Summary
Because increasing numbers of HIV vaccine candidates are being tested globally, it is essential to differentiate vaccine-from virus-induced antibodies. Most of the currently tested vaccines contain multiple viral components. As a result, many vaccine recipients give positive results in FDA-licensed HIV serodetection tests. We have identified conserved sequences in Env-gp41 and Gag-p6, which are recognized soon after infection but are not included in most HIV vaccine candidates. A new HIV serodetection assay, the HIVSELECTEST, was established that distinguishes between vaccine-induced antibodies and seroconversion due to true HIV infections. It is important to make this assay globally relevant, because many clinical trials are conducted around the world where most HIV infections are due to non-B subtype HIV-1. Therefore, the current study examined the reactivity of plasma samples from .3000 infections with diverse HIV subtypes worldwide. The HIVSELECTEST performed at .99% specificity and sensitivity. Both recent and established infections with clades A, B, C, D, E, F, G, J, and CRFs were detected. Antibodies elicited by other vaccinations or infections endemic to the clinical trial sites did not react in this assay. Therefore, HIV-SELECTEST could be an important differential diagnostic tool for HIV vaccine trials, blood banks, and population screening worldwide.
Keywords: HIV, AIDS, diagnosis, vaccine, clinical trials, phage display, peptides, epitope mapping
Persons infected with HIV-1 usually mount a strong humoral immune response to the virus, resulting in the production of specific antibodies. Consequently, serological detection of HIV antibodies has been the principal diagnostic tool used by clinicians and blood banks to detect HIV infections. Due to the increased complexity of current HIV vaccine candidates,1 participants in preventive HIV vaccine trials can generate antibodies that are reactive in serologic tests (Enzyme Immuno-Assay [EIA], rapid tests, and Western blots) that are licensed by the US Food and Drug Administration (FDA) and other national regulatory authorities.2–5 Thus, vaccine recipients could be erroneously diagnosed as being HIV-infected and may suffer from a range of economic and social harms.6 Accurate and rapid diagnosis of incident HIV infections during preventive vaccine trials using the current algorithm of tests is greatly compromised. This will become more problematic during large-scale Phase 3 trials with thousands of participants and even more so after several HIV vaccines are licensed. Furthermore, mass immunization with vaccines that are not expected to prevent infection but instead to reduce viral loads and disease progression will present a real challenge to identifying individuals who become infected after vaccination. The blood-banking industry is testing donated blood for the presence of HIV antibodies, and since 1999 has also begun additional testing of plasma for HIV RNA by nucleic acid amplification tests (NAT).7,8 Many HIV-uninfected individuals who are immunized with HIV vaccines will be rejected from blood donation at the first-line screen with HIV EIA or rapid tests. Furthermore, because some infected people control viremia to the point that NAT testing is negative, relying on NAT testing will no longer be sufficient for detection of HIV-infected donors, especially among those who were vaccinated.
Currently, there is no licensed HIV immunodiagnostic assay that differentiates between vaccine-induced antibodies and those produced after true HIV infection. Such a differential diagnostic tool will be of great value in future HIV preventive vaccine trials and for blood banks, should wide-spread vaccination take place.
To achieve this goal, a Gene-Fragment Phage Display Library (GFPDL) was constructed from the entire HIV-1 cDNA genome and was subjected to screening with sera from HIV-infected individuals near the time of seroconversion. This strategy led to the discovery of three novel early immunodominant epitopes: one in Gag p6 and two in the envelope-gp41 cytoplasmic tail. These epitopes are currently not included in most candidate HIV vaccines because they are unlikely to contribute to protective immunity. These three sequences also demonstrate a high degree of conservation among all group-M HIV subtypes. Based on the newly identified epitopes and the Los Alamos database, three consensus peptides were chemically synthesized and used for the development of a new HIV enzyme-linked immune absorbent assay (ELISA), termed HIV-SELECTEST. In the first proof-of-concept study, we demonstrated that participants in HIV vaccine trials scored negatively in the new assay but reacted positively in several licensed immunodiagnostic tests. Importantly, all intercurrent HIV infections with HIV-1 subtypes B and E during the course of several vaccine trials were detected by the HIV-SELECTEST within 3 months post-infection, as documented by RNA amplification assays.9
Before the new immunodiagnostic test could be implemented in international sites of future HIV vaccine trials, it is important to demonstrate the sensitivity of the HIV-SELECTEST to accurately diagnose HIV infections due to diverse viral subtypes that are currently known to exist. Additionally, the specificity of the assay must be established with plasma samples from developing countries, where other concomitant infections are endemic.
The current study was designed to establish the sensitivity and specificity of the HIV-SELECTEST with national and international samples obtained from cohorts of both early and established HIV infections. We also examined the potential for unwanted cross-reactivity (ie, false positives) due to other endemic infections and recent non-HIV vaccinations.
MATERIALS AND METHODS
Design and Procedures of the HIV-SELECTEST
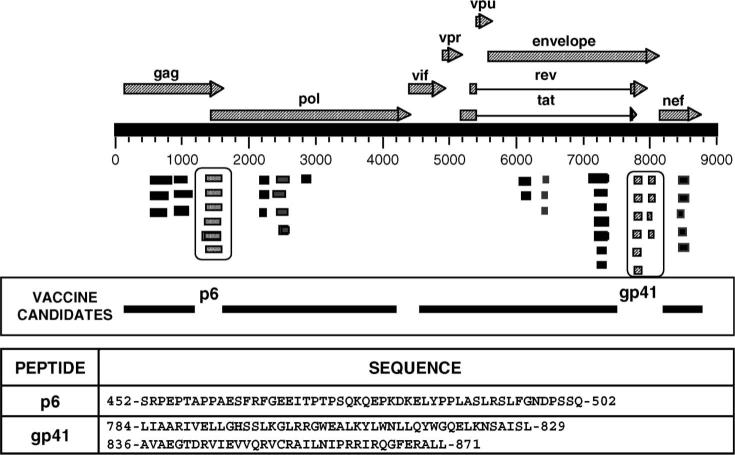
A GFPDL expressing all of the open reading frames of HIV-1 pNL4−3 was constructed and screened with plasma samples comprising the HIV-1 seroconversion panel PRB-910 from SeraCare BioServices (Gaithersburg, MD) as previously described.9 The inserts of the phage clones demonstrating high reactivity with HIV-1 seropositive sera were sequenced and mapped to different HIV proteins (Fig. 1). Three sequences that do not contain known HIV-protective epitopes (two in gp41 intracytoplasmic tail and one in p6; Los Alamos database; http://hiv-web.lanl.gov) were selected for the development a new HIV diagnostic assay, termed HIV-SELECTEST. The selected sequences from Gag p6 (452-SRPEPTAPPAESFRFGEEITPTPSQKQEPKDKELYPPLASLRSLFGNDPSSQ-502) and gp41 cytoplasmic region (SK1: 784 LIAARIVELLGHSSLKGLRRGWEALKYLWNLLQYWGQELKNSAISL-829 and SK2: 836-AVAEGTDRVIEVVQRVCRAILNIPRRIRQGFERALL-871) were chemically synthesized.
FIGURE 1.
Identification of HIV peptides for differential diagnosis of HIV infection during vaccine trials using gene-fragment phage display library. Various coding regions (ORFs) are depicted along their location in the HIV-1 genome. Alignment of HIV sequences displayed on the affinity-selected phage clones after the fourth round of panning with the HIV-1 genome sequence (pNL4−3 proviral clone) led to identification of the immunodominant regions recognized by antibodies generated soon after HIV infection. HIV genes/proteins that are part of current candidate HIV vaccines are also aligned to the HIV-1 genome. The peptide sequences that are being used in HIV-SELECTEST are shown. Numbers in each epitopic site indicate the position of the amino acid residues in the corresponding HIV-1–encoded proteins for the CON-OF-CONS alignment sequence in the Los Alamos database.
The optimal conditions for the p6 and gp41 ELISA were determined by screening a large number of HIV seronegative and seropositive sera (Fig. 2 and data not shown). Based on these analyses, p6 peptide was coated at 30 ng/100 mL/well and the gp41 peptides (SK1 and SK2) were coated at 150 ng/ 100 mL/well each (total 300 ng/well) on Immulon-2HB plates. After three washes with phosphate-buffered saline containing Tween-20 (PBST) (20 mM PBS, 0.1% Tween-20), the plates were blocked with PBST containing 2% whole milk (WMPBST). For testing, all specimens were diluted 1:100 in WMPBST and were added to peptide-coated wells for 1 hour at room temperature (RT). After 6 washes with PBST, the wells were reacted with HRP-conjugated goat anti-human IgG Fcspecific antibody (diluted 1:10,000) (Jackson ImmunoRe-search, West Grove, PA) at RT for 1 hour, followed by addition of O-phenylenediamine (OPD) substrate.
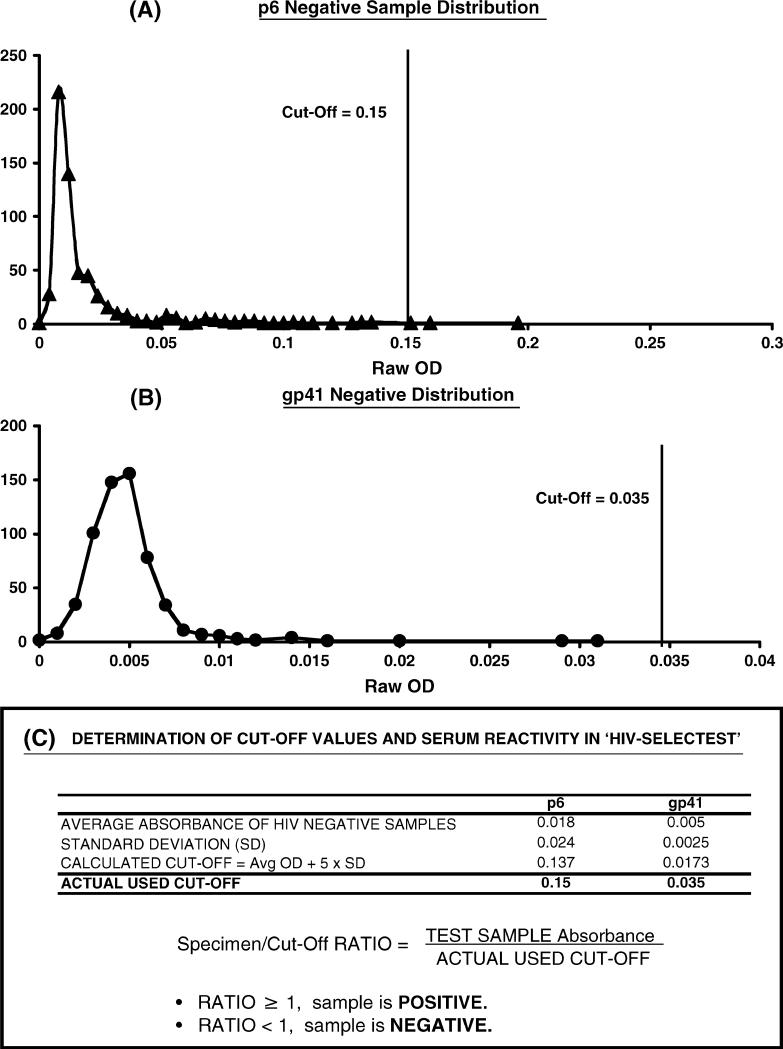
FIGURE 2.
Seroreactivity of HIV-negative samples with HIV-SELECTEST peptides and determination of cut-off values. ELISA conditions are described in Methods. Reactivity of 600 HIV seronegative serum/plasma samples at 1:100 dilution with the peptides, p6 (panel A) and gp41 (panel B) is shown. Values on the X axis depicts the test specimen optical density (OD) with the HIV peptide in specific ELISA, and the Y axis represents the number of serum/plasma samples that exhibited a specific ELISA absorbance reading as shown on the X axis. C, Cut-off value for each peptide was determined as the mean absorbance + 5 SD obtained with HIV-seronegative samples. All data are represented as ratios between test specimen optical densities (OD) to cut-off absorbance (CO).
The cut-off values (average optical absorbance of 600 HIV-negative samples at 450 nm + 5 standard deviations) were established for each peptide plate. Data are presented as the ratios of the absorbance of the test specimens to the cut-off values, whereby ratios of ≥ 1 are considered HIV-1 seropositive and ratios <1 are considered HIV-1 seronegative (Fig. 2).
Sources of HIV-Seropositive Specimens
HIV-1 seroconversion panels PRB-910, PRB-924, PRB-927, PRB-928, PRB-929, and PRB-931 were purchased from SeraCare BioServices, (Gaithersburg, MD). Panel ZM-6240 was purchased from Zeptometrix Corporation (Buffalo, NY). These panels contained plasma samples collected serially early after HIV-1 infection. At each blood collection, HIV RNA, p24, and antibodies were assessed by commercial diagnostic kits by the respective manufacturers, and this information was provided with the panels.
A large number of serum or plasma samples from HIV-infected individuals, either during early infections (,6 months) or established/chronic infections, were provided by various collaborators.
Western blot (WB)-confirmed positive blood donor specimens were obtained in studies conducted in the US, Brazil, and the Republic of South Africa.7,8 These samples were subcategorized into recent seroconversion and long standing infections using the Less-Sensitive (LS)-EIA strategy.10–12
Early infection and matched seronegative samples were obtained from the Zambia-Emory HIV Research Project, Lusaka, Zambia, a project funded by NIH and IAVI. Seroconverters were identified during follow-up of a 750 discordantcouple cohort in which seronegative partners are provided every 3 months with prevention counseling and condoms and are screened using rapid antibody tests. Seroconversion was ascertained using a two–rapid test algorithm and confirmed by ELISA at the Zambia Blood Transfusion Service.13
Plasma samples from HIV-infected subjects from Cameroon were obtained through an HIV seroprevalence study in rural villages in the equatorial rain forest regions and a blood-bank surveillance study in the cities of Yaounde and Douala. The genetic subtypes of the viruses infecting these individuals were determined for gag, pol, and/or env genes. The subtypes identified in some of the Cameroonian samples presented in Table 3 have previously been described.14–16
TABLE 3.
Detection of Diverse HIV-1 Subtype Infections in the HIV-SELECTEST
|
HIV-Selectest Reactivity (%) |
||||||
|---|---|---|---|---|---|---|
| Subtype | Total No. of Samples | Country of Origin | No. of Samples | p6 | gp41 | Combined* |
| A | 35 | Cameroon | 21 | 71 | 92 | 100 |
| Tanzania | 4 | 0 | 100 | 100 | ||
| Russia | 10 | 80 | 100 | 100 | ||
| US | 230 | 49 | 87 | 98 | ||
| B | 417 | Australia | 123 | 68 | 81 | 96 |
| Brazil | 50 | 86 | 88 | 98 | ||
| China | 14 | 43 | 93 | 100 | ||
| South Africa | 50 | 46 | 78 | 100 | ||
| Cameroon | 3 | 100 | 66 | 100 | ||
| Tanzania | 23 | 28 | 78 | 83 | ||
| C | 209 | India | 40 | 13 | 100 | 100 |
| Zambia | 39 | 80 | 67 | 97 | ||
| Ethiopia | 26 | 77 | 96 | 98 | ||
| China | 28 | 50 | 100 | 100 | ||
| D | 20 | Cameroon | 7 | 66 | 100 | 100 |
| Uganda | 13 | 39 | 92 | 93 | ||
| E | 240 | Thailand | 235 | 39 | 91 | 100 |
| Cameroon | 5 | 80 | 100 | 100 | ||
| F | 2 | Cameroon | 2 | 50 | 100 | 100 |
| G | 11 | Cameroon | 11 | 64 | 100 | 100 |
| J | 10 | Cameroon | 10 | 90 | 85 | 100 |
| AG | 150 | Cameroon | 150 | 80 | 95 | 100 |
| AJ | 7 | Cameroon | 7 | 57 | 100 | 100 |
| CRF | 28 | Cameroon | 28 | 63 | 96 | 100 |
| Unknown | 595 | Uganda | 318 | 51 | 94 | 100 |
| Kenya | 277 | 39 | 97 | 99 | ||
| Totals | 1724 | 53 | 91 | 99 | ||
Combined reactivity is a seropositive test in either the p6 or the gp41 peptide assay.
Sera were obtained from the Moscow Women's Health Study, an NIDA-supported Russian–US collaboration. Participants were enrolled in a study of HIV, HCV, and STI risks and vulnerabilities. All participants were current (2003−2005) sex workers, with a median work time of ∼8 months, suggesting recent infections. Few had been tested before enrollment. All were asymptomatic at the time of the blood draw.
The HIV infection status of a given sample was provided by the collaborating groups. In a fraction of samples, it was also determined in-house using the FDA-licensed Genetic Systems HIV-1/2 Plus O EIA (Bio-Rad Laboratories, Redmond, WA).
HIV-negative serum samples were obtained from National Institutes of Health Blood Bank, the Joint Clinical Research Center, Mengo, Uganda, and the Vaccine Research Center (VRC, NIAID, NIH, Bethesda, MD).
Sources of Specimens From Infections Other Than HIV
The TB-positive plasma samples were obtained with informed consent from smear-positive TB patients from Lala Ram Swarup Hospital for Tuberculosis, New Delhi, India.
Malaria specimens were obtained from smear-positive children who were screened for presence of Plasmodium falciparum histidine-rich protein-2 antigen using Core Malaria Pf (Core Diagnostics, Birmingham, UK). All the specimens tested positive using this assay and tested negative for HIV infection.
Specimens from blood donors with confirmed serology for HTLV I, hepatitis B (HBV), and hepatitis C (HCV) were obtained from the Retrovirus Epidemiology Donor Study (REDS-I).12
All specimens were unlinked from donor information. All studies were conducted under approval from the Research Involving Human Subjects Committee (RIHSC exemption no. 04−050B) at the Center for Biologics Evaluation and Research.
Sources of Postvaccination Specimens
Post-polio vaccination specimens were obtained from children (unidentified samples) that received full course of OPV, through collaboration with Dr. Olga Utnitskaya, State Center of Sanitary-Epidemiological Surveillance, Ekaterinburg, Russian Federation, and Dr. Alexander Ivanov (Division of Viral Products, CBER, FDA). Post-influenza vaccination samples were obtained from Dr. Roland Levandowski (Division of Viral Products, CBER, FDA), and post-Dryvax immune plasma were obtained from recently vaccinated CBER employees.
RESULTS
Establishment of HIV-SELECTEST
Based on the Los Alamos HIV sequence database, consensus peptides were designed to encompass the genetic variability among HIV-1 clades. The p6-derived and the two gp41-derived peptides were chemically synthesized and used for the development of a new ELISA, as previously described.9 Initially, each peptide was individually evaluated with 600 HIV-seronegative sera to determine specificity and to establish cut-off values (Fig. 2). Most of the seronegative samples showed low reactivity with the p6 peptide in ELISA, but there were a few outlier sera for which higher signal/noise ratios were observed (Fig. 2A). In contrast, both gp41 peptides displayed a similar very low reactivity with HIV-seronegative samples and could be combined in a single-well ELISA (Fig. 2B). The cut-off value (mean + 5 SD) for the gp41 peptide combination was 4-fold lower than the cut-off value for the p6 ELISA (Fig. 2C). In subsequent testing of > 1000 samples obtained from uninfected individuals, 100% specificity was demonstrated for the gp41 and 99.4% for the p6 peptide ELISA.
HIV-SELECTEST Detects HIV Antibodies Shortly After PCR-Confirmed Infections and Early Post-Seroconversion Periods
Initially, the intended use of the new HIV-SELECTEST will be in prophylactic HIV vaccine trials, where intercurrent HIV infections in vaccine trial participants may occur due to high-risk behavior and/or suboptimal vaccine-induced protection. Therefore, it was important to determine how long after infection antibodies against the gp41 and p6 epitopes are detected compared with other serological tests that contain multiple viral proteins/peptides. Five well-characterized seroconversion panels containing sequential blood draws collected around the time of acute viremia and seroconversion were obtained from SeraCare BioServices. Representative data from one of the five panels are shown in Table 1. HIV infection was confirmed by PCR on visit day 49, and seroconversion was confirmed in multiple FDA-licensed HIV-detection kits on day 92. Importantly, the same blood draw reacted positively in both p6 and gp41 ELISAs (Table 1). These data complement similar results obtained earlier with other seroconversion panels from SeraCare BioServices,9 demonstrating that HIV infections could be detected by the HIV-SELECTEST within 2 to 4 weeks after HIV-1 RNA detection by PCR, generally concurrent with previously licensed serological HIV-diagnostic kits.
TABLE 1.
Early Detection of HIV-1 Infection by HIV-SELECTEST in Seroconversion Panel
|
HIV-SELECTEST |
||||||
|---|---|---|---|---|---|---|
| Sample ID | Day Collected | p6* | gp41* | Abbott HIV 1/2† | FDA Lic. EIA Kits† | PCR2 |
| PRB904−01 | 0 | 0.0471 | 0.1143 | 0.1 | 0/8 | NEG |
| PRB904−02 | 21 | 0.0236 | 0.0857 | 0.2 | 0/8 | NEG |
| PRB904−03 | 49 | 0.0393 | 0.0857 | 0.2 | 0/8 | POS |
| PRB904−04 | 92 | 1.2150 | 1.2000 | 12.0 | 8/8 | POS |
| PRB904−05 | 99 | 2.9760 | 4.4857 | 12.2 | 8/8 | POS |
ELISA data are shown as the ratio of test specimen absorbance to cut-off value. Ratios ≥1.00 are considered HIV-seropositive, and sample ratios <1 are considered HIV-negative.
HIV early seroconversion panels (within 6 weeks after HIV infection), data for HIV RNA PCR quantification (Roche), and FDA-licensed serodiagnostic kits were provided by SeraCare BioServices (Gaithersburg, MD).
All of the incident infection panels that were initially tested were from HIV clade B infections. Because the majority of the planned HIV vaccine trials will be conducted at international sites with predominantly non-clade B circulating HIV strains, it was important to evaluate early infection samples from countries with diverse HIV subtypes. As summarized in Table 2, the HIV-SELECTEST showed similar detection rates with 493 plasma samples obtained from individuals who had recently seroconverted based on either prospective observations of exposed individuals (including a study of discordant couples in Zambia) or retrospective evaluations of cross-section confirmed HIV seropositive blood donors using the less-sensitive (LS) and FDA-licensed HIV serodetection EIAs. These included infections with HIV-1 strains of clades B, C, and E from multiple countries. This finding can be explained by the fact that peptides for the new assay were designed based on consensus sequences representing all HIV-1 group-M subtypes. The combined sensitivity of the p6 and gp41 HIV-SELECTEST (98.6%) was comparable to the sensitivity of current FDA-licensed HIV antibody detection assays for the samples tested.
TABLE 2.
HIV-SELECTEST Can Detect Early HIV Infection From Multiple HIV-1 Clades
|
HIV-SELECTEST Reactivity (%) |
||||||
|---|---|---|---|---|---|---|
| HIV-1 Subtype | Total No. of Samples | Country of Origin* | No. of Samples by Country | p6 | gp41 | Combined† |
| B | 260 | Australia | 22 | 78.2 | 66 | 95.4 |
| US | 213 | 49 | 80.2 | 98.6 | ||
| Brazil | 25 | 80 | 40 | 92 | ||
| C | 63 | South Africa | 25 | 48 | 72 | 100 |
| Zambia | 38 | 78 | 72 | 98 | ||
| E | 170 | Thailand | 170 | 42.4 | 85.6 | 100 |
| Totals | 493 | 51.9 | 78.7 | 98.7 | ||
HIV-SELECTEST Displays High Cross-Clade Reactivity With Established HIV-1 (Group M) Infections
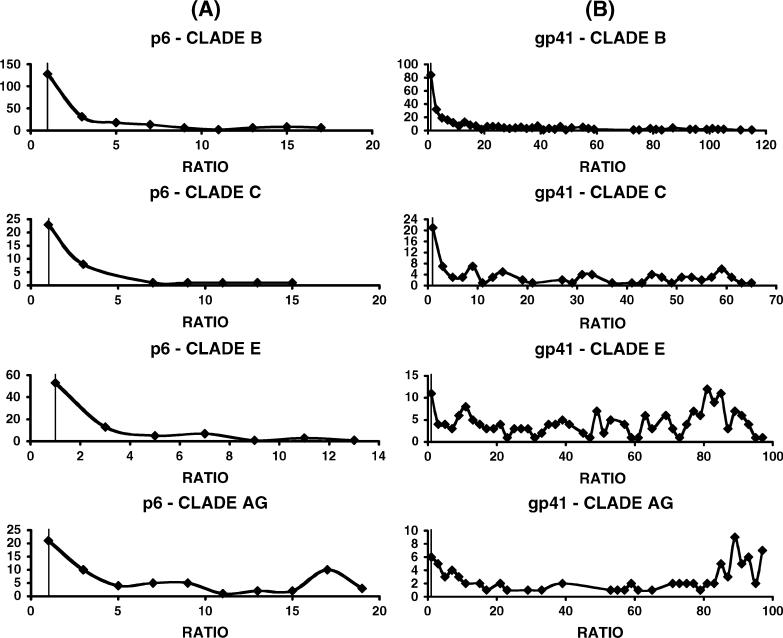
Once the assay sensitivity for detection of early HIV infections (1−6 months) was demonstrated, we set out to determine if the HIV-SELECTEST was also reactive with samples from established/chronic infections with diverse HIV-1 clades and subtypes. In earlier studies with clade B infections, we noticed that the reactivity against the p6 peptide (but not against the gp41 epitopes) diminished over time, suggesting discordance in the humoral responses against the two components of the HIV SELECTEST.9 Table 3 summarizes the response rates (% reactivity) with p6 and gp41 peptides for 1724 samples infected with diverse HIV-1 subtypes obtained from different countries. As can be seen, the response rates for all the specimens from HIV-1 group-M infections were approximately 53% for the p6, 91% for the gp41, and 99% for both peptides. Many samples were obtained from countries in which diverse circulating recombinant forms (CRFs) are found (eg, Cameroon). In addition, samples from Kenya and Uganda typically include unique recombinant forms (URFs). Figure 3 demonstrates the range of positive reactivity (ie, ratios of absorbance of samples/cut-off values for p6 and gp41) measured with specimens from patients infected with HIV-1 clades B, C, E, and AG viruses. In general, the ratios obtained with the gp41 peptide ELISA are higher than with the p6 peptide due to the lower background absorbance and cut-off values (Fig. 2). The HIV-SELECTEST gave similar distribution of ratios with plasma samples from all clades. In the case of clade B and clade C infections, our panels included larger numbers of early infections, which was reflected in the higher fractions of plasma with lower reactivity ratios in the gp41 peptide ELISA.
FIGURE 3.
Comparison of seroreactivity of samples obtained from individuals infected with different HIV clades with HIV-SELECTEST peptides. ELISA conditions were described in METHODS. Reactivity of serum/plasma samples at 1:100 dilution with the peptides, p6 (panel A) and gp41 (panel B) from early and chronically infected individuals with different HIV subtypes is shown as indicated in each graph. Values on the X axis depict the ratios between test specimen optical densities (OD) to cut-off values based on the mean OD of 600 negative controls. The Y axis represents the number of samples that exhibited a specific reactivity ratio as shown on the X axis.
Together, the combined sensitivity of the HIV-SELECTEST in detecting early and established HIV group M infections in multiple geographical sites with clades A, B, C, D, E, F, G, J, and CRFs is currently around 99% (Tables 2 and 3 and previously published data9).
HIV-SELECTEST Does Not React With Antibodies Generated by Non-HIV Vaccines and Other Infections
Antibodies generated by common vaccinations or by non-HIV endemic infections may produce false-positive results in HIV serodetection kits.17–19 It was important to look for such cross-reactivity, which may reduce the specificity of the HIV-SELECTEST, especially in developing countries where individuals may be exposed to a plethora of endemic infections. Blinded plasma panels from individuals vaccinated with inactivated influenza vaccine or live attenuated polio and smallpox vaccines at peak antibody responses did not show cross-reactivity with either the p6 or gp41 peptides used in the HIV-SELECTEST (Table 4). Furthermore, testing of specimens from patients infected with (and serologically reactive to) diverse infections known to be endemic in the developing countries showed no cross-reactivity in the gp41 peptide ELISA. In the case of p6, 1/20 samples positive for tuberculosis antibody demonstrated weak reactivity in the HIV-SELECTEST while samples from individuals infected with malaria, hepatitis B, hepatitis C, or HTLV-I did not (Table 5). These findings are in agreement with the overall 99.4% specificity observed for the p6 peptide with > 1800 non-HIV-infected serum samples tested to date. The gp41 peptide EIA currently performs with 100% specificity.
TABLE 4.
HIV-SELECTEST Does not React With Serum Samples From Non-HIV Vaccinations
|
HIV-Selectest Reactivity (%) |
|||
|---|---|---|---|
| Vaccine | Total No. of Samples | p6 | gp41 |
| Flu vaccine* | 37 | 0 | 0 |
| Polio vaccine† | 62 | 0 | 0 |
| Smallpox vaccine‡ | 22 | 0 | 0 |
The influenza-vaccine specimens were obtained from U.S. individuals immunized with Fluzone (Sanofi Pasteur, Swiftwater, PA, USA) in 2002 collected 2−3 weeks following immunization.
Samples were obtained from Russian children vaccinated with the oral polio vaccine.
Samples were obtained from US individuals immunized with Dryvax (Wyeth, Madison, NJ).
TABLE 5.
Reactivity of Serum Samples from Non-HIV Infections in HIV-SELECTEST
|
HIV-SELECTEST Positive |
|||
|---|---|---|---|
| Infection* | Total No. of Samples | p6 | gp41 |
| Malaria | 16 | 0 | 0 |
| Hepatitis B | 80 | 0 | 0 |
| Hepatitis C | 59 | 0 | 0 |
| Tuberculosis | 20 | 1 | 0 |
| HTLV-I | 52 | 0 | 0 |
Panels of HIV-seronegative individuals with the indicated infections were obtained from Bamako, Mali, Africa, India, and the US (REDS-I). Some panels were previously described in reference 12.
DISCUSSION
Concerted efforts are underway to develop preventive HIV vaccines to curtail the global AIDS epidemic. All of the new-generation vaccine candidates are complex products containing multiple HIV genes or proteins.20,21 Several of these vaccines will soon enter Phase 2B “proof-of-concept” trials in multiple countries and may progress into large-scale efficacy trials in regions with high HIV-infection rates. Because of the diversity of circulating strains, parallel clinical trials will be required to establish efficacy in different countries either with one multi-clade vaccine or with different vaccine products tailored to the predominant strains in a given country. It is expected that current vaccine candidates may not afford complete protection against HIV infection but may possibly improve the control of viremia, and disease progression will be delayed and that secondary transmissions to others will be reduced. To achieve the statistical power needed to demonstrate partial efficacy, it will be necessary to recruit thousands of volunteers into future Phase 3 HIV vaccine trials. Additionally, licensure of HIV preventive vaccines, even with a modest efficacy, undoubtedly will be followed by large-scale vaccination in countries with high HIV-transmission rates and will include health care personnel in most countries.22
Therefore, it is anticipated that, subsequent to vaccination, many uninfected individuals will test positive in all previously FDA-licensed HIV detection kits (including rapid tests) and could remain seropositive for many years. This assumption is supported by the observation that 30% to 100% of samples from uninfected participants in multiple HIV vaccine trials to date show positive reactivity in commonly used HIV diagnostic tests.2 This is a major public health concern because it will confound our ability to correctly identify those with true HIV infection and may lead to the inappropriate exclusion of HIV-uninfected vaccinees from the pool of blood donors, and also inadvertently lead to an array of social and economic hardships. Hence, further improvements in HIV diagnosis are urgently required. Clinics, blood banks, and vaccine trial sites need rapid and simple low-cost tests that will distinguish between vaccine-induced antibodies and true HIV infections that can be easily implemented for large-scale screening.23,24
The use of GFPDL to clone and express all the open reading frames of HIV afforded us the opportunity to identify all the epitopes that are recognized by antibodies shortly after HIV infection. Affinity selection of the phage-display library using recent seroconversion panels led to the identification of epitopes in gp41 and p6 that were selected for the development of the new differential diagnostic test, termed HIV-SELECTEST.9 Previously, we demonstrated that HIV vaccine–generated antibodies did not react in the HIV-SELECTEST while all intercurrent HIV infections were detected within 3 months of PCR-confirmed infection. However, it was noted that all of these trials took place either in the U.S. or in Thailand, and breakthrough infections were primarily with clade B and clade A/E strains, respectively.9
Future vaccine trials will take place in many sites world wide, such as Africa, South America, and India, where HIV transmission rates are high and the predominant strains belong to diverse subtypes, including new recombinants. Thus, it was important to establish the sensitivity of the HIV-SELECTEST to detect HIV antibodies elicited by infections with all circulating HIV clades.
In the current study, we examined either early-infection samples (< 6 months) or established HIV infections with viruses from clades A, B, C, D, E, F, G, J, and multiple recombinants (AG, AJ, and other CRFs). The combined detection rates (p6 + gp41) for the early infections with clades B, C, and E were 98.6% (Table 2), reflecting heterogeneity and unlinked humoral responses against the two viral components.
In patients with established and chronic HIV infections, the p6 EIA was reactive with 53% and the gp41 with 91% of all samples tested (Table 3). Importantly, while the combined sensitivity for detecting HIV-1 group-M infections is 99% (ie, positive reactivity in at least one peptide ELISA), irrespective of disease stage or infecting virus variant, the antibody responses against p6 and gp41 are not linked and seem to be subject to independent fluctuations during the course of infection.9 Therefore, at this stage, it is important to include both peptides in the HIV-SELECTEST. The utility of the assay during vaccine trials will depend also on the composition of the vaccine being tested. Most current vaccines do not contain the gp41 peptides. The p6 antigen is still present in some currently tested vaccine candidates. However, the p6 peptide sequence could be easily removed because it does not contain neutralizing antibody or CTL epitopes (Los Alamos database; http://hiv-web.lanl.gov) and is not likely to contribute to vaccine efficacy. Deletion of p6 from gag-pol–expressing vectors by scientists at the Vaccine Research Center (VRC, NIH) assured a 1:1 ratio of protein expression (compared with the 1:20 gag/pol protein ratio in virions).21 Thus, the removal of p6 is relatively simple (due to its location in the C terminus of gag) and may even be beneficial for vaccines containing both gag and pol genes. In our studies, immunization of rabbits with KLH-conjugated p6 and gp41 peptides generated a strong antibody response against these specific peptides. However, the polyclonal rabbit antibodies did not neutralize infection in vitro with either R5 or X4 HIV strains (data not shown). Together, this information supports the notion that the p6 and gp41 intracytoplasmic peptides used in the HIV-SELECTEST are not important for HIV vaccines.
All diagnostic assays may be subject to either interference or reduced specificity due to concurrent presence of antibodies against other pathogens or because of recent vaccinations. Thus far, we have demonstrated that recent vaccination (with influenza, polio, and smallpox vaccines) or other infections (malaria, HBV, HCV, HTLV-I, and tuberculosis) do not elicit antibodies that cross-react in the HIV-SELECTEST (Tables 4 and 5).
Together, these data provide strong proof-of-concept for the specificity and sensitivity of the new p6 and gp41 peptide-based EIA. The data further suggest that, if future vaccine candidates do not contain these epitopes, all uninfected vaccinees are expected to score negative in this assay, while antibodies generated after intercurrent HIV infections acquired in the course of HIV vaccine trials or at later times should be detected by the HIV-SELECTEST soon after infection.
This inexpensive and high-throughput assay could be added to the algorithm of detection tests used at clinical sites. This assay will be highly relevant for early diagnosis of intercurrent HIV infections in future preventive vaccine trials. It will also be of great value to blood banks in countries where a large portion of the population is likely to undergo vaccination against HIV. The addition of such a test may be a cost-saving factor during ongoing vaccine trials due to reduction in the number of questionable HIV-seropositive samples that need NAT testing. Importantly, the HIV-SELECTEST may help to reduce concerns regarding social and economic harms due to long-term seroconversion of uninfected participants in preventive HIV vaccine trials, resulting in improved recruitment into future trials. Future licensure of the assay would be based on further clinical evaluations under approved investigational new drug applications to be conducted by several interested partners.
In the coming years, a concerted effort will be made to convert the HIV-SELECTEST into a “rapid test” platform. Rapid tests are simpler to conduct and may be performed by untrained individuals. This will allow repeated testing of vaccine trial participants with short intervals during the vaccination regimen. It will enhance the probability of early diagnosis of breakthrough infections and will enable the collection of important virological and immunological end points soon after infection.
ACKNOWLEDGMENTS
We thank Dr. Judy Beeler, Dr. Keith Peden, and Dr. Carol Weiss for their thorough review and comments on the manuscript.
We thank the following investigators for contributions of blinded plasma panels: Dr. Olga Utnitskaya, State Center of Sanitary-Epidemiological Surveillance, Ekaterinburg, Russian Federation (OPV vaccines); Drs. Yiming Shao and Jianping Chen at the China CDC (NCAIDS), Beijing, China; Dr. Yongtao Sun at the Tangdu Hospital, Xi'an, China; and Drs. Eduard Sanders and Omu Anzala, Uganda. Dr. Julie Stachowiak and Alena Peryshkina of AIDS Infoshare, Moscow, were co-investigators with Dr. Beyrer on the Moscow Women's Health Study cohort.
All studies were conducted under approval from the Research Involving Human subjects Committee (RIHSC Exemption No. 04−050B) at the Center for Biologics Evaluation and Research.
This project was supported in part by an NIH “Bench-to-Bedside” grant in 2003−2004 and NHLBI grants for 2005−2006 (H. Golding and S. Khurana). Specimens from Cameroon were obtained with support of grants AI47053 and AI36085 from the NIAID (S. Zolla-Pazner and P. Nyambi) and in part from an IAG BY1-HB-5026−01 from NHBLI (I. Hewlett). Specimens from Zambia were obtained with support from IAVI and grants MH-766767, HD-40125, and AI-51231 from the National Institutes of Health (S. Allen and E. Hunter).
REFERENCES
- 1.Graham BS, Mascola JR. Lessons from failure: preparing for future HIV-1 vaccine efficacy trials. J Infect Dis. 2005;191:647–649. doi: 10.1086/428406. [DOI] [PubMed] [Google Scholar]
- 2.Ackers ML, Parekh B, Evans TG, et al. Human immunodeficiency virus (HIV) seropositivity among uninfected HIV vaccine recipients. J Infect Dis. 2003;187:879–886. doi: 10.1086/368169. [DOI] [PubMed] [Google Scholar]
- 3.Chuenchitra T, Wasi C, Louisirirojchanakul S, et al. Longitudinal study of humoral immune responses in HIV type 1 subtype CRF01_AE (E)-infected Thai patients with different rates of disease progression. AIDS Res Hum Retroviruses. 2003;19:293–305. doi: 10.1089/088922203764969492. [DOI] [PubMed] [Google Scholar]
- 4.Pitisuttithum P, Nitayaphan S, Thongcharoen P, et al. Safety and immunogenicity of combinations of recombinant subtype E and B human immunodeficiency virus type 1 envelope glycoprotein 120 vaccines in healthy Thai adults. J Infect Dis. 2003;188::219–227. doi: 10.1086/376506. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz DH, Mazumdar A, Winston S, et al. Utility of various commercially available human immunodeficiency virus (HIV) antibody diagnostic kits for use in conjunction with efficacy trials of HIV-1 vaccines. Clin Diagn Lab Immunol. 1995;2:268–271. doi: 10.1128/cdli.2.3.268-271.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheon AR, Wagner L, McElrath MJ, et al. Preventing discrimination against volunteers in prophylactic HIV vaccine trials: lessons from a phase II trial. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;19:519–526. doi: 10.1097/00042560-199812150-00012. [DOI] [PubMed] [Google Scholar]
- 7.Kleinman S, Busch MP, Hall L, et al. False-positive HIV-1 test results in a low-risk screening setting of voluntary blood donation. Retrovirus Epidemiology Donor Study. JAMA. 1998;280:1080–1085. doi: 10.1001/jama.280.12.1080. [DOI] [PubMed] [Google Scholar]
- 8.Busch MP, Glynn SA, Stramer SL, et al. A new strategy for estimating risks of transfusion-transmitted viral infections based on rates of detection of recently infected donors. Transfusion. 2005;45:254–264. doi: 10.1111/j.1537-2995.2004.04215.x. [DOI] [PubMed] [Google Scholar]
- 9.Khurana S, Needham J, Mathieson B, et al. Human immunodeficiency virus (HIV) vaccine trials: a novel assay for differential diagnosis of HIV infections in the face of vaccine-generated antibodies. J Virol. 2006;80:2092–2099. doi: 10.1128/JVI.80.5.2092-2099.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barreto CC, Sabino EC, Goncalez TT, et al. Prevalence, incidence, and residual risk of human immunodeficiency virus among community and replacement first-time blood donors in Sao Paulo, Brazil. Transfusion. 2005;45:1709–1714. doi: 10.1111/j.1537-2995.2005.00575.x. [DOI] [PubMed] [Google Scholar]
- 11.Heyns Adu P, Benjamin RJ, Swanevelder JP, et al. Prevalence of HIV-1 in blood donations following implementation of a structured blood safety policy in South Africa. JAMA. 2006;295:519–526. doi: 10.1001/jama.295.5.519. [DOI] [PubMed] [Google Scholar]
- 12.Busch MP, Laycock M, Kleinman SH, et al. Accuracy of supplementary serologic testing for human T-lymphotropic virus types I and II in US blood donors. Retrovirus Epidemiology Donor Study. Blood. 1994;83:1143–1148. [PubMed] [Google Scholar]
- 13.McKenna SL, Muyinda GK, Roth D, et al. Rapid HIV testing and counseling for voluntary testing centers in Africa. AIDS. 1997;11(Suppl 1):S103–S110. [PubMed] [Google Scholar]
- 14.Nyambi P, Heyndrickx L, Vereecken K, et al. Predominance of infection with HIV-1 circulating recombinant form CRF02_AG in major Cameroonian cities and towns. AIDS. 2002;16:295–296. doi: 10.1097/00002030-200201250-00022. [DOI] [PubMed] [Google Scholar]
- 15.Nyambi P, Zekeng L, Kenfack H, et al. HIV infection in rural villages of Cameroon. J Acquir Immune Defic Syndr. 2002;31:506–513. doi: 10.1097/00126334-200212150-00008. [DOI] [PubMed] [Google Scholar]
- 16.Zhong P, Burda S, Urbanski M, et al. HIV type 1 group M clades infecting subjects from rural villages in equatorial rain forests of Cameroon. J Acquir Immune Defic Syndr. 2002;31:495–505. doi: 10.1097/00126334-200212150-00007. [DOI] [PubMed] [Google Scholar]
- 17.Elm J, Desowitz R, Diwan A. Serological cross-reactivities between the retroviruses HIV and HTLV-1 and the malaria parasite Plasmodium falciparum. P N G Med J. 1998;41:15–22. [PubMed] [Google Scholar]
- 18.Dock NL, Lamberson HV, Jr, O'Brien TA, et al. Evaluation of atypical human immunodeficiency virus immunoblot reactivity in blood donors. Transfusion. 1988;28:412–418. doi: 10.1046/j.1537-2995.1988.28588337326.x. [DOI] [PubMed] [Google Scholar]
- 19.Papadopulos-Eleopulos E, Turner VF, Papadimitriou JM, et al. HIV antibodies: further questions and a plea for clarification. Curr Med Res Opin. 1997;13:627–634. doi: 10.1185/03007999709113336. [DOI] [PubMed] [Google Scholar]
- 20.McMichael AJ, Hanke T. HIV vaccines 1983–2003. Nat Med. 2003;9:874–880. doi: 10.1038/nm0703-874. [DOI] [PubMed] [Google Scholar]
- 21.Kong WP, Huang Y, Yang ZY, et al. Immunogenicity of multiple gene and clade human immunodeficiency virus type 1 DNA vaccines. J Virol. 2003;77:12764–12772. doi: 10.1128/JVI.77.23.12764-12772.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham BS, McElrath MJ, Connor RI, et al. Analysis of intercurrent human immunodeficiency virus type 1 infections in phase I and II trials of candidate AIDS vaccines. AIDS Vaccine Evaluation Group, and the Correlates of HIV Immune Protection Group. J Infect Dis. 1998;177:310–319. doi: 10.1086/514209. [DOI] [PubMed] [Google Scholar]
- 23.Weber J. Distinguishing between response to HIV vaccine and response to HIV. Lancet. 1997;350:230–231. doi: 10.1016/S0140-6736(97)22030-1. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz DH, Laeyendecker OB, Arango-Jaramillo S, et al. Extensive evaluation of a seronegative participant in an HIV-1 vaccine trial as a result of false-positive PCR. Lancet. 1997;350:256–259. doi: 10.1016/S0140-6736(97)01500-6. [DOI] [PubMed] [Google Scholar]