-deficient mice exhibit shorter thin filament lengths and reduced contractile function in skeletal muscle. J Cell Biol. 2006;173:905–916. doi: 10.1083/jcb.200603119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF) Cell Motil. 1982;2:1–8. doi: 10.1002/cm.970020102. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Bernstein BW, Sneider JM, Boyle JA, Minamide LS, Bamburg JR. In vitro activity differences between proteins of the ADF/cofilin family define two distinct subgroups. Biochemistry. 2004;43:7127–7142. doi: 10.1021/bi049797n. [DOI] [PubMed] [Google Scholar]
- Clarkson E, Costa CF, Machesky LM. Congenital myopathies: diseases of the actin cytoskeleton. J Pathol. 2004;204:407–417. doi: 10.1002/path.1648. [DOI] [PubMed] [Google Scholar]
- Cooper J. Actin dynamics: tropomyosin provides stability. Curr Biol. 2002;12:R523–R525. doi: 10.1016/s0960-9822(02)01028-x. [DOI] [PubMed] [Google Scholar]
- Danninger C, Gimona M. Live dynamics of GFP-calponin: isoform-specific modulation of the actin cytoskeleton and autoregulation by C-terminal sequences. J Cell Sci. 2000;113:3725–3736. doi: 10.1242/jcs.113.21.3725. [DOI] [PubMed] [Google Scholar]
- De Deyne PG. Formation of sarcomeres in developing myotubes: role of mechanical stretch and contractile activation. Am J Physiol Cell Physiol. 2000;279:C1801–C1811. doi: 10.1152/ajpcell.2000.279.6.C1801. [DOI] [PubMed] [Google Scholar]
- DesMarais V, Ichetovkin I, Condeelis J, Hitchcock-DeGregori SE. Spatial regulation of actin dynamics: a tropomyosin-free, actin-rich compartment at the leading edge. J Cell Sci. 2002;115:4649–4660. doi: 10.1242/jcs.00147. [DOI] [PubMed] [Google Scholar]
- Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- Friden J, Lieber RL. Structural and mechanical basis of exercise-induced muscle injury. Med Sci Sports Exerc. 1992;24:521–530. [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Lukoyanova N, Wriggers W, Egelman EH. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J Cell Biol. 2001;153:75–86. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Fattoum A, Walsh MP, Egelman EH. The CH-domain of calponin does not determine the modes of calponin binding to F-actin. J Mol Biol. 2006;359:478–485. doi: 10.1016/j.jmb.2006.03.044. [DOI] [PubMed] [Google Scholar]
- Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304:743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- Gimona M, Mital R. The single CH domain of calponin is neither sufficient nor necessary for F-actin binding. J Cell Sci. 1998;111:1813–1821. doi: 10.1242/jcs.111.13.1813. [DOI] [PubMed] [Google Scholar]
- Gimona M, Djinovic-Carugo K, Kranewitter WJ, Winder SJ. Functional plasticity of CH domains. FEBS Lett. 2002;513:98–106. doi: 10.1016/s0014-5793(01)03240-9. [DOI] [PubMed] [Google Scholar]
- Gimona M, Kaverina I, Resch GP, Vignal E, Burgstaller G. Calponin repeats regulate actin filament stability and formation of podosomes in smooth muscle cells. Mol Biol Cell. 2003;14:2482–2491. doi: 10.1091/mbc.E02-11-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetinck S, Waterston RH. The Caenorhabditis elegans muscle-affecting gene unc-87 encodes a novel thin filament-associated protein. J Cell Biol. 1994a;127:79–93. doi: 10.1083/jcb.127.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetinck S, Waterston RH. The Caenorhabditis elegans UNC-87 protein is essential for maintenance, but not assembly, of bodywall muscle. J Cell Biol. 1994b;127:71–78. doi: 10.1083/jcb.127.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- Goodman A, Goode BL, Matsudaira P, Fink GR. The Saccharomyces cerevisiae calponin/transgelin homolog Scp1 functions with fimbrin to regulate stability and organization of the actin cytoskeleton. Mol Biol Cell. 2003;14:2617–2629. doi: 10.1091/mbc.E03-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha M, Zhou M, Wang YL. Cortical actin turnover during cytokinesis requires myosin II. Curr Biol. 2005;15:732–736. doi: 10.1016/j.cub.2005.03.042. [DOI] [PubMed] [Google Scholar]
- Gungabissoon RA, Khan S, Hussey PJ, Maciver SK. Interaction of elongation factor 1α from Zea mays (ZmEF-1alpha) with F-actin and interplay with the maize actin severing protein, ZmADF3. Cell Motil Cytoskeleton. 2001;49:104–111. doi: 10.1002/cm.1024. [DOI] [PubMed] [Google Scholar]
- Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15:333–341. doi: 10.1016/j.tcb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Gupton SL, Anderson KL, Kole TP, Fischer RS, Ponti A, Hitchcock-DeGregori SE, Danuser G, Fowler VM, Wirtz D, Hanein D, et al. Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosin. J Cell Biol. 2005;168:619–631. doi: 10.1083/jcb.200406063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton CM, Taylor DW, Taylor KA. Novel structures for alpha-Actinin:F-Actin interactions and their implications for actin-membrane attachment and tension sensing in the cytoskeleton. J Mol Biol. 2007;368:92–104. doi: 10.1016/j.jmb.2007.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30:342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Paunola E, Vartiainen MK, Lappalainen P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol Biol Cell. 2005;16:649–664. doi: 10.1091/mbc.E04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichetovkin I, Han J, Pang KM, Knecht DA, Condeelis JS. Actin filaments are severed by both native and recombinant Dictyostelium cofilin but to different extents. Cell Motil Cytoskeleton. 2000;45:293–306. doi: 10.1002/(SICI)1097-0169(200004)45:4<293::AID-CM5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Iwasa JH, Mullins RD. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr Biol. 2007;17:395–406. doi: 10.1016/j.cub.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa H, Sugimoto K, Matsumoto H, Inoue T, Imadzu H, Takuwa K, Sakube Y. Genome structure, mapping and expression of the tropomyosin gene tmy-1 of Caenorhabditis elegans. J Mol Biol. 1995;251:603–613. doi: 10.1006/jmbi.1995.0459. [DOI] [PubMed] [Google Scholar]
- Kagawa M, Sato N, Obinata T. Effects of BTS (N-benzyl-p-toluene sulphonamide), an Inhibitor for myosin-actin Interaction, on myofibrillogenesis in skeletal muscle cells in culture. Zool Sci. 2006;23:969–975. doi: 10.2108/zsj.23.969. [DOI] [PubMed] [Google Scholar]
- Kiuchi T, Ohashi K, Kurita S, Mizuno K. Cofilin promotes stimulus-induced lamellipodium formation by generating an abundant supply of actin monomers. J Cell Biol. 2007;177:465–476. doi: 10.1083/jcb.200610005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranewitter WJ, Ylanne J, Gimona M. UNC-87 is an actin-bundling protein. J Biol Chem. 2001;276:6306–6312. doi: 10.1074/jbc.M009561200. [DOI] [PubMed] [Google Scholar]
- Kudryashov DS, Galkin VE, Orlova A, Phan M, Egelman EH, Reisler E. Cofilin cross-bridges adjacent actin protomers and replaces part of the longitudinal F-actin interface. J Mol Biol. 2006;358:785–797. doi: 10.1016/j.jmb.2006.02.029. [DOI] [PubMed] [Google Scholar]
- Lappalainen P, Drubin DG. Cofilin promotes rapid actin filament turnover in vivo. Nature. 1997;388:78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- Lener T, Burgstaller G, Gimona M. The role of calponin in the gene profile of metastatic cells: inhibition of metastatic cell motility by multiple calponin repeats. FEBS Lett. 2004;556:221–226. doi: 10.1016/s0014-5793(03)01401-7. [DOI] [PubMed] [Google Scholar]
- Littlefield R, Almenar-Queralt A, Fowler VM. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat Cell Biol. 2001;3:544–551. doi: 10.1038/35078517. [DOI] [PubMed] [Google Scholar]
- Maciver SK, Weeds AG. Actophorin preferentially binds monomeric ADP-actin over ATP-bound actin: consequences for cell locomotion. FEBS Lett. 1994;347:251–256. doi: 10.1016/0014-5793(94)00552-4. [DOI] [PubMed] [Google Scholar]
- Maciver SK, Wachsstock DH, Schwarz WH, Pollard TD. The actin filament severing protein actophorin promotes the formation of rigid bundles of actin filaments crosslinked with alpha-actinin. J Cell Biol. 1991a;115:1621–1628. doi: 10.1083/jcb.115.6.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver SK, Zot HG, Pollard TD. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J Cell Biol. 1991b;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver SK, Pope BJ, Whytock S, Weeds AG. The effect of two actin depolymerizing factors (ADF/cofilins) on actin filament turnover: pH sensitivity of F-actin binding by human ADF, but not of Acanthamoeba actophorin. Eur J Biochem. 1998;256:388–397. doi: 10.1046/j.1432-1327.1998.2560388.x. [DOI] [PubMed] [Google Scholar]
- McElhinny AS, Schwach C, Valichnac M, Mount-Patrick S, Gregorio CC. Nebulin regulates the assembly and lengths of the thin filaments in striated muscle. J Cell Biol. 2005;170:947–957. doi: 10.1083/jcb.200502158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough A. F-actin-binding proteins. Curr Opin Struct Biol. 1998;8:166–176. doi: 10.1016/s0959-440x(98)80034-1. [DOI] [PubMed] [Google Scholar]
- McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Heschl MF, Rosenbluth RE, Baillie DL. Genetic organization of the unc-60 region in Caenorhabditis elegans. Genetics. 1988;118:49–59. doi: 10.1093/genetics/118.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman DG, Fire A. Muscle: Structure, function, and development. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 417–470. [PubMed] [Google Scholar]
- Mohri K, Ono S. Actin filament disassembling activity of Caenorhabditis elegans actin-interacting protein 1 (UNC-78) is dependent on filament binding by a specific ADF/cofilin isoform. J Cell Sci. 2003;116:4107–4118. doi: 10.1242/jcs.00717. [DOI] [PubMed] [Google Scholar]
- Mohri K, Ono K, Yu R, Yamashiro S, Ono S. Enhancement of actin-depolymerizing factor/cofilin-dependent actin disassembly by actin-interacting protein 1 is required for organized actin filament assembly in the Caenorhabditis elegans body wall muscle. Mol Biol Cell. 2006;17:2190–2199. doi: 10.1091/mbc.E05-11-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouneimne G, DesMarais V, Sidani M, Scemes E, Wang W, Song X, Eddy R, Condeelis J. Spatial and temporal control of cofilin activity is required for directional sensing during chemotaxis. Curr Biol. 2006;16:2193–2205. doi: 10.1016/j.cub.2006.09.016. [DOI] [PubMed] [Google Scholar]
- Moza M, Mologni L, Trokovic R, Faulkner G, Partanen J, Carpen O. Targeted deletion of the muscular dystrophy gene myotilin does not perturb muscle structure or function in mice. Mol Cell Biol. 2007;27:244–252. doi: 10.1128/MCB.00561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy K, Wadsworth P. Myosin-II-dependent localization and dynamics of F-actin during cytokinesis. Curr Biol. 2005;15:724–731. doi: 10.1016/j.cub.2005.02.055. [DOI] [PubMed] [Google Scholar]
- Nishita M, Tomizawa C, Yamamoto M, Horita Y, Ohashi K, Mizuno K. Spatial and temporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional cell migration. J Cell Biol. 2005;171:349–359. doi: 10.1083/jcb.200504029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive M, Goldfarb LG, Shatunov A, Fischer D, Ferrer I. Myotilinopathy: refining the clinical and myopathological phenotype. Brain. 2005;128:2315–2326. doi: 10.1093/brain/awh576. [DOI] [PubMed] [Google Scholar]
- Ono K, Parast M, Alberico C, Benian GM, Ono S. Specific requirement for two ADF/cofilin isoforms in distinct actin-dependent processes in Caenorhabditis elegans. J Cell Sci. 2003;116:2073–2085. doi: 10.1242/jcs.00421. [DOI] [PubMed] [Google Scholar]
- Ono K, Yu R, Mohri K, Ono S. Caenorhabditis elegans kettin, a large immunoglobulin-like repeat protein, binds to filamentous actin and provides mechanical stability to the contractile apparatuses in body wall muscle. Mol Biol Cell. 2006;17:2722–2734. doi: 10.1091/mbc.E06-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. Purification and biochemical characterization of actin from Caenorhabditis elegans: its difference from rabbit muscle actin in the interaction with nematode ADF/cofilin. Cell Motil Cytoskeleton. 1999;43:128–136. doi: 10.1002/(SICI)1097-0169(1999)43:2<128::AID-CM4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Ono S. The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J Cell Biol. 2001;152:1313–1319. doi: 10.1083/jcb.152.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int Rev Cytol. 2007;258:1–82. doi: 10.1016/S0074-7696(07)58001-0. [DOI] [PubMed] [Google Scholar]
- Ono S, Benian GM. Two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins, encoded by the unc-60 gene, differentially regulate actin filament dynamics. J Biol Chem. 1998;273:3778–3783. doi: 10.1074/jbc.273.6.3778. [DOI] [PubMed] [Google Scholar]
- Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 2002;156:1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Baillie DL, Benian GM. UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J Cell Biol. 1999;145:491–502. doi: 10.1083/jcb.145.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, McGough A, Pope BJ, Tolbert VT, Bui A, Pohl J, Benian GM, Gernert KM, Weeds AG. The C-terminal tail of UNC-60B (ADF/cofilin) is critical for maintaining its stable association with F-actin and is implicated in the second actin-binding site. J Biol Chem. 2001;276:5952–5958. doi: 10.1074/jbc.M007563200. [DOI] [PubMed] [Google Scholar]
- Ono S, Mohri K, Ono K. Microscopic evidence that actin-interacting protein 1 actively disassembles actin-depolymerizing factor/cofilin-bound actin filaments. J Biol Chem. 2004;279:14207–14212. doi: 10.1074/jbc.M313418200. [DOI] [PubMed] [Google Scholar]
- Pacholsky D, Vakeel P, Himmel M, Lowe T, Stradal T, Rottner K, Furst DO, van der Ven PF. Xin repeats define a novel actin-binding motif. J Cell Sci. 2004;117:5257–5268. doi: 10.1242/jcs.01406. [DOI] [PubMed] [Google Scholar]
- Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85:164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Actin filament severing by cofilin. J Mol Biol. 2007;365:1350–1358. doi: 10.1016/j.jmb.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Salmikangas P, van der Ven PF, Lalowski M, Taivainen A, Zhao F, Suila H, Schroder R, Lappalainen P, Furst DO, Carpen O. Myotilin, the limb-girdle muscular dystrophy 1A (LGMD1A) protein, cross-links actin filaments and controls sarcomere assembly. Hum Mol Genet. 2003;12:189–203. doi: 10.1093/hmg/ddg020. [DOI] [PubMed] [Google Scholar]
- Witt CC, Burkart C, Labeit D, McNabb M, Wu Y, Granzier H, Labeit S. Nebulin regulates thin filament length, contractility, and Z-disk structure in vivo. EMBO J. 2006;25:3843–3855. doi: 10.1038/sj.emboj.7601242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Ono S. Dual roles of tropomyosin as an F-actin stabilizer and a regulator of muscle contraction in Caenorhabditis elegans body wall muscle. Cell Motil Cytoskeleton. 2006;63:659–672. doi: 10.1002/cm.20152. [DOI] [PMC free article] [PubMed] [Google Scholar]