Abstract
RFX4_v3 (regulatory factor X4 variant 3) is a brain-specific isoform of the transcription factor RFX4. Insertional mutagenesis in mice demonstrates that Rfx4_v3 is crucial for normal brain development. Many genes involved in critical processes during brain morphogenesis are dysregulated in Rfx4_v3 mutant brains. For example, Cx3cl1 is a CX3C-type chemokine that is abundant in brain and is a direct transcriptional target of RFX4_v3 through a specific promoter X-box (X-box 1), the responsive element for RFX proteins. To identify potential interacting partners for RFX4_v3, we performed yeast two-hybrid analysis. Nine candidate interactors were identified, including GPS2 (G-protein pathway suppressor 2). Indirect immunofluorescence demonstrated that GPS2 and RFX4_v3 co-localized to the nucleus. Both GPS2 and RFX4_v3 mRNAs were also present in most portions of the adult mouse brain as well as in brains at different ages, suggesting that the two proteins could bind to each other. Co-immunoprecipitation assays indicated that physical interactions between GPS2 and RFX4_v3 did indeed occur. Furthermore, GPS2 was recruited to the Cx3cl1 promoter by RFX4_v3 and potentiated RFX4_v3 transactivation on this promoter through X-box 1, suggesting that the protein-protein interaction was functionally relevant. GPS2 bound to both the carboxyl-terminal region (amino acids 575-735) and the middle region (amino acids 250-574) of the RFX4_v3 protein. RFX4_v3 amino acids 1-574 stimulated the Cx3cl1 promoter to a similar extent as the full-length RFX4_v3 protein; however, deletion of the carboxyl-terminal region of RFX4_v3 impaired the co-activating abilities of GPS2. Based on these data, we conclude that GPS2 interacts with RFX4_v3 to modulate transactivation of genes involved in brain morphogenesis, including Cx3Cl1.
Regulatory factor X (RFX)2 members belong to the winged helix subfamily of helix-turn-helix transcription factors, which share a highly conserved 76-amino acid winged helix DNA binding domain (DBD) that is distinct from any other DBD (1). These proteins regulate their target genes by binding to symmetrical X-box consensus sequences 5′-GTNRCCN0-3R-GYAAC-3′ (where N is any nucleotide, R is a purine, Y is a pyrimidine, and the two half sites GTNRCC and RGYAAC are separated by 0-3 base pairs). RFX family members have been identified in a broad range of eukaryotic organisms, including yeast, fungi, nematodes, fruit flies, and mammals (2).
In mammals, five RFX proteins have been isolated, named RFX1-RFX5. RFX1, RFX2, and RFX3 are structurally related proteins, which share the DBD, glutamine-rich (Q), and/or proline- and glutamine-rich (PQ) regions, and four additional evolutionarily conserved regions, A, B, C, and dimerization domains (2). These proteins form homodimers or heterodimers through their dimerization domains and regulate downstream gene expression. The functions of RFX1 and RFX2 remain elusive. RFX3 has been shown to direct nodal cilium development and left-right asymmetry specification (3). RFX5 is the most intensively studied family member. It does not contain the dimerization domain and forms a complex with other transcription factors to regulate major histocompatibility complex class II gene expression. Mutations in the RFX5 gene cause the bare lymphocyte syndrome (4).
The first two full-length RFX4 cDNAs were isolated in 2002 (2). These isoforms were expressed specifically in testis and named RFX4 transcript variant 1 (RFX4_v1) and RFX4 transcript variant 2 (RFX4_v2). The third variant of RFX4 was identified serendipitously in our laboratories by insertional mutagenesis in 2003. Transgenic mice were generated for cardiac specific overexpression of a human arachidonic acid epoxygenase gene (5), and one line of these mice developed an unexpected brain phenotype. The transgene was found to be inserted into an intron in the mouse Rfx4 locus and to prevent the expression of a novel brain-specific variant of Rfx4, termed Rfx4 transcript variant 3 (Rfx4_v3) (6). Several additional RFX4 splice variants have been identified more recently (7).
The Rfx4_v3 transcript variant is the only RFX4 isoform significantly expressed in fetal and adult brain, and its expression is restricted to brain. Studies in mice demonstrated that interruption of a single allele prevented formation of the subcommissural organ, a structure important for cerebrospinal fluid flow through the aqueduct of Sylvius, resulting in noncommunicating congenital hydrocephalus (6). Interruption of both alleles caused failure of dorsal midline brain structure formation and perinatal death (6). These studies implicated RFX4_v3 in early brain development as well as in the genesis of the subcommissural organ (6). Rfx4_v3 deficiency presumably caused brain abnormalities by altering the expression levels of key genes that are crucial for brain morphogenesis, such as the signaling components in the Wnt, bone morphogenetic protein, and retinoic acid (RA) pathways, and some important transcription factors (8). Besides its role in brain development, RFX4_v3 might also be involved in regulating the circadian clock and the pathogenesis of bipolar disorder (9, 10).
RFX4 proteins contain the characteristic DBD, B, C, and dimerization domains but lack the Q/PQ and A regions. Since PQ/Q and A regions play roles in transcriptional activation, it has been suggested that RFX4 has no transactivation capacity but instead functions as a transcriptional repressor (2). However, our experiments indicate that RFX4_v3 could function as a transcriptional activator under certain circumstances. Cx3cl1, a CX3C-type chemokine gene highly expressed in brain (11), is down-regulated in Rfx4_v3-/- brains (8). Both human and mouse Cx3cl1 proximal promoters contain highly conserved X-boxes. RFX4_v3 protein binds directly to the X-box 1 of the mouse Cx3cl1 promoter in vitro and in vivo and activates expression of this gene through this X-box (8). To date, Cx3cl1 is the only direct transcriptional target identified for the RFX4 protein.
To further elucidate the mechanisms whereby RFX4_v3 regulates gene expression, we conducted yeast two-hybrid screening to identify its interacting partners. One of the prey proteins identified was GPS2 (G-protein pathway suppressor 2). GPS2 is a nuclear protein that is ubiquitously expressed and could function as a transcriptional co-factor. In this study, we investigated the physical and functional relationship between RFX4_v3 and GPS2.
EXPERIMENTAL PROCEDURES
Nuclear Yeast Two-hybrid Screening—Automated yeast two-hybrid screening using ProNet technology was performed in conjunction with investigators at Myriad Genetics as previously described with some modifications (12). Briefly, the cDNA fragments encoding partial human or mouse RFX4_v3 proteins were obtained by PCR amplification and co-transformed with the linear bait vector pGBT.superB into the yeast strain PNY200 (MATα trp1-901 leu2-3,112 ura3-52 his3-200 ade2 gal4Δ gal80) to obtain the bait construct in vivo, in which the partial RFX4_v3 proteins were fused to the carboxyl-terminal end of the Gal4 DNA binding domain (residues 1-147). The cDNAs from each of three different sources were co-transformed with the prey vector pGAD.PN2 into the yeast strain BK100 (MATα trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80 LYS2::GAL-HIS3 GAL2-ADE2 met2::GAL7-lacZ) to generate three different prey libraries (human whole brain prey library, ∼60 million clones; human hippocampus prey library, ∼38 million clones; mouse embryo prey library, ∼2.9 million clones), in which prey proteins were fused to the carboxyl-terminal end of the Gal4 activation domain (residues 768-881). Approximately 25-30 million bait-containing haploid yeast PNY200 cells were mated with one of the prey libraries. Interacting bait-prey pairs were identified by selecting for the expression of two auxotrophic reporter genes, HIS3 and ADE2. Each interaction pair was confirmed by co-transforming purified bait and prey plasmid DNA into the naive yeast and assaying for expression of a third reporter gene lacZ. The specificity of the prey was investigated in a separate false positive test, where the prey was examined against a mixture of several heterologous baits.
Plasmid Constructions—The CMV.BGH3′/BS+ vector (13) was modified with insertion of either FLAG or hemagglutinin (HA) epitope tags downstream of the multiple cloning sites to create the pCMV-FLAG-BGH3′ or pCMV-HA-BGH3′ vectors, respectively. The full-length human RFX4_v3 cDNA was released from the RFX4_v3/MycHis vector (8) and subcloned into the pCMV-FLAG-BGH3′ or pCMV-HA-BGH3′ vectors. Six partial human RFX4_v3 cDNAs were amplified by PCR using the oligonucleotide primers shown in Table 1 and subsequently cloned into the pCMV-HA-BGH3′ vector. To direct nuclear translocation of RFX4_v3-(250-574), three copies of the simian virus 40 large T-antigen nuclear localization signal from the pEYFP-Nuc vector (Clontech) was cloned into the corresponding plasmid. To enhance expression levels and to direct nuclear localization of RFX4_v3-(1-300), RFX4_v3-(250-735), and RFX4_v3-(1-300/575-735), the enhanced yellow-green fluorescent protein (EYFP) with three copies of nuclear localization signal was cloned into each of these three plasmids, respectively. The human GPS2 cDNA (clone ID IOH10241) was purchased from Invitrogen, and the full-length GPS2 cDNA fragment was generated by PCR amplification (primers 5′-GATGGATCCCCCGCACTCCT-3′ and 5′-TGGTCTAGACTTGTGGTAGA-3′), and ligated into the pCMV-FLAG-BGH3′ or pCMV-HA-BGH3′ vectors. Correct sequences for all constructs were confirmed by direct sequencing using the ABI/Prism dRhodamine Terminator Cycle Sequencing Ready Reaction Kit. The plasmids carrying HA-tagged RFX1, RFX2, RFX3, and RFX4_v2 were kind gifts from Dr. James Davenport and have been described previously (2).
TABLE 1.
Primers used to amplify the partial RFX4_v3 cDNAs
Note that full-length human RFX4_v3 cDNA encodes 735 amino acids. The partial cDNAs are named according to the corresponding amino acids they encode.
| Partial RFX4_v3 cDNA | Primer sequences |
|---|---|
| RFX4_v3-(575-735) | 5′-GATGCTAGCATGAGGAACAC-3′ |
| 5′-TGGTCTAGATTTAGCCCATC-3′ | |
| RFX4_v3-(1-574) | 5′-GGAGACCCAAGCTTGGTACC-3′ |
| 5′-TGGTCTAGAACAGGGCAGCT-3′ | |
| RFX4_v3-(250-574) | 5′-GATGGATCCAACATTGTCGG-3′ |
| 5′-TGGTCTAGAACAGGGCAGCT-3′ | |
| RFX4_v3-(1-300) | 5′-GGAGACCCAAGCTTGGTACC-3′ |
| 5′-TGGGCTAGCGTCGTGGAGAG-3′ | |
| RFX4_v3-(250-735) | 5′-GATGGATCCAACATTGTCGG-3′ |
| 5′-TGGTCTAGATTTAGCCCATC-3′ | |
| RFX4_v3-(1-300/575-735) | 5′-GGAGACCCAAGCTTGGTACC-3′ |
| 5′-TGGGCTAGCGTCGTGGAGAG-3′ | |
| 5′-GATGCTAGCATGAGGAACAC-3′ | |
| 5′-TGGTCTAGATTTAGCCCATC-3′ |
TaqMan®Real Time PCR—Total RNA was isolated with the RNeasy minikit (Qiagen), treated with RQ1 RNase-free DNase (Promega), and reverse transcribed into first strand cDNA using the ABI/Prism High Capacity cDNA Archive Kit (Applied Biosystems). TaqMan®PCRs were conducted in triplicate. The amplifications were performed as follows: 2 min at 50 °C, 10 min at 95 °C, and then 40 cycles each at 95 °C for 15 s and 60 °C for 60 s in the ABI/Prism 7900 HT sequence detector system. For Rfx4_v3, the following primers and probe were used for the assay: forward primer, 5′-TCTCGCTCTCTCCTTCAGCTCTA-3′; reverse primer, 5′-GGTGGGTGAGAGGGCAAAA-3′; probe, 5′-CGCTTCTTCGCCTCTTTTCTTTCCACTAGTT-3′. The assay ID of the predesigned ABI primer/probe set for Gps2 is Mm_00517238_g1; for Cx3cl1, it is Mm00436454_m1; and for Igf1 (insulin-like growth factor 1), it is Mm00439561_m1. Results were normalized to an internal control transcript that encodes glyceraldehyde-3-phosphate dehydrogenase, using the TaqMan Rodent GAPDH control reagents (ABI/Prism).
Transient Transfection and Histochemical Staining—COS-1 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum and 100 units/ml penicillin/streptomycin. For immunostaining, COS-1 cells were plated on the glass coverslips (Ted Pella) and transfected with the FLAG- or HA-tagged constructs using Fugene 6 reagent (Roche Applied Science). Two days after transfection, cells were fixed with 3.7% formaldehyde, permeabilized with Triton X-100, and stained with mouse anti-FLAG monoclonal antibody M2 (Sigma) and rabbit anti-HA polyclonal antibody Y-11 (Santa Cruz Biotechnology), followed by fluorescein isothiocyanate-labeled goat anti-mouse secondary antibody Alexa Fluor 488 and rhodamine-conjugated goat anti-rabbit secondary antibody Alexa Fluor 594 (Invitrogen). The stained cells were mounted with VECTASHIELD mounting medium with 4′,6-diamidino-2-phenylindole (Vector Laboratories) to visualize the nuclei.
Co-immunoprecipitation and Western Blotting—Two days after transfection, cells were collected in cold phosphate-buffered saline, lysed in radioimmune precipitation buffer (150 mm NaCl, 1% (v/v) Nonidet P-40, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, 50 mm Tris-HCl, pH 7.5), and supplemented with EDTA-free protease inhibitor mixture (Roche Applied Science), and the supernatant was collected. The supernatant was incubated with anti-HA F-7 antibody (Santa Cruz Biotechnology) overnight and then with protein A-Sepharose 4B beads (Amersham Biosciences) for 1 h. After extensive washing, the bound proteins were eluted using the Laemmli sample buffer (Bio-Rad). The precipitated proteins were detected by Western blotting with the mouse anti-HA F-7 horseradish peroxidase (Santa Cruz Biotechnology) or anti-FLAG M2 horseradish peroxidase (Sigma) antibodies.
Reporter Assays—For reporter assays, COS-1 cells were grown in 12-well plates at 60% confluence and then transiently transfected with the RFX4_v3 expression vector (0.5 μg) and/or the GPS2 expression vector (0.5 μg) and a Cx3cl1 promoter plasmid (0.5 μg), using Superfect reagent (Qiagen). Plasmid pRL-SV40 (1 ng; Promega) was also co-transfected to normalize the transfection efficiency. Transfection assays were performed in triplicate. Forty-eight hours after transfection, the cells were washed once with phosphate-buffered saline and lysed in Passive Lysis Buffer (Promega). Luciferase activities in the lysates were measured with the Dual-Luciferase reporter assay system (Promega) according to the manufacturer's protocol.
Chromatin Immunoprecipitation (ChIP) Assays—To examine the binding of RFX4_v3 and GPS2 to the Cx3cl1 promoter, cells were transfected as described in the reporter assays. ChIP experiments were performed using the ChIP-IT™ express enzymatic Kit (Active Motif) according to the manufacturer's instructions. Immunoprecipitations were conducted using anti-HA F-7 antibody, anti-FLAG M2 antibody, or the negative control IgG (Active Motif). Immunoprecipitated DNA was then amplified by real time PCR using the ABI/Prism 7900 HT Sequence Detector System and Power SYBR Green PCR master mix. The primers used to amplify the mouse Cx3cl1 promoter were 5′-CCTAGGTTCTCCAGGGAAGG-3′ and 5′-GGGGAGAGGAAGAGCCTGTA-3′. A small fraction of each lysate prior to the immunoprecipitation was saved as “input DNA” and also amplified by real time PCR.
Statistics—Student's t test was used for all statistical analyses. Each experiment was performed at least in triplicate. Results were expressed as mean ± S.E. A p value of <0.05 was taken as the level of significance for all tests.
RESULTS
Identification of Proteins That Interact with RFX4_v3—To identify proteins that interact with RFX4_v3, we used partial human or mouse RFX4_v3 proteins (size range: 100-300 amino acids) fused to the Gal4 DNA-binding domain as baits to screen three different human or mouse prey libraries that might contain potential RFX4_v3 interactors. After yeast two-hybrid screening, 11 positive clones that represented nine genes were identified (Table 2). These preys can be classified into three groups. The first group contains proteins that are known or predicted to be present in the nucleus and includes the nuclear protein GPS2, the putative nuclear protein ZBTB1 (zinc finger- and BTB domain-containing 1) (14), calmodulin that translocates to the nucleus following Ca2+ stimulation (15), and PMSD2 (proteasome 26 S subunit, non-ATPase 2) that is localized in both the nucleus and cytoplasm (16). The second group contains proteins that are mainly expressed in the cytosol and includes NCDN (neurochondrin) (17), RICS (Rho GTPase-activating protein) (18), and FBF1 (tumor necrosis factor receptor superfamily, member 6 binding factor 1) (19). The third group contains proteins with no information on their subcellular localization and includes MLF2 (myeloid leukemia factor 2) and ZFP469 (zinc finger protein 469) (20). The proteins that are known to be exclusively expressed in the cytosol are unlikely to be true binding partners for RFX4_v3, since RFX4_v3 expression is thought to be limited to the nucleus (9).
TABLE 2.
Nine RFX4_v3-interacting proteins identified by yeast two-hybrid screening
Note that partial mouse RFX4_v3 protein (mRFX4_v3) or human RFX4_v3 protein (hRFX4_v3) was used as the bait to screen one of three cDNA libraries (shown in the last column). The amino acid regions of the bait and prey proteins are shown in the second and fourth columns, respectively.
| Bait | Bait amino acid coordinates | Prey | Prey amino acid coordinates | Library |
|---|---|---|---|---|
| mRFX4_v3 | 1-250 | CaM | 14-149 | Mouse embryo |
| mRFX4_v3 | 44-126 | ZBTB1 | 342-680 | Hippocampus |
| mRFX4_v3 | 175-510 | mFBF1 | 751-940/626-1031 | Mouse embryo/Mouse embryo |
| mRFX4_v3 | 175-510 | mPSMD2 | 602-908 | Mouse embryo |
| mRFX4_v3 | 551-735 | mMLF2 | 6-214 | Mouse embryo |
| hRFX4_v3 | 1-300 | NCDN | 499-712/489-712 | Brain/Hippocampus |
| hRFX4_v3 | 250-550 | ZFP469 | 2015-2183 | Brain |
| hRFX4_v3 | 575-735 | GPS2 | 10-327 | Brain |
| hRFX4_v3 | 575-735 | RICS | 1096-1374 | Hippocampus |
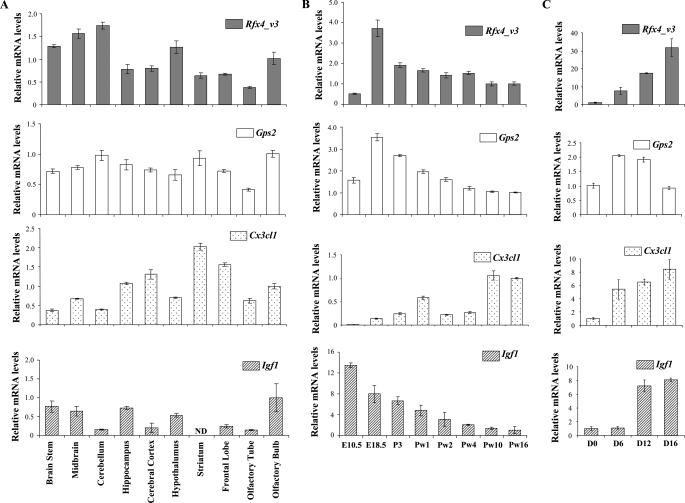
Expression of RFX4_v3 and GPS2—We further investigated the relationship between RFX4_v3 and GPS2, since GPS2 is reported to be a nuclear protein and could potentially play a role as a transcriptional cofactor. We first examined the expression patterns of both genes. RFX4_v3 is the only RFX4 variant that is significantly expressed in the brain, and it is not present in other tissues (6). In contrast, GPS2 has a broad tissue distribution (21). However, it remains unknown whether RFX4_v3 and GPS2 are expressed in a similar temporal and spatial pattern within the brain. Specific antibodies are not currently available to detect the endogenous RFX4_v3 or GPS2 proteins; hence, real time PCR experiments were performed to examine the tissue distribution of the corresponding transcripts. In adult mouse brain, both Rfx4_v3 and Gps2 mRNAs were detectable in all brain sections examined, including the brain stem, mid-brain, cerebellum, hippocampus, cerebral cortex, hypothalamus, striatum, frontal lobe, and olfactory bulb (Fig. 1A). Both transcripts were also present in the brain throughout development, with expression levels being highest at embryonic day 18.5 (Fig. 1B). Mouse embryonic stem cells (J1 cells) can differentiate into relatively homogeneous neuronal cells in response to RA treatment (22, 23). We found that both Rfx4_v3 and Gps2 transcripts were up-regulated in J1 cells during neuronal differentiation (Fig. 1C), whereas the Rfx4_v3 message was induced to a much greater extent than the Gps2 message. Together, these results demonstrate that both Rfx4_v3 and Gps2 transcripts have similar temporal-spatial distribution patterns within the brain, and they are present in differentiated neuronal cells, suggesting that they could meet under physiological conditions.
FIGURE 1.
Rfx4_v3, Gps2, Cx3cl1, and Igf1 expression in brain and J1 cells. A, tissue distribution of Rfx4_v3, Gps2, Cx3cl1, and Igf1 in adult mouse brain. For these genes, mRNA levels in the olfactory bulb are defined as 1.0. ND, not detectable. B, expression levels of Rfx4_v3, Gps2, Cx3cl1, and Igf1 in whole mouse brain at different ages. For these genes, mRNA levels at Pw16 are defined as 1.0. E, embryonic day; P, postnatal day; Pw, postnatal week. C, Rfx4_v3, Gps2, Cx3cl1, and Igf1 mRNA levels in J1 embryonic stem cells at different stages of neuronal differentiation. The day when RA was added to the culture to induce differentiation was designated as day 0. D, days of RA treatment. For these genes, mRNA levels at day 0 are defined as 1.0. All values are shown as mean ± S.E.
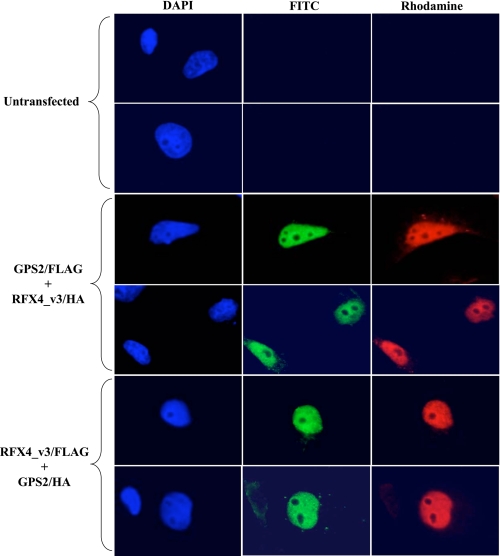
To further confirm that both RFX4_v3 and GPS2 proteins localized to overlapping cellular compartments, indirect immunofluorescence microscopy was performed in COS-1 cells transiently transfected with HA- or FLAG-tagged constructs. The subcellular localizations of both RFX4_v3 and GPS2 proteins were predominantly nuclear with nucleolar exclusion (Fig. 2), which is consistent with their function as transcription factors. We also observed punctate staining of RFX4_v3 and GPS2 in the cytoplasm and in perinuclear regions in some transfected cells, although this signal was much weaker than the nuclear one (data not shown).
FIGURE 2.
RFX4_v3 and GPS2 proteins are predominantly expressed in the nuclei. COS-1 cells were either untransfected, co-transfected with both RFX4_v3/HA and GPS2/FLAG constructs, or co-transfected with both RFX4_v3/FLAG and GPS2/HA constructs. Cells were stained with mouse anti-FLAG antibody and rabbit anti-HA antibody, followed by fluorescein isothiocyanate (FITC)-conjugated anti-mouse (green) and rhodamine-conjugated anti-rabbit (red) secondary antibodies. Cells were covered with 4′,6-diamidino-2-phenylindole (DAPI) containing mounting media to visualize the nuclei (blue).
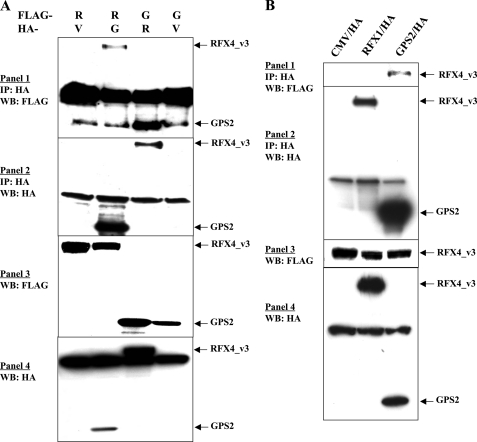
RFX4_v3 and GPS2 Interact with Each Other—To confirm that physical interactions between RFX4_v3 and GPS2 do indeed occur, co-immunoprecipitation experiments were performed in COS-1 cells transfected with mammalian expression constructs containing HA-tagged full-length RFX4_v3 and FLAG-tagged full-length GPS2 or HA-tagged GPS2 and FLAG-tagged RFX4_v3 (Fig. 3A). We found that HA-tagged RFX4_v3 or GPS2 could help to precipitate FLAG-tagged GPS2 or RFX4_v3, respectively, whereas empty HA vector or the HA-tagged nuclear protein RFX1 could not pull down either protein (Figs. 3, A and B, and 7). These data demonstrate that RFX4_v3 and GPS2 are in the same protein complex. Since co-transformation of the purified RFX4_v3 bait plasmid and the GPS2 prey plasmid into naive yeast results in activation of the lacZ reporter gene, the interaction between the two proteins is more likely to be a direct one, and GPS2 is a likely binding partner for RFX4_v3. Therefore, RFX4_v3 and GPS2 physically interact with each other in both yeast and mammalian cells.
FIGURE 3.
Physical interaction of RFX4_v3 with GPS2. A, COS-1 cells were co-transfected with plasmids for RFX4_v3/FLAG and GPS2/HA or for GPS2/FLAG and RFX4_v3/HA, as indicated at the top. V, empty CMV/HA vector that is used as the negative control; R, RFX4_v3; G, GPS2. Immunoprecipitation (IP) was performed with anti-HA monoclonal antibody F-7. Precipitated proteins were examined by Western blotting (WB) using anti-FLAG antibody (1) or anti-HA antibody (2). Western blotting of the whole cell extracts with either the anti-FLAG antibody (3) or anti-HA antibody (4) was used to show the tagged protein expression in the cell extracts. Note that a nonspecific band co-migrates with GPS2 in 1. B, similar experiments were performed when COS-1 cells were co-transfected with FLAG-tagged RFX4_v3 and either empty CMV/HA vector, HA-tagged RFX1, or HA-tagged GPS2 construct.
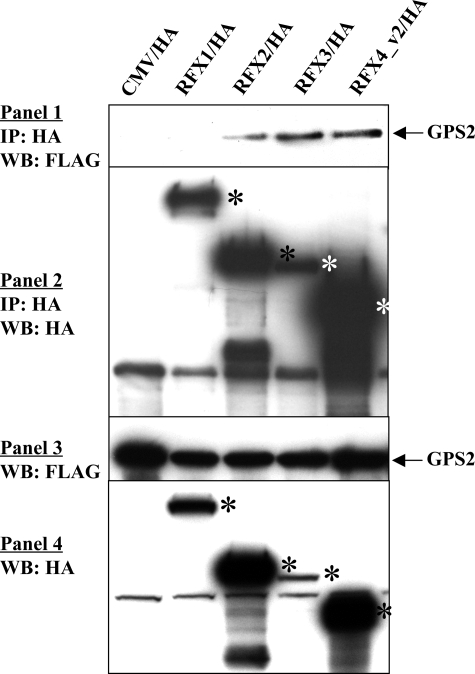
FIGURE 7.
Physical interaction of GPS2 with other RFX members and RFX4 isoform. COS-1 cells were co-transfected with FLAG-tagged GPS2 and HA-tagged full-length RFX1, RFX2, RFX3, or RFX4_v2 expression constructs. Immunoprecipitation (IP) was performed with anti-HA monoclonal antibody F-7. Precipitated proteins were examined by Western blotting (WB) using anti-FLAG antibody (1) or anti-HA antibody (2). Western blotting of the whole cell extracts with either the anti-FLAG antibody (3) or anti-HA antibody (4) was used to show tagged protein expression in the cell extracts. The empty CMV/HA vector was used as the negative control. *, RFX protein bands.
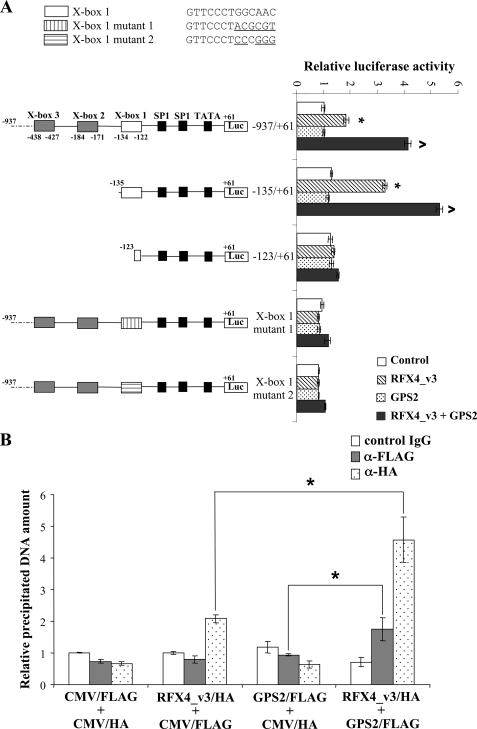
GPS2 Stimulates RFX4_v3-dependent Transcription—Cx3cl1, a CX3C-type chemokine that is abundant in the cortex, hippocampus, basal ganglia, and olfactory bulb of the brain, has been proposed to play an important role in the response of the brain to injury and infection (24, 25). Cx3cl1 expression levels are decreased in Rfx4_v3 null brains at embryonic day 10.5 (8). Both human and mouse Cx3cl1 proximal promoters contain highly conserved X-box sequences, known cis-acting elements for RFX protein binding. There are three potential RFX binding sites within 500 bp of the transcriptional start site (TSS) in the mouse Cx3cl1 gene (X-box 1 is located 122 bp upstream of the TSS, X-box 2 is located 171 bp upstream of the TSS, and X-box 3 is located 427 bp upstream of the TSS), and there is one potential RFX binding site within the proximal promoter of the human CX3CL1 gene (located 150 bp upstream of the TSS) (8). Moreover, mouse X-box 1 and the human X-box are located at similar positions and are highly homologous (8). Our previous experiments indicated that the RFX4_v3 protein could induce mouse Cx3cl1 promoter activities and that this stimulation was through X-box 1, since either deletion or mutation of this X-box completely abolished RFX4_v3 activation of the Cx3cl1 promoter (8).
Since GPS2 physically interacted with RFX4_v3, we examined whether GPS2 was involved in RFX4_v3-mediated transcriptional activation of Cx3cl1. COS-1 cells were co-transfected with various mouse Cx3cl1 promoter constructs, and either control vector, a vector containing the RFX4_v3 cDNA alone, a vector containing the GPS2 cDNA alone, or both RFX4_v3- and GPS2-containing vectors (Fig. 4A). Transfection with GPS2 alone had no effects on Cx3cl1 promoter activities. However, transfection with GPS2 stimulated transactivation of Cx3cl1 by RFX4_v3. For either of the promoter constructs that contained an intact X-box 1 site (-937/+61 or -135/+61), RFX4_v3 alone up-regulated transcription ∼2-3-fold, whereas GPS2 and RFX4_v3 co-transfection stimulated promoter activities ∼4-6-fold (Fig. 4A). When X-box 1 was deleted (-123/+61 construct, which contained only 2 nucleotides of X-box 1) or mutated (X-box 1 mutant 1 or X-box 1 mutant 2), both the RFX4_v3 activation and the GPS2 potentiation of the RFX4_v3 activation were markedly diminished, indicating that GPS2 stimulated RFX4_v3-dependent transcription on the Cx3cl1 promoter through X-box 1 as well (Fig. 4A). There were two possible explanations for the stimulating effects of GPS2 on the RFX4_v3 activation of the Cx3cl1 promoter: 1) GPS2 interacted with RFX4_v3 and functioned as a transcriptional co-activator; or 2) co-transfection with GPS2 somehow resulted in the accumulation of the RFX4_v3 protein, and higher RFX4_v3 protein levels led to increased transcription. Since co-expression with GPS2 did not increase the expression levels of RFX4_v3 protein (data not shown), the latter possibility was excluded, and we conclude that GPS2 might function as a transcriptional cofactor.
FIGURE 4.
GPS2 potentiates RFX4_v3 activation of the Cx3cl1 promoter through X-box 1. A, five luciferase constructs containing different lengths of the 5′-upstream region of mouse Cx3cl1 promoter (-937/+61, 937 bp upstream and 61 bp downstream relative to the TSS; -135/+61, 135 bp upstream and 61 bp downstream relative to the TSS; and -123/+61, 123 bp upstream and 61 bp downstream relative to the TSS) or containing mutant X-box 1 sequences (X-box 1 mutant 1 and X-box 1 mutant 2) were co-transfected into COS-1 cells with control vector (white bars), a vector containing the RFX4_v3 cDNA (striped bars), a vector containing the GPS2 cDNA (dotted bars), or both RFX4_v3- and GPS2-containing vectors (gray bars). The firefly luciferase activities were normalized to Renilla luciferase activities, and the luciferase activity from a construct containing -937/+61 of the Cx3cl1 promoter and co-transfected with the control vector is defined as 1.0. All values shown are means ± S.E.; *, p < 0.05 versus control vector; ^, p < 0.05 versus RFX4_v3-containing vector of the same luciferase construct. Native and mutant Cx3cl1 X-box 1 sequences are shown at the top of the figure (mutated nucleotides are underlined). B, COS-1 cells were transiently transfected with Cx3cl1 promoter in combination with either the empty control CMV/FLAG and CMV/HA vectors, RFX4_v3/HA expression vector and empty CMV/FLAG vector, GPS2/FLAG expression vector and empty CMV/HA vector, or both the RFX4_v3/HA and GPS2/FLAG expression vectors. Two days after transfection, lysates from the cells were subjected to ChIP analysis with either the negative control IgG, the anti-FLAG antibody (α-FLAG), or the anti-HA antibody (α-HA). The immunoprecipitated DNA was amplified by real time PCR. Each precipitated DNA amount was normalized to the respective “input DNA” amount, and the relative DNA amount was calculated as a ratio of the DNA isolated from the cells transfected with the empty control vectors and precipitated with control IgG. All values shown are means ± S.E.; *, p < 0.05.
To further confirm that GPS2 is recruited to the Cx3cl1 promoter by RFX4_v3 and acts as a transcriptional co-activator, ChIP assays were performed. As shown in Fig. 4B, when cells were transfected with the empty control vectors, comparable amounts of Cx3cl1 promoter fragments were precipitated with the negative control antibody, anti-FLAG, or anti-HA antibody. Upon overexpression of the RFX4_v3 protein alone, precipitation of Cx3cl1 promoter fragments by the anti-HA antibody was enriched about 2-fold compared with the negative control antibody or the anti-FLAG antibody, indicating that RFX4_v3 could bind to the mouse Cx3cl1 promoter. However, overexpression of the GPS2 protein alone could not enrich the promoter fragments, suggesting that GPS2 did not bind to the promoter by itself. In cells co-transfected with RFX4_v3 and GPS2, GPS2 immunoprecipitated with the Cx3cl1 promoter fragments, supporting the notion that RFX4_v3 helped to bring the GPS2 protein to the Cx3cl1 promoter; RFX4_v3 could immunoprecipitate more promoter fragments when GPS2 was co-expressed (RFX4_v3 versus RFX4_v3 + GPS2: 2-fold versus 4.5-fold), suggesting that GPS2 might enhance RFX4_v3 binding to the Cx3cl1 promoter. The anti-FLAG antibody precipitated less promoter DNA than the anti-HA antibody in cells co-transfected with both proteins. There are several possible explanations for this phenomenon: 1) RFX4_v3 binds to the Cx3cl1 promoter directly, whereas GPS2 binds to the promoter indirectly through its interaction with RFX4_v3; 2) not all the RFX4_v3 proteins are associated with GPS2 under our experimental conditions; and/or 3) the anti-FLAG antibody is simply not as efficient as the anti-HA antibody in the ChIP assays. Together, the results of luciferase reporter and ChIP assays indicate that GPS2 enhances the ability of RFX4_v3 to activate the Cx3cl1 promoter.
We further examined the spatial and temporal expression patterns of Cx3cl1 in the brain as well as its expression in differentiated neurons. There were both similarities and differences when we compared the expression profile of Cx3cl1 with those of Rfx4_v3 and Gps2 (Fig. 1). If 1) the Cx3cl1 gene is solely regulated by RFX4_v3 and GPS2 proteins, 2) RFX4_v3 and GPS2 protein levels are directly correlated with their message levels, and 3) the Cx3cl1 gene is stimulated by RFX4_v3 and GPS2 in a dose-dependent manner, then the Cx3cl1 expression pattern should be very similar to those of Rfx4_v3 and Gps2. We know that at least the first assumption is incorrect, since in the Rfx4_v3 null mouse brain, Cx3cl1 message is down-regulated by ∼50%, not totally abolished, suggesting that Cx3cl1 is regulated by other transcription factors. Igf1 levels are not affected in the Rfx4_v3 null mouse brain, and its proximal promoter has no conserved X-box, indicating that Igf1 is not a direct target of RFX4_v3 (8). The temporal patterns of Igf1 mRNA expression were different from those of Rfx4_v3, Gps2, and Cx3cl1, and its message levels decreased with brain development (Fig. 1). However, in the different portions of the adult mouse brain, as well as in the differentiated neurons, there were similarities between Cx3cl1 and Igf1 expression patterns, suggesting that it is not possible to judge whether a gene is regulated by RFX4_v3 and/or GPS2 by simply examining its expression profile.
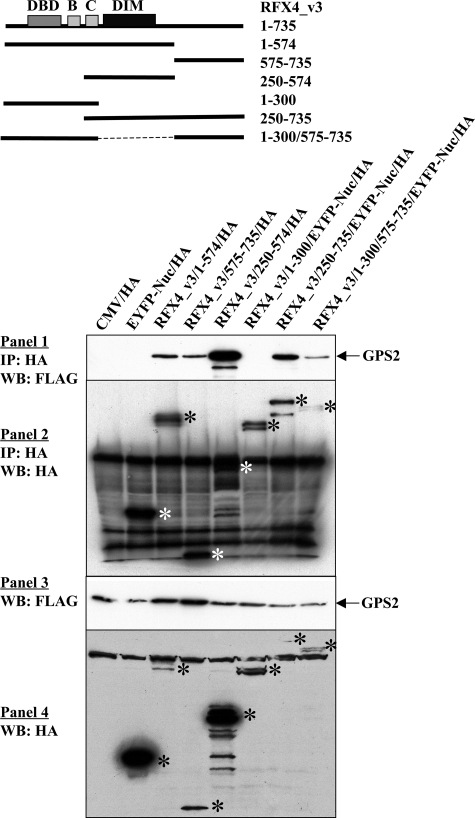
Regions of RFX4_v3 That Interact with GPS2—The RFX4_v3 protein contains a long carboxyl-terminal region that is devoid of conserved domains that has no known function. However, in the yeast two-hybrid screening, this carboxyl-terminal region (truncated RFX4_v3 protein containing amino acids 575-735; RFX4_v3-(575-735)) was found to interact with GPS2. To verify this finding, and to further investigate whether other regions of the RFX4_v3 protein also had the ability to bind the full-length GPS2, co-immunoprecipitation experiments were carried out with a series of truncated proteins (Fig. 5). The RFX4_v3-(575-735) truncated protein was mainly expressed in the nucleus, whereas the RFX4_v3-(1-574) truncated protein was present in both the nucleus and the cytoplasm, with higher levels in the nucleus (data not shown). As illustrated in Fig. 5, both of these truncated RFX4_v3 proteins interacted with GPS2, suggesting that there were at least two distinct regions within the RFX4_v3 protein that could interact with GPS2. The RFX4_v3-(1-574) protein contains the DBD (amino acids 61-136), B (amino acids 199-234), C (amino acids 261-300), and dimerization domains (amino acids 317-489). To further investigate which region within these 574 amino acids was responsible for the GPS2 binding, two shorter truncated proteins were constructed: RFX4_v3-(250-574) (containing amino acids 250-574) and RFX4_v3-(1-300) (containing amino acids 1-300). GPS2 interacted with protein RFX4_v3-(250-574), but not protein RFX4_v3-(1-300) (Fig. 5), suggesting that the dimerization domain of RFX4_v3 and/or its proximal region were capable of GPS2 binding. It is not surprising that the truncated proteins RFX4_v3-(250-735) (containing amino acids 250-735) and RFX4_v3-(1-300/575-735) (containing amino acids 1-300 and 575-735) could interact with GPS2 as well (Fig. 5), since they each contained at least one GPS2 binding region.
FIGURE 5.
Regions of RFX4_v3 that bind GPS2. COS-1 cells were co-transfected with FLAG-tagged GPS2 and either HA-tagged full-length or HA-tagged truncated RFX4_v3 constructs. Immunoprecipitation (IP) was performed with anti-HA monoclonal antibody F-7. Precipitated proteins were examined by Western blotting (WB) using anti-FLAG antibody (1) or anti-HA antibody (2). Western blotting of the whole cell extracts with either the anti-FLAG antibody (3) or anti-HA antibody (4) was used to show tagged protein expression in the cell extracts. Both the empty vector CMV/HA and the EYFP-Nuc expression plasmid were used as the negative controls, since some truncated proteins were fused to the EYFP-Nuc protein. *, full-length or truncated RFX4_v3 protein bands.
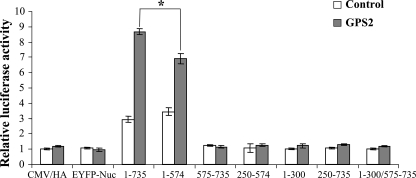
Carboxyl-terminal Region of RFX4_v3 Is Involved in GPS2 Potentiation on the Cx3cl1 Promoter—The truncated RFX4_v3 proteins (RFX4_v3-(1-574), RFX4_v3-(575-735), RFX4_v3-(250-574), RFX4_v3-(250-735), and RFX4_v3-(1-300/575-735)) interacted with GPS2, so we further tested whether these interactions had effects on GPS2-stimulated RFX4_v3 activation. It is not surprising that the truncated proteins RFX4_v3-(575-735), RFX4_v3-(250-574), or RFX4_v3-(250-735), alone or in combination with GPS2, had minimal effects on Cx3cl1 promoter activity (Fig. 6). These proteins do not contain the RFX4_v3 DNA binding domain and therefore cannot bind to the RFX binding sites within the Cx3cl1 promoter. In contrast, RFX4_v3-(1-574) has both the DNA binding and dimerization domains, and it induced promoter activities to a similar extent as the full-length RFX4_v3 protein (p = 0.20 for RFX4_v3-(1-735) versus RFX4_v3-(1-574)) (Fig. 6). However, GPS2 enhanced the ability of RFX4_v3-(1-574) to stimulate the Cx3cl1 promoter, albeit to a lesser extent than the full-length RFX4_v3 protein (p < 0.05 for RFX4_v3-(1-735) + GPS2 versus RFX4_v3-(1-574) + GPS2). This suggested that the carboxyl-terminal region of RFX4_v3 may not be required for RFX_v3-mediated transactivation but may be involved in cooperation with other cofactors, such as GPS2. The truncated proteins RFX4_v3-(1-300) and RFX4_v3-(1-300/575-735) could not activate Cx3cl1 promoter (Fig. 6), indicating that the dimerization domain of the RFX4_v3 protein might be critical for the activity of RFX4_v3 on the Cx3cl1 promoter. Moreover, GPS2 could not enhance the activities of these two truncated proteins on the Cx3cl1 promoter (Fig. 6).
FIGURE 6.
Deletion of carboxyl-terminal region of RFX4_v3 does not affect its ability to stimulate the Cx3cl1 promoter but interferes with the co-activation function of GPS2. COS-1 cells were co-transfected with the -937/+61 Cx3cl1 promoter construct, and either the full-length or truncated RFX4_v3 constructs, with (gray bars) or without (white bars) the GPS2 construct. Both the empty vector with HA tag (CMV/HA) and the EYFP-Nuc expression plasmid were used as negative controls, since some truncated proteins were fused to the EYFP-Nuc protein. Firefly luciferase activities were normalized to Renilla luciferase activities, and the luciferase activity of the empty vector CMV/HA alone is defined as 1.0. All values shown are means + S.E.; *, p < 0.05.
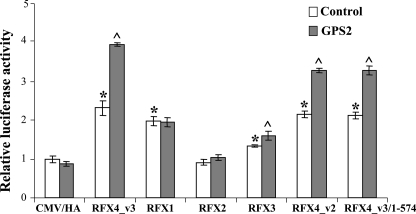
GPS2 Interaction and Co-activation of Other RFXs—Since RFX4_v3 contains the typical RFX-type DBD, B, C, and dimerization domains, and its dimerization domain and/or the proximal region have been shown to interact with GPS2, we examined whether three other mammalian RFX family members that have the dimerization domains (RFX1, RFX2, and RFX3) are capable of GPS2 binding. As shown in Fig. 7, GPS2 formed a stable complex with RFX2 and RFX3, respectively, whereas under the same experimental conditions, GPS2 did not interact with RFX1. We further examined the functional significance of these interactions on the Cx3cl1 promoter (Fig. 8). RFX1 could stimulate the Cx3cl1 promoter to a similar extent as RFX4_v3; however, GPS2 could not further enhance RFX1 activation, which is consistent with the finding that GPS2 did not bind to RFX1. RFX2 overexpression had no effect on Cx3cl1 promoter activity; nor did GPS2 co-expression activate the promoter. RFX3 induced Cx3cl1 promoter activity; although the induction was statistically significant, it was small compared with RFX4_v3 or RFX1 activation. GPS2 potentiated RFX3 activation; however, the co-activation effect was also relatively small.
FIGURE 8.
GPS2 co-activation with other RFX members and RFX4 isoform. COS-1 cells were co-transfected with the -937/+61 Cx3cl1 promoter construct and the RFX protein as indicated, with (gray bars) or without (white bars) the GPS2 construct. The empty CMV/HA vector was used as a negative control. Firefly luciferase activities were normalized to Renilla luciferase activities, and the luciferase activity of the empty vector CMV/HA alone is defined as 1.0. All values shown are means ± S.E.; *, p < 0.05 versus CMV/HA control vector without GPS2; ^, p < 0.05 versus the same RFX vector without GPS2.
RFX4_v2 is an RFX4 isoform that is abundantly expressed in the testis. RFX4_v2 contains 563 amino acids, which encode the DBD, B, C, and dimerization domains. Amino acids 22-554 of RFX4_v2 are identical to amino acids 13-545 of RFX4_v3. Therefore, based on the sequence identity, RFX4_v2 is similar to the truncated RFX4_v3 protein RFX4_v3-(1-574). Co-immunoprecipitation experiments confirmed RFX4_v2 interaction with GPS2 (Fig. 7). Promoter studies indicated that RFX4_v2, like RFX4_v3-(1-574), could activate the Cx3cl1 promoter to a similar extent as the full-length RFX4_v3, and the potentiation effect of GPS2 on RFX4_v2 was not as potent as the effect of GPS2 on RFX4_v3 (Fig. 8). Since Cx3cl1 message is present in the testis (data not shown), our results suggest that RFX4_v2 and GPS2 might be involved in the regulation of Cx3cl1 gene expression in that tissue.
DISCUSSION
GPS2 is a 327-amino acid nuclear protein with a predicted molecular mass of 37 kDa. Although the precise biological functions of GPS2 are largely unknown, several lines of evidence suggest its potential involvement in diverse cellular functions. It was initially isolated via its ability to suppress lethal G protein subunit-activating mutations in the pheromone response pathway of the yeast Saccharomyces cerevisiae (26). Pheromone binds to its cognate receptor and triggers a Gβ, γ-mediated kinase cascade that shares structural similarities with mammalian mitogen-activated protein kinase pathways. Consistent with its role in conserved signaling pathways, overexpression of GPS2 in mammalian cells strongly suppresses a RAS/mitogen-activated protein kinase-mediated signal and interferes with JNK1 (c-Jun NH2-terminal kinase 1) activation by serum factors or tumor necrosis factor α (21, 26). Studies have shown that GPS2 interacts with the human T cell lymphotrophic virus type I (HTLV-I) Tax oncoprotein and suppresses its ability to activate JNK1 (21). The N-CoR and HDAC3 (histone deacetylase 3) complex participates in diverse repression pathways and contains about 10-12 associated proteins. GPS2 is one subunit of this complex (27, 28). GPS2 inhibition of c-Jun NH2-terminal kinase activation is mediated through an associated N-CoR-dependent corepressor function (28). Furthermore, studies have also suggested a role for GPS2 in mediating cellular responses to DNA damage (29), and GPS2 might function in concert with a hMSH4-hMSH5 heterocomplex during the process of homologous recombination (30).
Our studies demonstrate the relationship between RFX4_v3 and GPS2. First, both RFX4_v3 and GPS2 are present throughout the brain and in differentiated neuronal cells, suggesting that their interaction is physiologically relevant. Second, yeast two-hybrid screening and co-immunoprecipitation experiments in mammalian cells demonstrate the physical interaction between these two proteins. Third, GPS2 is able to enhance the activating abilities of RFX4_v3 on the Cx3cl1 promoter, suggesting that GPS2 is a co-activator for RFX4_v3-dependent transcriptional events.
Transcription cofactors may exert variable effects on transcriptional regulation, depending on their interacting partners. As stated above, GPS2 can suppress transcription by association with N-CoR and HDAC3 repressor complexes. However, GPS2 has also been reported to interact with several transcription factors and enhance their transactivation. For example, GPS2 binds the papillomavirus E2 protein activation domain and is necessary for stimulation of E2 transcriptional activity (31). GPS2 is also associated with the p53 tumor suppressor and facilitates the p53 response by augmenting p53-dependent transcription (29). GPS2 can also stimulate the transcription function of a c-Jun AD-Gal4 DBD fusion protein (31). The mechanisms of GPS2 functioning as a transcription co-activator are not well understood. GPS2 might activate E2-dependent transcription by directly interacting with both E2 and p300 (32). Complex formation among E2, GPS2, and p300 may function by bringing p300 (with histone acetyltransferase activity) close to the transcription initiation sites. Histone acetylation of nearby nucleosomes is thought to remodel the chromatin structure and enhance access of the transcriptional machinery to DNA. Therefore, one possible mechanism by which GPS2 potentiates RFX4_v3 transactivation is that GPS2 might recruit other transcriptional activators to the promoters of RFX4_v3 target genes. Based on our ChIP results, another possible explanation for GPS2 co-activation of RFX4_v3 activities is that interaction of RFX4_v3 with GPS2 might enhance the binding of RFX4_v3 protein to the Cx3cl1 promoter. GPS2 alone was also shown to be sufficient to activate transcription from several artificial promoters (33); however, it had no ability to either activate or repress the transcription of the Cx3cl1 gene by itself.
RFX4 proteins lack the Q/PQ and A regions, which play roles in transcriptional activation and are believed to function as a transcriptional repressor (2). However, our experiments indicate that RFX4_v3 can activate Cx3cl1 gene expression. The activation domain of RFX4_v3 protein has not yet been identified. It seems that the activation region is located within the first 574 amino acids, since RFX4_v3-(1-574) can stimulate Cx3cl1 gene transcription to a similar extent as the full-length RFX4_v3 protein. Further carboxyl-terminal deletion of the RFX4_v3-(1-574) protein to generate RFX4_v3-(1-300) totally abolishes RFX4_v3 activation on the Cx3cl1 promoter, suggesting that the dimerization domain may be essential for RFX4_v3 activities, since amino acids 301-574 mainly encode the dimerization domain. The carboxyl-terminal region of RFX4_v3 (amino acids 575-735) has no effect on RFX4_v3 transactivating abilities. However, this region was involved in the GPS2 potentiation of RFX4_v3 activation. There are several possible explanations for this phenomenon. RFX4_v3-(1-574) alone was sufficient for GPS2 binding; however, additional binding between GPS2 and the RFX4_v3 carboxyl-terminal region could 1) enhance the interaction between two proteins or 2) lead to conformational changes of either or both proteins. These changes might facilitate complex formation with other transcription factor(s) or alternatively increase the activating abilities of RFX4_v3 itself. Additional experiments will be needed to further examine these possibilities.
Besides the interaction with RFX4_v3, GPS2 has the ability to bind other mammalian RFX family members, including RFX2 and RFX3, but not RFX1. It has been suggested that the dimerization domain might be involved in the interaction. RFX4_v2 forms a heterodimer with RFX2 and RFX3, but not RFX1, and forms a homodimer with RFX4_v2 itself, theoretically through the dimerization domains (2). It is interesting to note that both GPS2 and RFX4_v2 do not bind RFX1, although according to the sequence alignment, the dimerization domains of RFX1-RFX3 are closely related, and the dimerization domain of RFX4 is slightly different from the other three dimerization domains.
GPS2 is a relatively well characterized transcriptional cofactor; therefore, we chose to focus our initial efforts on the interaction between GPS2 and RFX4_v3. However, eight other potential binding partners for RFX4_v3 were uncovered by the yeast two-hybrid screening, and several may be worthy of further consideration. For example, calmodulin is associated with numerous neuronal functions, including dendrite growth (34) and synaptic plasticity (35). There is a putative calmodulin binding site between amino acids 114 and 168 of the RFX4_v3 protein. This is consistent with our yeast two-hybrid results, which show that the first 250 amino acids of RFX4_v3 could interact with calmodulin. Proteasome function is linked with neurodegenerative disorders (36), and one proteasome subunit PMSD2 might interact with RFX4_v3. ZBTB1 contains both zinc finger and BTB domains. BTB domains from several zinc finger proteins have been shown to mediate transcriptional repression and to interact with components of histone deacetylase co-repressor complexes, including N-CoR and SMRT (37). NCDN, RICS, and FBF1 are believed to be cytosolic proteins, but it will be interesting to examine whether they could potentially translocate to the nucleus and interact with RFX4_v3 under certain circumstances. MLF2 and ZFP469 are novel proteins with no information on their cellular localization or physiological function. For each of these potential RFX4_v3 binding partners, examination of cellular localization, validation of physical interactions with RFX4_v3, and further investigation of the functional relevance of these interactions with respect to regulation of RFX4_v3-mediated gene transcription will provide for a better understanding of the role of RFX4_v3 in brain development.
Acknowledgments
We thank Dr. Georges Frech for oversight of the yeast two-hybrid project at Myriad Genetics and Dr. Sean Liour for providing RNA of J1 cells under neuronal differentiation. We are grateful to Drs. Anton Jetten and Christina Teng for critical comments during preparation of the manuscript.
This research was supported by the Intramural Research Program of NIEHS, National Institutes of Health. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: RFX, regulatory factor X; DBD, DNA binding domain; EYFP, yellow-green fluorescence protein; HA, hemagglutinin; PQ, proline- and glutamine-rich; Q, glutamine-rich; RA, retinoic acid; TSS, transcription start site; ChIP, chromatin immunoprecipitation; CMV, cytomegalovirus; N-CoR, nuclear receptor corepressor.
References
- 1.Gajiwala, K. S., Chen, H., Cornille, F., Roques, B. P., Reith, W., Mach, B., and Burley, S. K. (2000) Nature 403 916-921 [DOI] [PubMed] [Google Scholar]
- 2.Morotomi-Yano, K., Yano, K., Saito, H., Sun, Z., Iwama, A., and Miki, Y. (2002) J. Biol. Chem. 277 836-842 [DOI] [PubMed] [Google Scholar]
- 3.Bonnafe, E., Touka, M., AitLounis, A., Baas, D., Barras, E., Ucla, C., Moreau, A., Flamant, F., Dubruille, R., Couble, P., Collignon, J., Durand, B., and Reith, W. (2004) Mol. Cell Biol. 24 4417-4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reith, W., and Mach, B. (2001) Annu. Rev. Immunol. 19 331-373 [DOI] [PubMed] [Google Scholar]
- 5.Seubert, J., Yang, B., Bradbury, J. A., Graves, J., Degraff, L. M., Gabel, S., Gooch, R., Foley, J., Newman, J., Mao, L., Rockman, H. A., Hammock, B. D., Murphy, E., and Zeldin, D. C. (2004) Circ. Res. 95 506-514 [DOI] [PubMed] [Google Scholar]
- 6.Blackshear, P. J., Graves, J. P., Stumpo, D. J., Cobos, I., Rubenstein, J. L., and Zeldin, D. C. (2003) Development 130 4539-4552 [DOI] [PubMed] [Google Scholar]
- 7.Matsushita, H., Uenaka, A., Ono, T., Hasegawa, K., Sato, S., Koizumi, F., Nakagawa, K., Toda, M., Shingo, T., Ichikawa, T., Noguchi, Y., Tamiya, T., Furuta, T., Kawase, T., Date, I., and Nakayama, E. (2005) Cancer Sci. 96 801-809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang, D., Stumpo, D. J., Graves, J. P., DeGraff, L. M., Grissom, S. F., Collins, J. B., Li, L., Zeldin, D. C., and Blackshear, P. J. (2006) J. Neurochem. 98 860-875 [DOI] [PubMed] [Google Scholar]
- 9.Araki, R., Takahashi, H., Fukumura, R., Sun, F., Umeda, N., Sujino, M., Inouye, S. T., Saito, T., and Abe, M. (2004) J. Biol. Chem. 279 10237-10242 [DOI] [PubMed] [Google Scholar]
- 10.Glaser, B., Kirov, G., Bray, N. J., Green, E., O'Donovan, M. C., Craddock, N., and Owen, M. J. (2005) Mol. Psychiatry 10 920-927 [DOI] [PubMed] [Google Scholar]
- 11.Tarozzo, G., Bortolazzi, S., Crochemore, C., Chen, S. C., Lira, A. S., Abrams, J. S., and Beltramo, M. (2003) J. Neurosci. Res. 73 81-88 [DOI] [PubMed] [Google Scholar]
- 12.Garrus, J. E., von Schwedler, U. K., Pornillos, O. W., Morham, S. G., Zavitz, K. H., Wang, H. E., Wettstein, D. A., Stray, K. M., Cote, M., Rich, R. L., Myszka, D. G., and Sundquist, W. I. (2001) Cell 107 55-65 [DOI] [PubMed] [Google Scholar]
- 13.Lai, W. S., Carballo, E., Strum, J. R., Kennington, E. A., Phillips, R. S., and Blackshear, P. J. (1999) Mol. Cell Biol. 19 4311-4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strausberg, R. L., Feingold, E. A., Grouse, L. H., Derge, J. G., Klausner, R. D., Collins, F. S., Wagner, L., Shenmen, C. M., Schuler, G. D., Altschul, S. F., Zeeberg, B., Buetow, K. H., Schaefer, C. F., Bhat, N. K., Hopkins, R. F., Jordan, H., Moore, T., Max, S. I., Wang, J., Hsieh, F., Diatchenko, L., Marusina, K., Farmer, A. A., Rubin, G. M., Hong, L., Stapleton, M., Soares, M. B., Bonaldo, M. F., Casavant, T. L., Scheetz, T. E., Brownstein, M. J., Usdin, T. B., Toshiyuki, S., Carninci, P., Prange, C., Raha, S. S., Loquellano, N. A., Peters, G. J., Abramson, R. D., Mullahy, S. J., Bosak, S. A., McEwan, P. J., McKernan, K. J., Malek, J. A., Gunaratne, P. H., Richards, S., Worley, K. C., Hale, S., Garcia, A. M., Gay, L. J., Hulyk, S. W., Villalon, D. K., Muzny, D. M., Sodergren, E. J., Lu, X., Gibbs, R. A., Fahey, J., Helton, E., Ketteman, M., Madan, A., Rodrigues, S., Sanchez, A., Whiting, M., Young, A. C., Shevchenko, Y., Bouffard, G. G., Blakesley, R. W., Touchman, J. W., Green, E. D., Dickson, M. C., Rodriguez, A. C., Grimwood, J., Schmutz, J., Myers, R. M., Butterfield, Y. S., Krzywinski, M. I., Skalska, U., Smailus, D. E., Schnerch, A., Schein, J. E., Jones, S. J., and Marra, M. A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 16899-1690312477932 [Google Scholar]
- 15.Teruel, M. N., Chen, W., Persechini, A., and Meyer, T. (2000) Curr. Biol. 10 86-94 [DOI] [PubMed] [Google Scholar]
- 16.Peters, J. M., Franke, W. W., and Kleinschmidt, J. A. (1994) J. Biol. Chem. 269 7709-7718 [PubMed] [Google Scholar]
- 17.Dateki, M., Horii, T., Kasuya, Y., Mochizuki, R., Nagao, Y., Ishida, J., Sugiyama, F., Tanimoto, K., Yagami, K., Imai, H., and Fukamizu, A. (2005) J. Biol. Chem. 280 20503-20508 [DOI] [PubMed] [Google Scholar]
- 18.Moon, S. Y., Zang, H., and Zheng, Y. (2003) J. Biol. Chem. 278 4151-4159 [DOI] [PubMed] [Google Scholar]
- 19.Schmidt, T., Karsunky, H., Frass, B., Baum, W., Denzel, A., and Moroy, T. (2000) Biochim. Biophys. Acta 1493 249-254 [DOI] [PubMed] [Google Scholar]
- 20.Katayama, S., Tomaru, Y., Kasukawa, T., Waki, K., Nakanishi, M., Nakamura, M., Nishida, H., Yap, C. C., Suzuki, M., Kawai, J., Suzuki, H., Carninci, P., Hayashizaki, Y., Wells, C., Frith, M., Ravasi, T., Pang, K. C., Hallinan, J., Mattick, J., Hume, D. A., Lipovich, L., Batalov, S., Engstrom, P. G., Mizuno, Y., Faghihi, M. A., Sandelin, A., Chalk, A. M., Mottagui-Tabar, S., Liang, Z., Lenhard, B., and Wahlestedt, C. (2005) Science 309 1564-1566 [DOI] [PubMed] [Google Scholar]
- 21.Jin, D. Y., Teramoto, H., Giam, C. Z., Chun, R. F., Gutkind, J. S., and Jeang, K. T. (1997) J. Biol. Chem. 272 25816-25823 [DOI] [PubMed] [Google Scholar]
- 22.Liour, S. S., and Yu, R. K. (2003) Glia 42 109-117 [DOI] [PubMed] [Google Scholar]
- 23.Bibel, M., Richter, J., Schrenk, K., Tucker, K. L., Staiger, V., Korte, M., Goetz, M., and Barde, Y. A. (2004) Nat. Neurosci. 7 1003-1009 [DOI] [PubMed] [Google Scholar]
- 24.Soriano, S. G., Amaravadi, L. S., Wang, Y. F., Zhou, H., Yu, G. X., Tonra, J. R., Fairchild-Huntress, V., Fang, Q., Dunmore, J. H., Huszar, D., and Pan, Y. (2002) J. Neuroimmunol. 125 59-65 [DOI] [PubMed] [Google Scholar]
- 25.Tong, N., Perry, S. W., Zhang, Q., James, H. J., Guo, H., Brooks, A., Bal, H., Kinnear, S. A., Fine, S., Epstein, L. G., Dairaghi, D., Schall, T. J., Gendelman, H. E., Dewhurst, S., Sharer, L. R., and Gelbard, H. A. (2000) J. Immunol. 164 1333-1339 [DOI] [PubMed] [Google Scholar]
- 26.Spain, B. H., Bowdish, K. S., Pacal, A. R., Staub, S. F., Koo, D., Chang, C. Y., Xie, W., and Colicelli, J. (1996) Mol. Cell Biol. 16 6698-6706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon, H. G., Chan, D. W., Huang, Z. Q., Li, J., Fondell, J. D., Qin, J., and Wong, J. (2003) EMBO J. 22 1336-1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang, J., Kalkum, M., Chait, B. T., and Roeder, R. G. (2002) Mol. Cell 9 611-623 [DOI] [PubMed] [Google Scholar]
- 29.Peng, Y. C., Kuo, F., Breiding, D. E., Wang, Y. F., Mansur, C. P., and Androphy, E. J. (2001) Mol. Cell Biol. 21 5913-5924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, T. H., Yi, W., Griswold, M. D., Zhu, F., and Her, C. (2006) DNA Repair (Amst.) 5 32-42 [DOI] [PubMed] [Google Scholar]
- 31.Breiding, D. E., Sverdrup, F., Grossel, M. J., Moscufo, N., Boonchai, W., and Androphy, E. J. (1997) Mol. Cell Biol. 17 7208-7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng, Y. C., Breiding, D. E., Sverdrup, F., Richard, J., and Androphy, E. J. (2000) J. Virol. 74 5872-5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Degenhardt, Y. Y., and Silverstein, S. J. (2001) J. Virol. 75 151-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alvania, R. S., Chen, X., and Ginty, D. D. (2006) Neuron 50 813-815 [DOI] [PubMed] [Google Scholar]
- 35.Colbran, R. J., and Brown, A. M. (2004) Curr. Opin. Neurobiol. 14 318-327 [DOI] [PubMed] [Google Scholar]
- 36.Hol, E. M., Fischer, D. F., Ovaa, H., and Scheper, W. (2006) Expert Rev. Neurother. 6 1337-1347 [DOI] [PubMed] [Google Scholar]
- 37.Melnick, A., Carlile, G., Ahmad, K. F., Kiang, C. L., Corcoran, C., Bardwell, V., Prive, G. G., and Licht, J. D. (2002) Mol. Cell Biol. 22 1804-1818 [DOI] [PMC free article] [PubMed] [Google Scholar]