Abstract
Considerable evidence suggests that oxidative stress may be involved in the pathogenesis of Transmissible Spongiform Encephalopathies (TSEs). To investigate the involvement of iron metabolism in TSEs, we examined the expression levels of iron regulatory proteins (IRPs), ferritins, and binding activities of IRPs to iron-responsive element (IRE) in scrapie-infected mice. We found that the IRPs–IRE-binding activities and ferritins were increased in the astrocytes of hippocampus and cerebral cortex in the brains of scrapie-infected mice. These results suggest that alteration of iron metabolism contributes to development of neurodegeneration and that some protective mechanisms against iron-induced oxidative damage may occur during the pathogenesis of TSEs.
Keywords: Transmissible Spongiform Encephalopathies (TSEs), Oxidative stress, Neurodegeneration, Iron regulatory proteins (IRPs), Ferritin, Astrocytosis
Transmissible Spongiform Encephalopathies (TSEs) or prion diseases are a group of fatal neurodegenerative disorders in humans and animals [4]. Although the pathogenic mechanisms of TSEs are not fully understood, aberrant metabolism of prion protein (PrP), specifically conversion of a host cellular PrP (PrPC) into an abnormal, protease-resistant isoform (PrPSc), is believed to be an important molecular mechanism of TSEs [21].
Several neurodegenerative disorders, such as Alzheimer’s disease (AD) and Parkinson’s disease (PD), are associated with oxidative stress [10,22]. Cumulated evidence reveals that PrPC might play a role as an antioxidant; therefore, the brains of TSEs are likely to be more susceptible to oxidative stress due to the conversion of PrPC into PrPSc [10,13].
Iron is physiologically essential for growth and survival, playing important roles in many biological processes, such as electron and oxygen transport, and DNA replication [10]. However, free iron can be toxic to cells due to the ability of iron to catalyze free radical formations that can destroy lipid membranes and other cellular constituents [14]. Intracellular iron homeostasis is mainly controlled by cytoplasmic IRPs (IRP1 and IRP2), which are known as iron-sensing proteins; thus, iron regulatory proteins play a central role in iron metabolism [7,14]. Although the two IRPs have similar IRE-binding affinities and are regulated by iron valence and its association with other molecules within cells, their expression levels are different in various cell types, indicating the possibility that the two IRPs may play different roles or are involved in different mechanisms in the regulation of iron metabolism [19]. The iron storage protein, ferritin, also plays a key role in iron metabolism. Its ability to sequester iron gives ferritin the dual functions: iron detoxification and iron reserve [2,9].
In this study, the brains of control and scrapie-infected mice were analyzed for several key components involved in iron metabolism: IRP1, IRP2, and H- and L-ferritins. We demonstrated that there is disturbance of iron metabolism in brains of scrapie-positive mice.
Male C57BL/6J mice, 4–6 weeks of age, were obtained from the Experimental Animal Center of Hallym University. ME7 scrapie strain was provided by Dr. Alan Dickinson (Neuropathogenesis Unit, Edinburgh, UK). Mice were divided into two groups: control and scrapie-infected. Mice were inoculated intracerebrally with 30 μl of 1% (w/v) brain homogenates in 0.01 M PBS prepared from C57BL/6J mice in terminal stage of ME7 scrapie infection [11]. Animal maintenance, injection, monitoring and euthanasia were carried out in accordance with the NIH guidelines.
Western blot analysis was performed to detect the protein expression levels of IRP1, IRP2, H- and L-ferritin in the brains of control and scrapie-infected mice. Brain homogenates were prepared in homogenization buffer (1.15% KCl, 10 mM phosphate, 5 mM EDTA, pH 7.4) [11,15]. Equal amounts (50 μg) of proteins were used to detect each protein. Antibodies used were as follows: anti-IRP1 serum (1:5000), anti-IRP2 serum (1:3000), anti-ferritin serum (1:3000; DAKO, Denmark), and anti-recombinant H-ferritin serum (1:3000) [8]. The expression of a housekeeping gene, β-actin protein, was measured with anti-β-actin antibody (1:10,000; Sigma). Each experiment was repeated at least three times.
Serial tissue sections (3 μm thick) were placed on slides coated with poly-L lysine, then deparaffinzed and rehydrated. After inhibiting endogenous peroxidase activity with methanol containing 3% H2O2, the slides were treated with 10% normal goat serum to block non-specific binding. Then the slides were incubated with anti-IRP1 (1:200), anti-IRP2 (1:200), anti-ferritin (1:1000), anti-recombinant H-ferritin (1:400) or antiglial fibrillary acidic protein (GFAP) (1:100) serum. The sections were incubated with biotinylated secondary antibodies. Then the slides were incubated with avidin–biotin complex and 3,3′-diaminobenzidine (DAB) was used as a chromogen. The sections were counterstained with Harris hematoxylin. As a negative control of the immunostaining, scrapie-positive sections were prepared and stained in the same manner as that described above, except for the omission of primary antibodies. Sections from mice infected with normal mouse brain were used as controls.
A 237 bp PCR product of mouse L-ferritin 5′UTR region was ligated into pBluesript II SK (+) vector via BamHI and XhoI sites. After amplification, the cDNA was cut by SmaI and purified by gel elution kit (QIAGEN, Germany). A 135 bp IRE RNA probe was made and radiolabeled by T3 RNA polymerase and 32P-CTP (NEN, USA). After purification by Sephadex G-50 column (Roche, Germany), the specific radioactivity of the IRE RNA probe (5 × 104 cpm/μl) was counted by Beckman scintillation counter. RNA band shift assay and electrophoresis of the RNA–protein complexes were performed as described previously with slight modifications [23].
All results were representative of at least three separate experiments. Statistical analysis was performed using the Student’s t-test. Values of p < 0.05 or p < 0.01 were considered statistically significant.
Densitometry (Fig. 1C) of Western blots of IRP1 (Fig. 1A) and IRP2 (Fig. 1B) indicate that protein expression levels were increased by 2.0-fold and 2.1-fold, respectively, in the brains of scrapie-infected mice compared with levels in controls (Fig. 1C; p < 0.05). Intense immunoreactive staining of IRP1 and IRP2 was found in the brains of scrapie-infected mice, particularly in the hippocampus and cerebral cortex (Fig. 2B, E, H and K); in contrast, there was only weak IRP1 and IRP2 immunoreactive staining in the brains of control mice (Fig. 2A, D, G and J). Immunoreactive staining for IRP1 and IRP2 was found predominantly in the cytoplasm of the cells, which morphologically appeared to be reactive astrocytes. As evidence of this, IRP1 and IRP2 immunoreactivity was co-localized with GFAP staining in serial sections of brains of scrapie-infected mice (Fig. 2C, F, I and L), indicating that IRP1- and IRP2-immunolabled cells were reactive astrocytes.
Fig. 1.
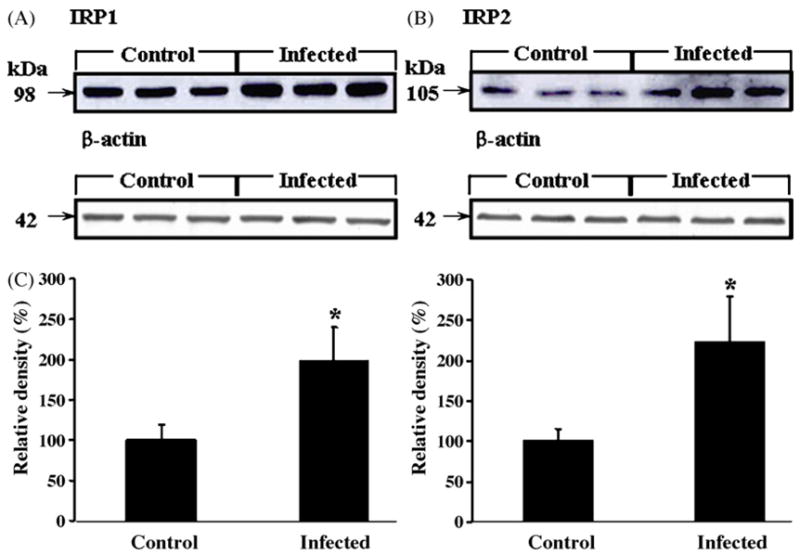
Western blots of protein expression levels of IRP1 (A) and IRP2 (B) in the brains of control (n = 18) and scrapie-infected mice (n = 18). The IRP1 and IRP2 protein expression levels in (A and B) were measured by densitometry (C); expression levels were increased by 2.0-fold and 2.1-fold, respectively, in the brains of scrapie-infected mice compared with control mice. *Values significantly different from control (p < 0.05). Expression of the housekeeping gene, β-actin, was measured in (A and B).
Fig. 2.
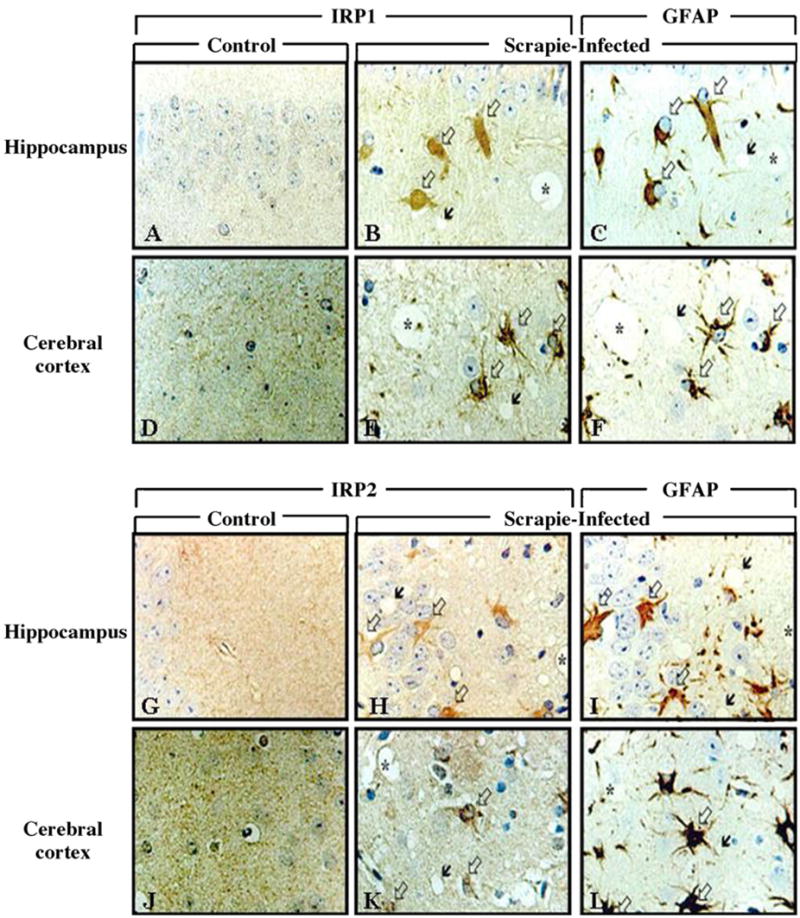
Immunohistochemical localization of IRP1 and IRP2 in the brains of control and scrapie-infected mice. Intense immunolabeling of IRP1 and IRP2 appear in the hippocampus and cerebral cortex of scrapie-infected mice. Black arrows: vacuoles, white arrows: immunolabeled cells with IRP1 (B and E) and IRP2 (H and K). In (C, F, I and L) white arrows indicate immunolabeling of GFAP and black arrows indicate vacuoles. In sequential sections, immunoreactivity of IRP1 and IRP2 is colocalized with GFAP-positive reactive astrocytes. Sequential sections are B and C, E and F, H and I, K and L. Asterisk (*) indicates landmark blood vessels in the adjacent tissue section.
To determine the relative IRE-binding activity of IRP1 and IRP2, we performed RNA gel shift assays using brain preparations from control and scrapie-infected mice. As shown in Fig. 3, IRE-binding activity of IRP2 is higher than that of IRP1 both in control and scrapie-infected mice (lanes 2–5). This finding was also observed in brain samples in the presence of high levels of 2-ME which can reveal total RNA binding capacity of IRPs (lanes 6–9). Significantly, spontaneous IRE-binding activity (active form) of both IRP1 and IRP2 in the scrapie-infected mice (lanes 4 and 5) is higher than that of the control mice (lanes 2 and 3). In addition, total IRE-binding capacity of both IRP1 and IRP2 in scrapie-infected brains (lanes 8 and 9) is higher than that in control brains (lanes 6 and 7). To confirm that the detected bands were true IRE–IRP1 and IRE–IRP2 complexes, we performed competition assays using 2-ME-treated brain samples. Competition with unlabeled IRE markedly reduced the formation of radioactive IRE–protein complexes (lanes 10 and 11).
Fig. 3.
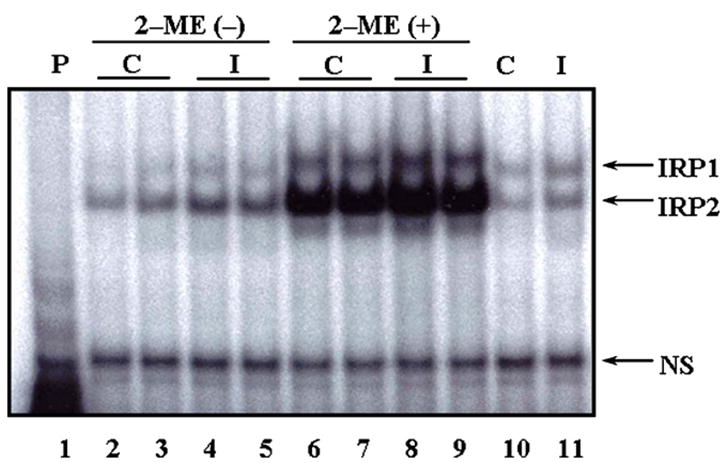
IRE-binding activities of IRP1 and IRP2 in the brains of control (n = 12) and ME7 scrapie-infected C57BL mice (n = 12) by REMSA. IRE-binding activities of IRP1 and IRP2 are increased in scrapie-infected brains (lanes 4, 5, 8 and 9), compared with control brains (lanes 2, 3, 6 and 7). Lane 1: IRE probe only; lanes 2, 3, 6, 7 and 10: control; lanes 4, 5, 8, 9 and 11: scrapie-infected; lanes 6–9: full activation of IRPs detected by addition of 2% 2-ME; lanes 10 and 11: competition assay using 2-ME-treated brain samples as unlabeled IRE added to IRE probe: compare lanes 10 and 11 to 6, 7 and 8, 9, respectively. P: probe; C: control; I: infected; NS: non-specific bands.
H/L-ferritin (Fig. 4A) and H-ferritin (Fig. 4B) protein expression levels were examined by Western blot in Fig. 4A and B, respectively. The expression of both proteins was significantly increased in the brains of scrapie-infected mice compared to the levels in brains of control mice (Fig. 4C; p < 0.05 and p < 0.01).
Fig. 4.
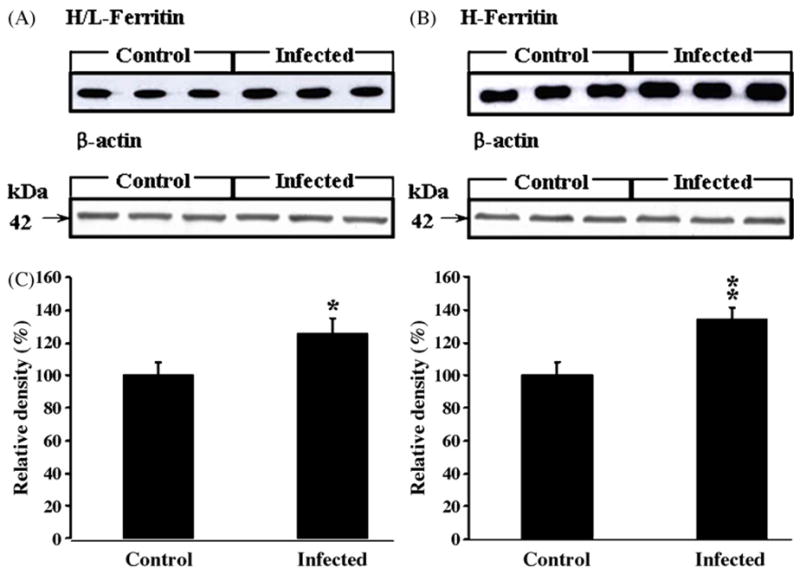
Protein expression levels of H/L- (A) and H-ferritin (B) in the brains of control (n = 18) and scrapie-infected mice (n = 18). The H/L- and H-ferritin protein expression in (A and B) were measured by densitometry in (C); expression levels were increased significantly in the brains of scrapie-infected mice compared with controls. *Values significantly different from control (*p < 0.05; **p < 0.01). Lanes 1–3: control; lanes 4–6: scrapie-infected mice. Expression of the housekeeping gene, β-actin, was measured in (A and B).
By immunohistochemistry, intense expression of H/L-ferritin and H-ferritin was seen in scrapie-infected group (Fig. 5). Intense ferritin immunoreactivity was co-localized with GFAP-positive cells in the brains of scrapie-infected mice. In contrast, there was only weak ferritin immunoreactivity in the brain cells of the control group.
Fig. 5.
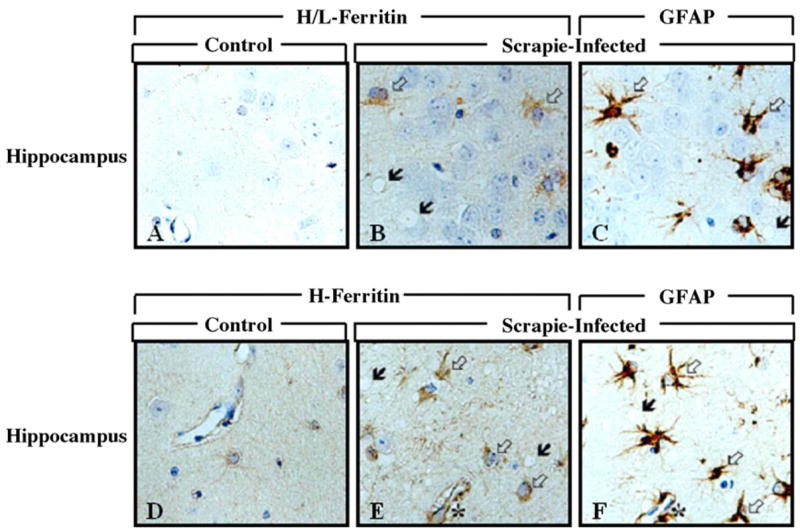
Immunohistochemical localization of H/L- and H-ferritin in the hippocampus of control and scrapie-infected C57BL mice. Black arrows: vacoulation; white arrows: immunolabeled cells with H/L- (B) and H-ferritin (E). In (C and F) white arrows indicate GFAP immunolabeled cells and black arrows indicate vacuoles. In sequential sections, immunoreactivity for H/L- and H-ferritin is colocalized with GFAP-positive astrocytes. Sequential reactions are B and C, E and F. Asterisk (*) indicates landmark blood vessels in the adjacent tissue sections.
In this study, we demonstrated that the expression levels of several key proteins involved in iron metabolism, IRP1, IRP2, H- and H/L-ferritins, were significantly increased in the brains of scrapie-infected mice compared to the brains of control mice. It has been reported that IRP1 is expressed constitutively in cells [3,16] and regulated posttranslationally by switching between cytoplasmic aconitase and IRE-binding form [12,16]. On the other hand, IRP1 gene expression was downregulated by NO, indicating that IRP1 can also be modulated by both transcriptional and translational pathways [1,6,17]. We previously reported that total iron and ferric iron (Fe3+) content were elevated in the brains of scrapie-infected mice [14]; although the origin of increased iron content is not known, it could result from the alteration of blood-brain barrier caused by scrapie infection or from an increase of inflammatory cytokines, such as IL-β and TNF-α [13,24]. Our results together with previous findings suggest that disturbance of iron metabolism and related oxidative stress are closely associated with neurodegeneration in TSEs.
Intracellular IRPs are either intrinsically active or latent with respect to their ability to bind to IRE-containing RNA. A variety of factors including valence and association of iron and oxidative stress can affect functions of IRPs and alter the distribution of both IRP1 and IRP2 with regard to active and latent forms; these factors can also influence the total amount of intracellular IRPs. Therefore, it was important to determine the proportion of active and latent forms of IRP1 and IRP2 as well as their protein expression levels in the brains of scrapie-infected mice in order to examine their effects on the pathogenic process of neurodegeneration in TSEs. RNA gel shift assay showed that IRP1 and IRP2 may function together in response to scrapie infection. Since IRP2 changed more dramatically than IRP1 in scrapie infection, IRP2 may act as a principal regulator of iron metabolism in both physiological and pathological conditions, whereas IRP1 may act as a supplementary regulator.
H-and H/L-ferritins, in the brains of scrapie-infected mice, are seen in the cytoplasm of reactive astrocytes in brains of scrapie-infected mice. Because ferritin, especially H-ferritin is known to reduce iron-induced oxidative stress by capturing and decreasing intracellular free iron, increased ferritin in reactive astrocytes of our model could play a role in protecting brain from oxidative damage. The increase in ferritin synthesis is mediated by heme oxygenase (HO), an enzyme induced by oxidative stress that liberates iron from heme [12]. The released iron stimulates ferritin synthesis through the IRE–IRP machinery and the newly synthesized ferritin reduces the amount of iron available for ROS production and cellular damage. Our previous report showing that there is increased expression of HO-1 in reactive astrocytes of scrapie-infected rodents [5] further supports the concept that ferritin synthesis may be closely associated with protection against iron-induced oxidative stress.
It was expected that increased IRP/IRE-binding activity would decrease ferritin synthesis. However, in the present study, the upregulation of IRP1 and IRP2 RNA binding activity in scrapie-infected brain did not lead to a decrease in ferritin. The elevation of iron level in scrapie brain [14] could counteract the effect of increased IRP1 and IRP2 binding activity. It has been shown that ferritin H chain is transcriptionally regulated by oxidative stress [18,20]. In this regard, it is likely that the increased oxidative stress in scrapie brain led to the increase in ferritin level by affecting its transcriptional process. It is possible that, under particular patho-physiological conditions, iron release from ferritin may be unregulated. Thus, there are many conflicting factors that can lead to either deterioration or protection from oxidative damage in the brains of the ME7 scrapie-infected model. Although the precise role of IRPs and ferritin in TSEs is uncertain, it is feasible that the redox state in the microenvironment in brain may be one of the key modulators that determine their effects as pro-oxidants or anti-oxidants. Considering the fact that the topographical distribution of the pathological lesions was generally in accord with the location of components involved in iron metabolism, i.e., IRP1, IRP2 and ferritins, it is probable that disturbances of iron metabolism and related oxidative stress contribute to the pathogenic mechanism of neurodegeneration in TSEs, probably through the action of activated astrocytes. Therefore, disclosing the location of IRPs and ferritins inside the activated astrocytes maybe explain the aberrant metabolism of iron in TSE, thus remain as a further study.
Acknowledgments
This study was supported by a grant of the Korea Research Foundation Grant funded by the Korean Government (MOEHRD)(KRF-2006-311-E00034) and the International Research Internship Program of the Korea Science and Engineering Foundation (KOSEF)(M07-2004-000-10005-0).
References
- 1.Bouton C, Chauveau MJ, Lazereg S, Drapier JC. Recycling of RNA binding iron regulatory protein 1 into an aconitase after nitric oxide removal depends on mitochondrial ATP. J Biol Chem. 2002;277:31220–31227. doi: 10.1074/jbc.M203276200. [DOI] [PubMed] [Google Scholar]
- 2.Cairo G, Rappocciolo E, Tacchini L, Schiaffonati L. Expression of the genes for ferritin H and L subunits in rat liver and heart. Evidence for tissue specific regulations at pre- and post-translational levels. Biochem J. 1991;275:813–816. doi: 10.1042/bj2750813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cairo G, Ronchi R, Recalcati S, Campanella A, Minotti G. Nitric oxide and peroxynitrite activate the iron regulatory protein-1 of J774A.1 macrophages by direct disassembly of the Fe-S cluster of cytoplasmic aconitase. Biochemistry. 2002;41:7435–7442. doi: 10.1021/bi025756k. [DOI] [PubMed] [Google Scholar]
- 4.Carp RI, Ye X, Kascsak RJ, Rubenstein R. The nature of the scrapie agent. Biological characteristics of scrapie in different scrapie strain-host combinations. Ann NY Acad Sci. 1995;724:221–234. doi: 10.1111/j.1749-6632.1994.tb38912.x. [DOI] [PubMed] [Google Scholar]
- 5.Choi YG, Kim JI, Lee HP, Jin JK, Choi EK, Carp RI, Kim YS. Induction of heme oxygenase-1 in the brains of scrapie-infected mice. Neurosci Lett. 2000;289:173–176. doi: 10.1016/s0304-3940(00)01277-5. [DOI] [PubMed] [Google Scholar]
- 6.Drapier JC, Hirling H, Wietzerbin J, Kaldy P, Kuhn LC. Reciprocal modulation of aconitase activity and RNA-binding activity of iron regulatory factor by nitric oxide. Adv Exp Med Biol. 1994;356:141–148. doi: 10.1007/978-1-4615-2554-7_16. [DOI] [PubMed] [Google Scholar]
- 7.Gerlach M, Ben-Schachar D, Reiderer P, Youdim MBH. Altered brain metabolism of iron as a cause of neurodegenerative diseases. J Neurochem. 1994;63:793–807. doi: 10.1046/j.1471-4159.1994.63030793.x. [DOI] [PubMed] [Google Scholar]
- 8.Guo B, Yu Y, Leibold EA. Iron regulates cytoplasmic levels of a novel iron-reponsive element-binding protein without aconitase activity. J Biol Chem. 1994;269:24252–24260. [PubMed] [Google Scholar]
- 9.Harrison PM, Arosio P. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta. 1996;1275:161–203. doi: 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]
- 10.Hur K, Kim JI, Choi SI, Choi EK, Carp RI, Kim YS. The pathogenic mechanisms of prion diseases. Mech Ageing Dev. 2002;123:1637–1647. doi: 10.1016/s0047-6374(02)00099-4. [DOI] [PubMed] [Google Scholar]
- 11.Ju WK, Park KJ, Choi EK, Kim J, Carp RI, Wisniewski HM, Kim YS. Expression of inducible nitric oxide synthase in the brains of scrapie-infected mice. J Neurovirol. 1998;4:445–450. doi: 10.3109/13550289809114544. [DOI] [PubMed] [Google Scholar]
- 12.Kiemer AK, Fornges AC, Pantopoulos K, Bilzer M, Andriopoulos B, Gerwig T, Kenngott S, Gerbes AL, Vollmar AM. ANP-induced decrease of iron regulatory protein activity is independent of HO-1 induction. Am J Physiol Gastrointest Liver Physiol. 2004;287:G518–G526. doi: 10.1152/ajpgi.00514.2003. [DOI] [PubMed] [Google Scholar]
- 13.Kim JI, Choi SI, Kim NH, Jin JK, Choi EK, Carp RI, Kim YS. Oxidative stress and neurodegeneration in prion diseases. Ann NY Acad Sci. 2001;928:182–186. doi: 10.1111/j.1749-6632.2001.tb05648.x. [DOI] [PubMed] [Google Scholar]
- 14.Kim NH, Park SJ, Jin JK, Kwon MS, Choi EK, Carp RI, Kim YS. Increased ferric iron content and iron-induced oxidative stress in the brains or scrapie-infected mice. Brain Res. 2000;884:98–103. doi: 10.1016/s0006-8993(00)02907-3. [DOI] [PubMed] [Google Scholar]
- 15.Lee DW, Sohn HO, Lim HB, Lee YG, Kim YS, Carp RI, Wisniewski HM. Alternation of free radical metabolism in the brain of mice infected with scrapie agent. Free Radic Res. 1999;30:499–507. doi: 10.1080/10715769900300541. [DOI] [PubMed] [Google Scholar]
- 16.Muckenthaler M, Gunkel N, Frishman D, Cyrklaff A, Tomancak P, Hentze MW. Iron-regulatory protein-1 (IRP-1) is highly conserved in two invertebrate species. Charaterisation of IRP-1 homologous in Drosophila melanogaster and Caenorhabditis elegans. Eur J Biochem. 1998;254:230–237. doi: 10.1046/j.1432-1327.1998.2540230.x. [DOI] [PubMed] [Google Scholar]
- 17.Oliveira L, Drapier JC. Down-regulation of iron regulatory protein 1 gene expression by nitric oxide. Proc Natl Acad Sci. 2000;97:6550–6555. doi: 10.1073/pnas.120571797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orino K, Lehman L, Tsuji Y, Ayaki H, Torti SV, Torti FM. Ferritin and the response to oxidative stress. Biochem J. 2001;357:241–247. doi: 10.1042/0264-6021:3570241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pantopoulos K. Iron metabolism and the IRE/IRP regulatory system: an update. Ann NY Acad Sci. 2004;1012:1–13. doi: 10.1196/annals.1306.001. [DOI] [PubMed] [Google Scholar]
- 20.Pham CG, Bubici C, Zazzeroni F, Papa S, Jones J, Alvarez K, Jayawardena S, De Smaele E, Cong R, Beaumont C, Torti FM, Torti SV, Franzoso G. Ferritin heavy chain upregulation by NF- kappaB inhibits TNFalpha-induced apoptosis by suppressing reactive oxygen species. Cell. 2004;119:529–542. doi: 10.1016/j.cell.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 21.Prusiner SB. Molecular biology of prion diseases. Science. 1991;252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 22.Tabner BJ, Turnbull S, El-Agnaf OMA, Allsop D. Production of reactive oxygen species from aggregating proteins implicated in Alzheimer’s disease, Parkinson’s disease and other neurodegenerative diseases. Curr Top Med Chem. 2001;1:507–517. doi: 10.2174/1568026013394822. [DOI] [PubMed] [Google Scholar]
- 23.Thomson AM, Rogers JT, Leedman PJ. Iron-regulatory proteins, iron-responsive elements and ferritin mRNA translation. Int J Biochem Cell Biol. 1999;31:1139–1152. doi: 10.1016/s1357-2725(99)00080-1. [DOI] [PubMed] [Google Scholar]
- 24.Wisniewski HM, Lossinsky AS, Moretz RC, Vorbrodt AW, Lassman H, Carp RI. Increased blood-brain barrier permeability in scrapie-infected mice. J Neuropathol Exp Neurol. 1983;42:615–626. doi: 10.1097/00005072-198311000-00002. [DOI] [PubMed] [Google Scholar]


