Abstract
A crucial step in exploiting the information inherent in genome sequences is to assign to each protein sequence its three-dimensional fold and biological function. Here we describe fold assignment for the proteins encoded by the small genome of Mycoplasma genitalium. The assignment was carried out by our computer server (http://www.doe-mbi.ucla.edu/people/frsvr/frsvr.html), which assigns folds to amino acid sequences by comparing sequence-derived predictions with known structures. Of the total of 468 protein ORFs, 103 (22%) can be assigned a known protein fold with high confidence, as cross-validated with tests on known structures. Of these sequences, 75 (16%) show enough sequence similarity to proteins of known structure that they can also be detected by traditional sequence–sequence comparison methods. That is, the difference of 28 sequences (6%) are assignable by the sequence–structure method of the server but not by current sequence–sequence methods. Of the remaining 78% of sequences in the genome, 18% belong to membrane proteins and the remaining 60% cannot be assigned either because these sequences correspond to no presently known fold or because of insensitivity of the method. At the current rate of determination of new folds by x-ray and NMR methods, extrapolation suggests that folds will be assigned to most soluble proteins in the next decade.
Keywords: protein fold recognition, computer analysis of genome sequences
In this era of genome sequencing, the vast protein sequence information accumulating in our databases offers challenges to understanding protein structure, function, and evolution. Here we ask two focused questions about the computational assignment of three-dimensional (3D) folds to protein sequences: (i) Using a computerized method of fold assignment (1), for what percentage of the genome sequences can a 3D fold be inferred? (ii) Does our method of fold assignment permit assignment of more folds than do sequence similarity searches?
From previous work, we would expect that more than 10% of genome sequences can be assigned a 3D fold. Other investigators have reported that roughly 10% of genome sequences show clear sequence similarity to proteins of known structure (e.g., refs. 2 and 3). Because clear sequence similarity implies structural similarity (4), at least for these 10% of sequences a known fold can be assigned. But there is often structural similarity even when sequence similarity is within the “twilight zone” (usually meaning a sequence identity <25%) (5). In some cases this similarity can be recognized using sequence–structure compatibility searches (6), also known as threading, 3D profiles, fold recognition, and fold assignment (7–9). Here we have chosen the smallest known genome of any free-living organism, that of Mycoplasma genitalium (MG) (10), as a test of the capabilities of our automatic fold recognition server and as a case study to identify the difficulties facing automated fold assignment.
MATERIALS AND METHODS
The MG Sequences.
The 468 MG sequences were obtained from The Institute for Genome Research (TIGR) through its Web address: http://www.tigr.org/tdb/mdb/mgdb/mgdb.html. Three types of annotation (based on searches in the sequence database) accompany each TIGR sequence (10): (i) functional assignment—a clear sequence similarity with a protein of known function from another organism was found (317 sequences, 67.7%); (ii) hypothetical protein—sequence similarity with a protein of another organism but of unknown function (55 sequences, 11.7%); and (iii) no annotation—no similarity in the sequence database (96 sequences, 20.5%).
The Fold Assignment Method.
The results reported here are based on two variants of the fold-assignment method by Fischer and Eisenberg (1). This method matches sequences to structures using sequence-derived predictions and the “global-local” alignment algorithm (1, 21). These variants are implemented as part of our fold recognition server frsvr (http://www.doe-mbi.ucla.edu/people/frsvr/frsvr.html). Each genome sequence is compared for compatibility with each of the 3D folds in a library of known structures, and the fold scoring the highest in sequence–structure compatibility is the assigned fold if the compatibility score is above a threshold value. In computing the compatibility, first the secondary structure of the sequence is predicted, using the PHD program of Rost and Sander (11). The compatibility function (equation 1 in ref. 1) has two terms. The first term reflects the extent of agreement of the secondary structure predicted from the sequence and the observed secondary structure of the fold. The second term reflects the similarity of the genome sequence to the sequence of the assigned fold, using a standard 20 × 20 sequence comparison table, in this case the table of Gonnet et al. (12). The two variants of the method of Fischer and Eisenberg (1) are: (i) SDP (for sequence-derived-prediction method), which uses only the original amino acid sequence of the genome sequence, and (ii) SDPMA (for SDP with multiple alignment), which uses a multiple alignment of the genome sequence with homologous sequences (if any; equation 2 in ref. 1).
Details of the fold assignment procedure are as follows. The library of known structures used here to match genome sequences was derived from the Protein Data Bank (PDB) (13) in April 1997. It contains 1,632 entries (corresponding to 1,291 full protein chains and 341 domains), where no two entries of similar lengths share more than 50% sequence identity.
For each genome sequence the fold recognition server frsvr carries out the following steps. (i) It searches the sequence database (SwissProt, PIR, and the sequences of the structures in PDB) to select homologous sequences [blast (14) and fasta (15)]. (ii) It builds a multiple alignment from the selected sequences (pileup in GCG Genetics Computer Group, Madison, WI). (iii) From the multiple alignment, it predicts the secondary structure (11) of the genome sequence. (iv) It identifies transmembrane α-helices with the aid of the hydrophobic moment plot (moment; ref. 16). (v) It executes the fold-assignment method of Fischer and Eisenberg (1), with variants SDP and SDPMA. (vi) It runs three sequence-based methods, motifs (17), profilesearch (GCG Genetics Computer Group, Madison, WI), and “gon” (1), plus two other (optional) fold-assignment methods, H3P2 (18) and topits (19). The results from these programs are stored for documentation purposes and are not used in our fold-assignment process.
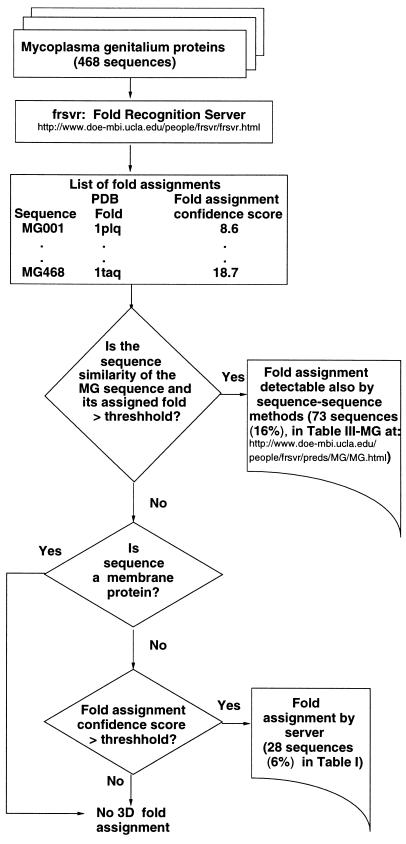
Each genome sequence is submitted to the server only once, all the above steps are executed automatically, and the results are processed, stored, and made accessible via the Web (http://www.doe-mbi.ucla.edu/people/frsvr/preds/MG/MG.html). The above steps are represented by the box labeled “Fold Recognition Server” in Fig. 1.
Figure 1.
Flow diagram for assignment of 3D folds to the Mycoplasma genitalium sequences with the aid of the UCLA–DOE fold-recognition server (http://www.doe-mbi.ucla.edu/people/frsvr/frsvr.html). The automatic assignments are of two categories: (i) fold assignment detectable by fold recognition and by sequence–sequence methods (table III-MG, available at http://www.doe-mbi.ucla.edu/people/frsvr/preds/MG/MG.html) and (ii) fold assignment detectable only by fold-recognition methods (Table 1).
Fold-Assignment Confidence Score.
For each genome sequence, SDP and SDPMA report the fold having the highest sequence–structure compatibility score. This z-score, computed as defined in ref. 1, is the number of standard deviations above the mean compatibility score in the library of known structures. We combine the results of SDP and SDPMA to form a single fold assignment with a fold-assignment confidence score Z attached to it as follows. Let z1 (z2) be the z-score attached to the highest scoring fold f1 (f2) obtained with SDP (SDPMA). If f1 and f2 belong to different fold classes (as defined by SCOP; ref. 20), then the fold assigned is f1 and Z equals the minimum of z1 and z2. Otherwise (f1 and f2 belong to the same fold class), if z2 > z1, then the fold assigned is f2 and Z equals z2; else the fold assigned is f1 and Z equals the average of z1 and z2.
Determining the Confidence Threshold Zth.
For the automatic application of a predictive method, it is necessary to draw a threshold value Zth for which an assignment with a score Z > Zth is likely to be correct. If the threshold is too high, only true positives score above it, but this occurs in only a small number of cases. As the threshold is lowered, the number of assignments with above-threshold scores increases, but false positives begin to appear. In automatic fold assignment we seek the threshold that minimizes the number of false positives but maximizes the number of valid fold assignments of sequences that cannot be identified by sequence comparison alone.
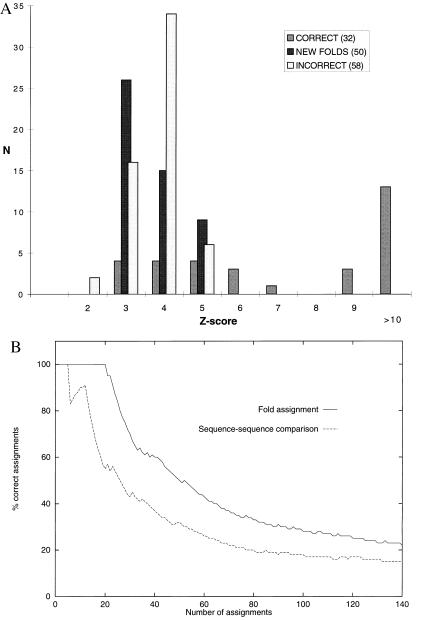
The threshold Zth was determined as follows. We first created a fold library of structures released before 1996 (containing 989 chains). Then we compiled a nonredundant representative set of 140 protein sequences whose structures were determined during 1996 and whose sequences are not obviously related to any protein in the library. Next, we submitted each of these 140 sequences to the server and evaluated the value of Z, above which all assignments are correct. The library and the representative set are available at the URL http://www.doe-mbi.ucla.edu/fischer/largescaledata.html. Out of the 140 sequences, 50 sequences correspond to new folds (not observed until 1996) and 90 correspond to folds observed before 1996. All 50 sequences corresponding to new 1996 folds received a score Z < 6; 20 of the 90 sequences that correspond to folds previously observed were assigned the correct fold with Z > 6 (Fig. 2A). No other assignment had a score above 6. Of the remaining 70 assignments with Z < 6, only 12 were correct. We conclude that for this test, the value of Z = 6 separates correct from incorrect assignments. This value confirms our previous experience from computational benchmarks (1, 18, 21) and from a blind prediction experiment (8). For the automatic assignment of this paper, we use a conservative value of Zth = 7. Fig. 2B shows that automatic fold assignment with these test sequences works better than sequence comparison alone.
Figure 2.
Results of the test of our method on 140 sequences of known structures not obviously related to proteins in the fold library. (A) Only correct assignments are obtained with Z-scores above 6. The histogram shows the distribution of Z-scores of the highest-ranking fold for each sequence. Of the 140 test sequences, 90 correspond to known folds and 50 correspond to “new” folds not in the fold library. The shaded bars correspond to the 32 correct assignments, of which 20 had scores above 6. The open bars correspond to the 58 incorrect assignments, all of which had scores below 6. The solid bars correspond to the 50 new folds in the test, all of which with scores below 6. (B) Fold assignment is superior to sequence comparison. The figure shows the results of fold assignment obtained by the server (top line) with those obtained by the sequence-comparison method “gon” (1) (bottom line). The x-axis corresponds to the 140 assignments, ordered by decreasing Z-score. The y-axis shows the percentage of correct assignments.
Sequences Assignable also by Sequence Comparison Methods.
A number of the server’s assignments correspond to MG sequences with clear sequence similarity to their assigned fold and thus can also be detected using sequence comparison methods alone [e.g., blast (14) or Smith–Waterman (22)]. We identify those 3D fold assignments as follows. Given an MG sequence a and the sequence f of the highest compatibility fold suggested by the server, we say f can be assigned to a by sequence–sequence methods if the optimal Smith–Waterman sequence alignment of a and f fulfills the following: (i) The sequence identity is at least 24%. (ii) The z-score obtained from a random distribution of scores is at least 8.0. This distribution is obtained by generating at least 130 random sequences with the same composition and length as a and optimally aligning them to f.
Membrane Proteins.
Our fold-assignment methods are not yet able to assign folds to trans-membrane domains because only a few 3D folds are currently known. To identify those MG sequences with putative trans-membrane helices we used the program moment (16) and identified 91 (19.4%) MG sequences. Assignments were attempted for membrane sequences only if the server assigned a globular fold with a score Z > 10 for a nonmembrane segment (see below).
RESULTS
All 468 MG sequences were processed by the server, and 103 3D fold assignments were detected above thresholds; 75 of the 103 assignments made by the server are also detectable by sequence–sequence methods alone (see Materials and Methods). These are available from http://www.doe-mbi.ucla.edu/people/frsvr/preds/MG/MG.html, under the name “Table III-MG.” The server assigned scores above 10 for 74 of the 75 assignments. These 75 fold assignments correspond to roughly 16% (75/468) of the MG genome, a figure higher than the 8–10% previously reported (2, 3). This difference is due in part to (i) similarities to newly determined structures and (ii) our criteria of assignability by sequence–sequence methods, which includes all assignments identified by blast (scores <10−5) plus nine additional ones. The remaining 28 fold assignments with Z > Zth do not fulfill our criteria of assignability by sequence–sequence methods (see Materials and Methods) and are listed in Table 1. As examples we discuss seven of the fold assignments listed in Table 1.
Table 1.
Fold assignment by clear sequence–structure compatibility but not by sequence comparison alone
| Z-score | Code | TIGR Characterization | ID% | PDB | Name |
|---|---|---|---|---|---|
| 33.0 | MG053 | Phosphomannomutase (cpsG) | 22 | 3pmga | Phosphoglucomutase |
| 29.2 | MG324 | Aminopeptidase P (pepP) | 17 | 1chma | Creatine amidinohydrolase |
| 20.8 | MG004 | DNA gyrase subunit A (gyrA) | 23 | 1bgw | Topoisomerase |
| 18.3 | MG204 | Topoisomerase IV subunit A (parC) | 23 | 1bgw | Topoisomerase |
| 17.7 | MG310 | Proline iminopeptidase (pip) | 18 | 1broa | Bromoperoxidase A2 |
| 16.5 | MG327 | Magnesium-chelatase 30 kDa subunit | 15 | 1broa | Bromoperoxidase A2 |
| 14.6 | MG365 | Methionyl-tRNA formyltransferase | 18 | 1cde | P. ribosylglycinamide formyltransferase |
| 14.4 | MG375 | Threonyl-tRNA synthetase (thrSv) | 17 | 1htta | Histidyl-trna synthetase |
| 14.0 | MG020 | Proline iminopeptidase (pip) | 22 | 1broa | Bromoperoxidase A2 |
| 13.7 | MG344 | Lipase–esterase (lip1) | 21 | 1broa | Bromoperoxidase A2 |
| 13.2 | MG274 | Pyruvate dehydrogenase E1-alpha | 24 | 1trka | Transketolase |
| 11.4 | MG006 | Thymidylate kinase (CDC8) | 20 | 1uky | Uridylate kinase |
| 10.8 | MG412 | 19 | 1pbp | Phosphate-binding protein | |
| 9.7 | MG353 | 21 | 1huea | HU protein | |
| 9.1 | MG470 | SpoOJ regulator | 14 | 1nipb | Nitrogenase iron protein |
| 8.9 | MG030 | Uracil phosphoribosyltransferase | 20 | 1hgxa | Hypoxanthine-guanine-xanthine phosphoribosyltransferase |
| 8.8 | MG335 | Hypothetical protein | 23 | 1gky | Guanylate kinase |
| 8.8 | MG367 | Ribonuclease III (rnc) | 16 | 1finb | Cyclin A |
| 8.7 | MG287 | Nodulation protein F (nodF) | 26 | 1acp | Acyl carrier protein |
| 8.6 | MG001 | DNA polymerase III beta subunit | 19 | 2pola | Pol III |
| 8.4 | MG394 | Serine hydroxymethyltransferase | 19 | 1tpla | Tyrosine phenol-lyase |
| 8.4 | MG194 | Phenylalanyl-tRNA synthetase beta | 23 | 1asya | Aspartyl trna synthetase |
| 8.4 | MG383 | Sporulation protein (outB) | 23 | 1gpma | Gmp synthetase |
| 8.2 | MG283 | Prolyl-tRNA synthetase (proS) | 13 | 1htta | Histidyl-trna synthetase |
| 8.2 | MG330 | Cytidylate kinase (cmk) | 18 | 3adk | Adenylate kinase |
| 7.9 | MG323 | Hypothetical protein | 22 | 1hyha | L-2-Hydroxyisocaproate dehydrogenase |
| 7.9 | MG149 | 31 | 1hcd | Hisactophilin | |
| 7.8 | MG088 | Ribosomal protein S7 (rpS7) | 20 | 1guha | Glutathione S-transferase |
The table lists the 28 fold assignments automatically detected by the fold-recognition server (scores above the threshold Zth = 7). These assignments do not fulfill our criteria of assignability by sequence–sequence methods (see Materials and Methods). Z-score, confidence score (Z) attached to the assignment; code, identification code of a sequence in the genome of Mycoplasma genitalium assigned by TIGR (10); TIGR characterization, name (if any) of a homologous sequence from another organism found in the sequence database, as annotated by TIGR; ID %, sequence identity percentage between the MG sequence and the assigned fold; PDB, PDB (13) code of the assigned fold; name, name of the protein as given in the PDB entry. Note: Our server identified one MG sequence >500 residues with Z > Zth, which is not shown in the table. Because large sequences may contain more than one domain and are prone to be high-scoring false positives, we applied the following additional filter for all large sequences (>500 residues) scoring above Zth. First, we cut the large sequence into shorter pieces of various lengths, and then we execute separate sequence–structure searches for each of the shorter pieces. If the Z of the fold assignment of each of the shorter pieces is below Zth, then we reject the assignment.
Two Sequences Assigned the Nucleotide Kinase Fold.
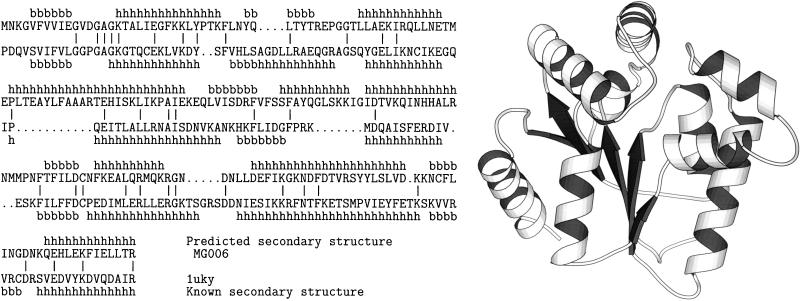
The fold-assignment procedure is illustrated by two MG sequences in Table 1 that have been assigned the nucleotide kinase fold. The first is MG006, functionally characterized by TIGR as thymidilate kinase, because a significant sequence similarity was found between MG006 and a thymidilate kinase sequence from Saccharomyces cerevisiae. However, MG006 shows no significant sequence similarity to any protein of known structure using blast (14) and Smith–Waterman (22). But the fold-assignment method of this paper assigns MG006 the fold of uridylate kinase (PDB code, 1uky) with a confidence score of Z = 11.4 (see Table 1 and Fig. 3). The sequence identity of the sequence–structure alignment between MG006 and 1uky is only 20%. The second MG sequence in Table 1 to which the nucleotide kinase fold was assigned is MG330, characterized by TIGR as cytidylate kinase. The PDB fold assigned by our server to MG330 is adenylate kinase (PDB code, 3 adk), with a score Z = 8.2. The sequence identity of the sequence–structure alignment is 18%. We conclude that the 3D structures of cytidylate kinase and thymidilate kinase from MG are similar to the nucleotide kinase fold observed in the PDB entries 1uky and 3 adk.
Figure 3.
Sequence–structure alignment of Mycoplasma genitalium sequence MG006 with the fold of uridylate kinase. The top line in the alignment shows the predicted secondary structure (11) of the sequence. The second line is the amino acid sequence of MG006. The third line is the amino acid sequence of the fold (uridylate kinase). The fourth line is the observed secondary structure of the fold. The bars represent identical amino acid matches. Sequence identity in the alignment is 20%.
α/β-Hydrolase Folds.
Other examples of the fold assignments in Table 1 are given by four MG sequences that have been assigned the fold of an haloperoxidase (PDB code, 1bro), a member of the α/β-hydrolase family (23). These are: MG310 and MG020, characterized by TIGR as proline iminopeptidases; MG344, characterized as a lipase–esterase; and MG327, characterized as a magnesium–chelatase subunit. The sequence identity of the sequence–structure alignments ranges from 15 to 22%. The server’s fold assignments are well above our confidence threshold Zth, with Z-scores ranging from 13.7 for MG344 to 17.7 for MG310. Inspection of the sequence–structure alignments reveals that the catalytic triad residues of 1bro are matched to identical residues in the MG sequences. We conclude that the 3D structures of these four MG proteins are members of the α/β hydrolase fold.
Fold Assignment to Uncharacterized MG Sequences.
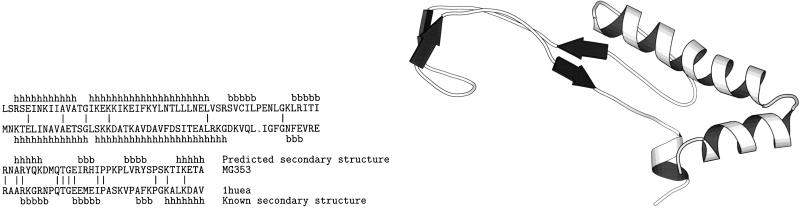
Another type of outcome from our server is the fold assignment of MG sequences that are not characterized functionally because no similar sequence exists in the sequence databases. Accordingly, there is no information regarding functional similarity that can support or deny our assignments. One such sequence is MG353. The server found a high (Z = 9.7) sequence–structure compatibility with a DNA-binding protein having a histone-like fold (PDB code, 1hue). The sequence identity of the sequence–structure alignment is only 21%. Fig. 4 shows that the alignment has only one gap, and the compatibility of the predicted secondary structure with the observed one is also high, supporting the plausibility of this assignment.
Figure 4.
Sequence–structure alignment of Mycoplasma genitalium sequence MG353 with the histone-like fold of 1huea. See legend of Fig. 3 for explanation. Notice that the predicted (first line) and observed (fourth line) secondary structures match relatively well, except for a (possibly overpredicted) β-strand and a match of a predicted α-helix to a β-strand. This illustrates the resiliency of the method to inexact secondary structure predictions. Sequence identity in the alignment is 21%.
In total, our fold-assignment method (1), as executed by the server, succeeded to assign folds to 103 MG sequences (Table 1 and table III-MG), which correspond to 22% of the sequences of the MG genome and to 34% of the MG sequences functionally characterized by TIGR (10).
DISCUSSION
Requirements for the Automatic Assignment of Folds to Genome Sequences.
Our procedure for automatic assignment of folds to genome sequences requires that a compatibility score (Z) be attached to each fold assignment and a confidence threshold (Zth) be set, above which we accept a fold assignment as correct. Here we set a high threshold, so that based on experience with benchmarks of known structures (1, 18, 21) and on blind prediction experiments (8), an assignment with Z > Zth has always been a correct assignment. Thus, for the sake of increased selectivity (fewer false positives), we may have lost some sensitivity (more false negatives).
Automatic vs. Human Fold Assignments.
One of the goals of this work was to explore automatic fold assignment. Using our high threshold (Zth), the method fails to report those correct assignments with Z < Zth (false negatives). Even so, automatic fold assignment can be helpful for assignments with below-threshold scores if used as a first screen. An expert can analyze the below-threshold assignments, possibly applying other methods and biological knowledge. For example, we found 16 MG sequences with Z < Zth for which the functional characterization given by TIGR is similar to the function of the highest scoring fold detected by the server (Table 2). Although these are subthreshold assignments, they are likely to be correct. Devising algorithms to assess these subthreshold assignments is needed, because this manual procedure cannot routinely be applied for the large number of genome sequences.
Table 2.
Possible fold assignments detected by fold recognition (rank 1) and with the aid of human inspection
| Z-score | Code | TIGR Characterization | ID% | PDB | Name |
|---|---|---|---|---|---|
| 6.6 | MG382 | Uridine kinase (udk) | 22 | 1uky | Uridylate kinase |
| 5.9 | MG387 | GTP-binding protein era homolog | 15 | 5p21 | C-H-ras P21 |
| 5.7 | MG015 | Transport ATP-binding protein | 20 | 3adk | Adenylate kinase |
| 5.5 | MG090 | Ribosomal protein S6 (rpS6) | 22 | 1ris | Ribosomal protein |
| 4.9 | MG058 | Phosphoribosylpyrophosphate synthetase | 19 | 1sto | Orotate phosphoribosyltransferase |
| 4.8 | MG039 | Glycerol-3-phosphate dehydrogenase | 20 | 3lada | Dihydrolipoamide dehydrogenase |
| 4.7 | MG273 | Pyruvate dehydrogenase E1-beta | 13 | 1trka | Transketolase |
| 4.3 | MG253 | Cysteinyl-tRNA synthetase (cysS) | 16 | 1gln | Glutamyl-trna synthetase |
| 4.3 | MG024 | GTP-binding protein (gtp1) | 21 | 1gky | Guanylate kinase |
| 4.1 | MG014 | Transport ATP-binding protein | 20 | 1gky | Guanylate kinase |
| 4.1 | MG467 | Heterocyst maturation protein | 21 | 1uky | Uridylate kinase |
| 3.9 | MG079 | Oligopeptide transport ATP-binding protein | 15 | 3adk | Adenylate kinase |
| 3.9 | MG126 | Tryptophanyl-tRNA synthetase | 21 | 2ts1 | Tyrosyl-trna synthetase |
| 3.9 | MG023 | Fructose-bisphosphate aldolase | 19 | 1gox | Glycolate oxidase |
| 3.7 | MG266 | Leucyl-tRNA synthetase (leuS) | 15 | 1gtsa | Glutaminyl-trna synthetase |
| 3.6 | MG251 | Glycyl-tRNA synthetase | 20 | 1htta | Histidyl-trna synthetase |
The table lists 16 possible fold assignments with below-threshold scores detected with the aid of human inspection by comparing the functional characterization given by TIGR (10) and the function of the highest-scoring fold detected by the server. Legend is as in Table 1. In addition to the assignments listed above, we identified the following possibly correct assignments at rank 2. MG276, adenine phosphoribosyltransferase (possible fold: 1 hmpa, hypoxanthine guanine phosphoribosyltransferase); and MG340, DNA-directed RNA polymerase beta (possible fold: 1kln, DNA Polymerase I). Fourteen tRNA synthetase sequences in MG were assigned a fold (Tables 1 and 2; table III-MG). Three additional ones, MG021 (methionyl), MG334 (valyl), and MG378 (argynyl), had the fold of 1gln, glutamyl-trna synthetase, in second rank.
Fraction of MG Proteins Having Previously Observed Folds.
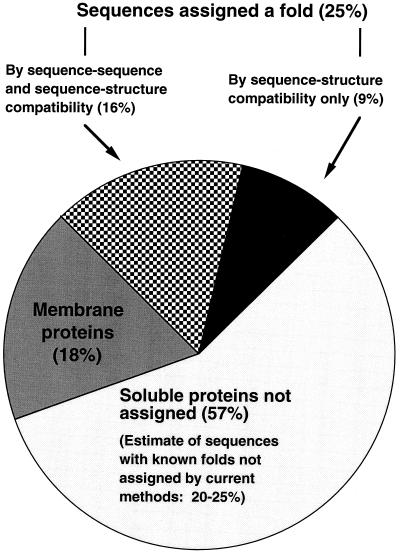
Fig. 5 shows that (excluding putative membrane proteins) our fold-assignment methods did not assign a fold for about 57% of the MG sequences. Some of these sequences certainly correspond to folds not yet observed by structural biologists. However, because the sensitivity of our methods is still poor, some of the sequences left unassigned correspond to proteins with an already known fold (false negatives). Based on computational benchmarks (1, 18, 21) and our experience in blind predictions (8), we estimate that our methods failed to identify an existing compatible fold for 20–25% of the MG sequences. Thus, considering the 25% of the MG sequences for which we did assign a fold, we estimate that 45–50% of the MG sequences may correspond to proteins with already-observed folds.
Figure 5.
Assignment of Mycoplasma genitalium amino acid sequences to 3D protein folds. Twenty-five percent of the MG sequences were assigned a fold. Sixteen percent of these assignments are also detectable by sequence–sequence methods, and 9% are detectable only by fold-recognition methods (6% automatically and 3% with the aid of human intervention). Of the 91 MG sequences predicted to contain trans-membrane helices, 5 were assigned a fold (table III-MG) and 86 (18%) were left unassigned. The remaining 57% of the MG genome was left unassigned. However, 20–25% of the genome is estimated to correspond to proteins having already-observed folds, even though our methods could not assign folds to them (see text). Thus, we estimate that 45–50% of the genome corresponds to proteins with already-observed folds.
Sequences not assigned folds may be targets of opportunity for structural studies. Although, as noted in the preceding paragraph, a fraction of the MG sequences for which our method fails to assign a fold will belong to a fold that is known, the others will, in fact, represent new folds. These new folds are attractive targets for structural studies, because, among other things, new folds accelerate the process of assigning folds to other genome sequences. Table IV-MG, available on the web (URL given above), lists 38 proteins that received low Z-scores during the assignment process and that also have many homologous sequences in the sequence database. Thus, knowing their structures would substantially broaden knowledge of structural biology.
How Many More 3D Structures Are Needed?
How many more 3D structures do we need in our library of known structures to assign folds to most of the nonmembrane MG sequences (i.e., 82% of the genome; see Fig. 5)? In this paper we find that 25% of the MG sequences can be assigned a fold. If we assume that an increase in the size of our library of representative folds will yield a linear increase in the number of genome sequences assignable, then a 3.3-fold increase (82/25) in the number of known structures in our library will result in the fold assignment of most soluble proteins. The current rate of structure determination results in an annual increase in the size of our library of 30% (989 in 1996 and 1,291 in 1997). If this rate of determination of new structures continues throughout the next 5 years, our library will grow by a factor of 3.7. The Human Genome Project is expected to be completed before the year 2004, so, provided that the above assumptions hold, the variety of known structures at that time could be adequate for assigning folds to the majority of human soluble proteins. This goal will not be met if the rate of determination of new structures drops, but even so, fold assignment methods are likely to contribute significantly to our knowledge of protein structure.
Validity of the Fold Assignments.
Our expectation of valid fold assignments is based on three lines of evidence. (i) For assignments in Tables 1 and 2, the confidence score (Z) exceeds the threshold value (Zth) found in tests to give valid assignments. (ii) For most of the functionally characterized MG sequences, the folds assigned have functions similar to the functions of the homologous sequences in other organisms. (iii) In some of the cases analyzed, the quality of the alignment or the matching of active site residues is compatible with the assignment. However, only the eventual determination of their 3D structures can confirm or reject these assignments.
Acknowledgments
We thank Burkhard Rost and Chris Sander for use of their PHD program; Danny W. Rice, Scott Le Grand, Todd Yeates, and James Bowie for discussions; and the Department of Energy and the UC Star program for support.
ABBREVIATIONS
- 3D
three-dimensional
- MG
Mycoplasma genitalium
- frsvr
fold-recognition server
- TIGR
The Institute for Genome Research
Note Added in Proof
New protein structures added to the PDB since April 1997 have permitted five additional fold assignments, as given in http://www.doe-mbi.ucla.edu/people/frsvr/preds/MG/MG.html.
References
- 1.Fischer D, Eisenberg D. Protein Sci. 1996;5:947–955. doi: 10.1002/pro.5560050516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moult J. Curr Opin Biotechnol. 1996;7:422–427. doi: 10.1016/s0958-1669(96)80118-2. [DOI] [PubMed] [Google Scholar]
- 3.Casari G, Ouzounis C, Valencia A, Sander C. In: Pacific Symposium on Biocomputing. Hunter L, Klein T L, editors. Singapore: World Scientific; 1996. pp. 707–709. [Google Scholar]
- 4.Doolittle R F. Of Urfs and Orfs: A Primer on How to Analyze Derived Amino Acid Sequences. Mill Valley, CA: University Science Books; 1986. [Google Scholar]
- 5.Orengo C A. Curr Opin Struct Biol. 1994;4:429–440. [Google Scholar]
- 6.Bowie J U, Luthy R, Eisenberg D. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 7.Fischer D, Rice D W, Bowie J U, Eisenberg D. FASEB J. 1996;10:126–136. doi: 10.1096/fasebj.10.1.8566533. [DOI] [PubMed] [Google Scholar]
- 8.Rice, D. W., Fischer, D., Weiss, R. & Eisenberg, D. (1997) Proteins, in press. [DOI] [PubMed]
- 9.Braxenthaler M, Sippl M J. In: Protein Folds. Bohr H, Brunak S, editors. Boca Raton, FL: CRC; 1995. pp. 80–84. [Google Scholar]
- 10.Fraser C M, Gocayne J D, White O, Adams M D, Clayton A, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 11.Rost B, Sander C. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 12.Gonnet G H, Cohen M A, Benner S A. Science. 1992;256:1433–1445. doi: 10.1126/science.1604319. [DOI] [PubMed] [Google Scholar]
- 13.Bernstein F C, Koetzle T F, Williams G J B, Meyer E F, Brice M D, Rodgers J R, Kennard O, Shimanouchi T, Tasumi M. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Pearson W R, Lipman D J. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenberg D, Schwarz E, Komaromy M, Wall R. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 17.Bairoch A. Nucleic Acids Res. 1992;20:2013–2018. doi: 10.1093/nar/20.suppl.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rice D W, Eisenberg D. J Mol Biol. 1997;267:1026–1038. doi: 10.1006/jmbi.1997.0924. [DOI] [PubMed] [Google Scholar]
- 19.Rost B. In: Proceedings of the Conference on Intelligent Systems in Molecular Biology, ISMB-95. Rawlings C, Clark D, Altman R, Hunter L, Lengauer T, Wodak S, editors. Menlo Park, CA: AAI Press; 1995. pp. 314–321. [Google Scholar]
- 20.Murzin A G, Brenner S E, Hubbard T, Chothia C. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 21.Fischer D, Elofsson A, Rice D W, Eisenberg D. In: Pacific Symposium on Biocomputing. Hunter L, Klein T L, editors. Singapore: World Scientific; 1996. pp. 300–318. [PubMed] [Google Scholar]
- 22.Smith T F, Waterman M S. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 23.Ollis D L, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken S M, Harel M, Remington S J, Silman I, Schrag J, Sussman J L, Verschueren K H G, Goldman A. Protein Eng. 1992;3:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]