Summary
Glutaminyl-tRNA synthetase and asparaginyl-tRNA synthetase evolved from glutamyl-tRNA synthetase and aspartyl-tRNA synthetase, respectively, after the split in the last universal communal ancestor (LUCA). Glutaminyl-tRNAGln and asparaginyl-tRNAAsn were likely formed in LUCA by amidation of the mischarged species, glutamyl-tRNAGln and aspartyl-tRNAAsn, by tRNA-dependent amidotransferases as is still the case in most bacteria and all known archaea. The amidotransferase GatCAB is found in both domains of life while the heterodimeric amidotransferase, GatDE, is found only in Archaea. The GatB and GatE subunits belong to a unique protein family with Pet112 that is encoded in the nuclear genomes of numerous eukaryotes. GatE was thought to have evolved from GatB after the emergence of the modern lines of decent. Our phylogenetic analysis though places the split between GatE and GatB prior to the phylogenetic divide between Bacteria and Archaea and Pet112 to be of mitochondrial origin. In addition, GatD appears to have emerged prior to the bacterial-archaeal phylogenetic divide. Thus, while GatDE is an archaeal signature protein it likely was present in LUCA together with GatCAB. Archaea retained both amidotransferases while Bacteria emerged with only GatCAB. The presence of GatDE has favored a unique archaeal tRNAGln that may be preventing acquisition of glutaminyl-tRNA synthetase in Archaea. Archaeal GatCAB on the other hand has not favored a distinct tRNAAsn suggesting tRNAAsn recognition is not a major barrier to the retention of asparaginyl-tRNA synthetase in more Archaea.
Keywords: tRNA-dependent amidotransferase, GatCAB, GatDE, Pet112, LUCA
Introduction
Aminoacyl-tRNA synthetases (aaRSs) are the enzymes primarily responsible for accurately pairing an amino acid with its cognate tRNA, a requirement for high fidelity protein synthesis.1 In most bacteria2 and all known archaea,3 glutaminyl-tRNA synthetase (GlnRS) is absent and Gln -tRNAGln is formed via an indirect route. These organisms take advantage of an aaRS with relaxed tRNA specificity non-discriminating glutamyl-tRNA synthetase (ND-GluRS) to form Glu-tRNAGln.4 The Glu moiety on the tRNAGln is then converted into Gln via an amidation reaction catalyzed by a glutamyl-tRNAGln amidotransferase (Glu-AdT) in these species.5 Similarly, in the many bacteria and archaea that lack AsnRS2, 3, 6 a non-discriminating aspartyl-tRNA synthetase (ND-AspRS)7 and an aspartyl-tRNAAsn amidotransferase (Asp-AdT) are used to synthesize Asn-tRNAAsn.8, 9
Two different tRNA-dependent amidotransferases (AdTs) are found in nature, GatCAB10 and GatDE.3 The latter is an archaeal signature protein11 and acts only as a Glu-AdT.3 GatCAB is present in most bacteria and archaea.2, 3, 6 In vitro, the bacterial GatCAB can transamidate both Glu-tRNAGln and Asp-tRNAAsn.2, 12-15 Its role in vivo is determined by which mischarged tRNA species (Glu-tRNAGln and/or Asp-tRNAAsn) is present.2, 12-14, 16, 17 Archaeal GatCAB is unable to use archaeal Glu-tRNAGln as a substrate (see accompanying article), is only present in Archaea lacking an AsnRS,3, 6 and is thought to act as an Asp-AdT in vivo.
Both AdTs catalyze three distinct reactions in order to transamidate their mischarged tRNA substrates, Glu-tRNAGln and/or Asp-tRNAAsn.18 1) They phosphorylate the γ- or β- carboxyl group of the Glu or Asp moiety attached to the tRNA in an ATP-dependent manner to form an activated intermediate.19 This kinase activity is the province of the B and E-subunits of the respective holoenzymes.20-22 2) The AdTs have a glutaminase subunit (GatA and GatD, respectively) to liberate ammonia from an amide donor such as Gln or Asn.2, 20, 23, 24 GatD belongs to the type I L-asparaginase (AnsA) family3, 20, 23 while GatA is an amidase.2, 10, 21, 24 GatC, a small protein of approximately 100 amino acids, is required by GatA for proper folding10 and binding to GatB.21 3) The AdTs amidate the activated intermediate using the ammonia liberated by the glutaminase subunit.
In the last universal communal ancestor (LUCA), the diverse community of ancient organisms that the modern three domains of life are hypothesized to have evolved from,25 it is thought that the indirect paths for Gln-tRNAGln and Asn-tRNAAsn synthesis were extant since the specific aaRSs for these amino acids (GlnRS and AsnRS) evolved at a later stage.6, 26-29 Given GatCAB is present in both Archaea and Bacteria while GatDE is archaeal specific, it was speculated the AdT in LUCA was GatCAB with GatDE evolving later in early archaea.30 To address this hypothesis and to gain a better understanding of the evolution of the two AdTs, we examined the phylogenetic relationship between the kinase subunits of the two AdTs (GatB and GatE). We constructed phylogenetic trees for four additional components of tRNA-dependent Gln and Asn synthesis: GatA, AnsA and the GatD family, tRNAGln, and tRNAAsn. This comparative phylogentic approach reveals the co-evolution of the subunits of GatDE and GatCAB as well as with their tRNA substrates and resolves the evolutionary connection between the two distantly related AdTs. Our results suggest that although GatDE is an archaeal signature protein it evolved before the archaeal-bacterial phylogenetic divide and was present in LUCA.
Results
Ancient divergence between GatB and GatE
GatB and GatE belong to a unique enzyme family that includes Pet112 (Fig. 1).3, 21-23, 31 The latter enzymes are encoded in the nuclear genomes of many eukaryotes.31 In Saccharomyces cerevisiae, Pet112 is essential for mitochondrial translation.31
Figure 1. Domain architecture of GatB, Pet112, GatE and YqeY.
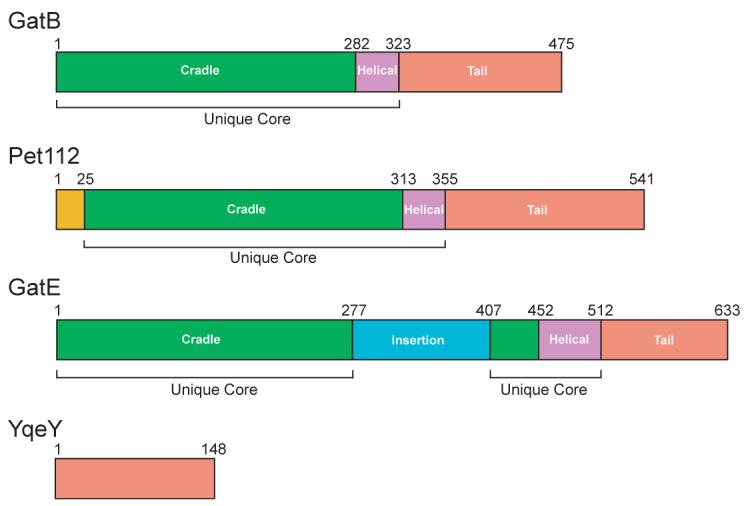
The structural domains of the GatB, Pet112, and GatE are colored with the cradle (green), helical (violet), and tail (salmon) domains denoted. Given the homology between YqeY and the tail domains, the former is also colored salmon. In addition, the N-terminal extension of Pet112 is colored gold. The insertion domain found only in GatE is in teal. The cradle and helical domains make up the unique core of the GatB/GatE/Pet112 family and are denoted accordingly. The numbering of GatB, Pet112, GatE and YqeY are of the Staphylococcus aureus, the S. cerevisiae, the Pyrococcus abyssi, and the B. subtilis enzymes, respectively.
Because GatDE is confined to the Archaea, it was assumed GatE arose from a duplication of either an archaeal GatB or a bacterial GatB that was horizontally transferred to the Archaea.30 Pet112 on the other hand is predicted to be of mitochondrial origin.31, 32 To ascertain the evolutionary relationship between GatB, GatE, and Pet112 we calculated their phylogenetic relationship using a protein sequence alignment of the unique core (cradle and helical domains, Fig. 1) of the enzymes. Large insertions, which could bias the results, were manually removed prior to analysis including the GatE specific insertion domain, which is theorized to enable GatDE to bind GluRS but not AspRS (see Materials and Methods).22 The GatB sequences from the Mollicutes were excluded from the final analysis due to the faster evolutionary rates of these pathogenic organisms to minimize artifacts from long branch attractions.33
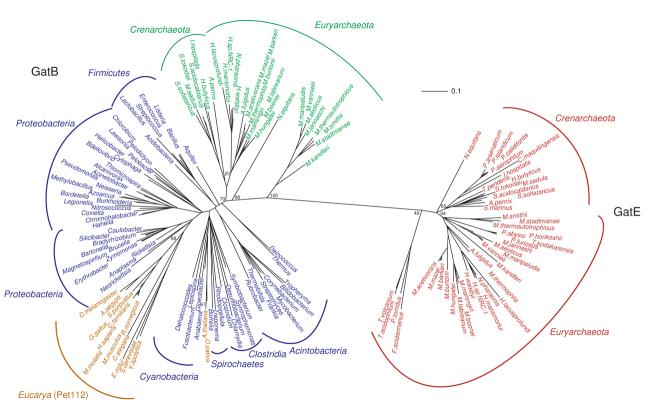
Since no other proteins share homology with the unique core of GatE, Pet112 and GatB, an unrooted phylogeny is presented (Fig. 2). Reciprocal rooting of the tree34 with either the Sulfolobus solfataricus GatB or the Methanosarcina mazei GatE produces identical trees demonstrating our result was not an artifact of which sequence served as the starting point for the phylogenetic analysis. If GatE evolved from GatB after the phylogenetic split of LUCA, one would expect the phylogenetic pattern to show these enzymes to be specifically related to a particular GatB branch much like GlnRS enzymes group with the eukaryotic GluRS proteins.26, 35 Instead, GatEs are highly distinct and separated from the GatBs by a long branch (Fig. 2) suggesting a scenario in which the GatB/GatE split occurred prior to the to phylogenetic split between Bacteria and Archaea. The same has been predicted for the origins of SepRS from PheRS.29 Among the GatBs, the split between archaeal and bacterial enzymes is seen (Fig. 2). In the bacterial GatBs, most of the major taxa grouped in accordance with standard taxonomy. The association of Clostridia GatBs with the Spirochaetes as opposed to other Firmicutes suggests horizontal gene transfer (HGT) amongst these bacterial phyla.
Figure 2. Unrooted phylogenetic tree of the core of archaeal GatE (red) sequences with GatB sequences from bacterial (blue), archaeal (green) and eukaryotic (gold) sources.
Representative bacterial and eukaryotic enzymes are shown. Scale bar represents 0.1 changes/site. Only bootstrap values for the branch points discussed in the text are shown for clarity. A tree with the additional bootstrap values can be found in the supplementary material. The Methanosarcina mazei GatE was used as the outgroup for this reconstruction. See Materials and Methods for details.
GlnRS is absent in chloroplasts and therein Gln-tRNAGln is formed via the indirect route.36, 37 Not surprisingly, the GatBs from Arapadopsis thalina and Oryza sativa clustered with the Cynanobacteria in agreement with the cynanobacterial origins of the organelle.38 The Pet112 family forms a group with the α-Proteobacteria, with a specific relationship with the Rickettsia, this result indicates Pet112 is not a component of the nuclear line of decent but is of mitochondrial origin.39, 40
The function of Pet112, the mitochondrial GatB-like protein, in eukaryotes such as S. cerevisae is unresolved. Our alignment of Pet112 with GatB and GatE shows conservation of the kinase active site (Supplemental Fig. 1), which has been characterized in the Methanothermobacter thermautotrophicus GatE.22 This suggests Pet112 has been retained as a kinase.
Phylogentic arrangement of archaeal GatB was as expected with two noticeable exceptions (Fig. 2). Within the main archaeal GatB branch there is a clear divide between the Euryarchaea and Crenarchaea. The Nanoarchaeum equitans GatB grouped outside of the main archaeal branch. N. equitans exist in a symbiotic relationship with Ignicoccus hospitalis.41 Such a result therefore maybe an artifact of long branch attractions caused by the accelerated rate of evolution in N. equitans. The GatBs from Methanobacteria, Methanocci, and Methanopyrus also grouped away from the other archaeal GatBs suggesting a possible faster evolutionary rate in the unique core of these enzymes for reasons unknown (discussed further below).
The euryarchaeal-crenarchaeal divide is present in archaeal GatEs with one exception. The Thermoplasmata occupy a basal position in the GatE tree. Interestingly, the organisms have a discriminating AspRS and do not form Asp-tRNAAsn.6 As a consequence, GatDE in these organisms do not have to distinguish Glu-tRNAGln from Asp-tRNAAsn. The removal of this constraint may have allowed these sequences to drift further from the other euryarchaeal GatE sequences.
The phylogentic analysis based on the GatB/GatE core domain suggested but does not conclusively show that GatE diverged from GatB before the emergence of the main lines of decent. The unique core of the GatB/E family lacks a root, as no homologs of the core of the enzymes are known.3, 21-23 YqeY is the closest known relative of GatB and GatE. It shows homology to the tail domain of both subunits.21-23 YqeY is found in a diverse array of bacteria across multiple phyla but their function remains unknown.42 A YqeY domain appended to the C-terminus of the Deinococcus radiodurans GlnRS is important for the enzyme to productively bind to tRNAGln.42 The tail domains in the AdT subunits play a similar role by enabling specific binding to their tRNA substrates.21, 22
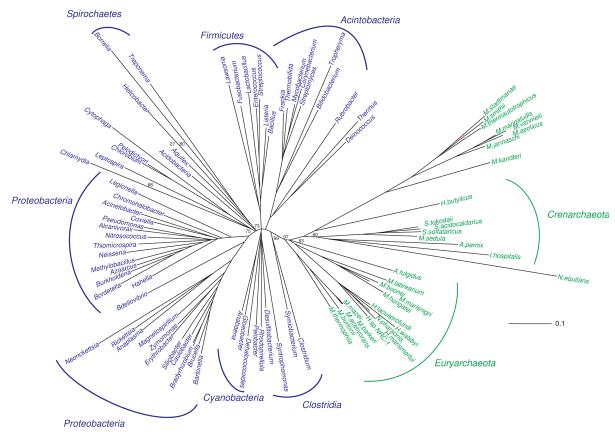
Because YqeY is homologous to but more distantly related to GatB and GatE than they are to each other, it serves as an ideal outgroup for the tail domain phylogeny. Since, the GatE/GatB tail domain phylogeny is in general congruent with the phylogeny of the core of the enzymes, the tail domain is an adequate phylogenetic marker for the entire molecule. Thus, the tail domain rooting should properly position the root for the GatB/GatE divergence (Fig. 3). The sequences from the tail domains of GatBs from the Mollicutes, mitochondrial and chloroplast GatBs, N. equitans GatB and GatE, and the GatE proteins from the Thermoplasmata and Thermoproteales were excluded from the analysis due to apparent accelerated rates of change in this short segment of the enzymes. As discussed above, the faster clocks for the N. equitans and Mollicutes was not unexpected nor is it unusual to see accelerated rates in organellar proteins.
Figure 3. Rooted phylogenetic tree of the tail domain of archaeal GatE (red) sequences with GatB sequences from bacterial (blue) and archaeal (green) sources.
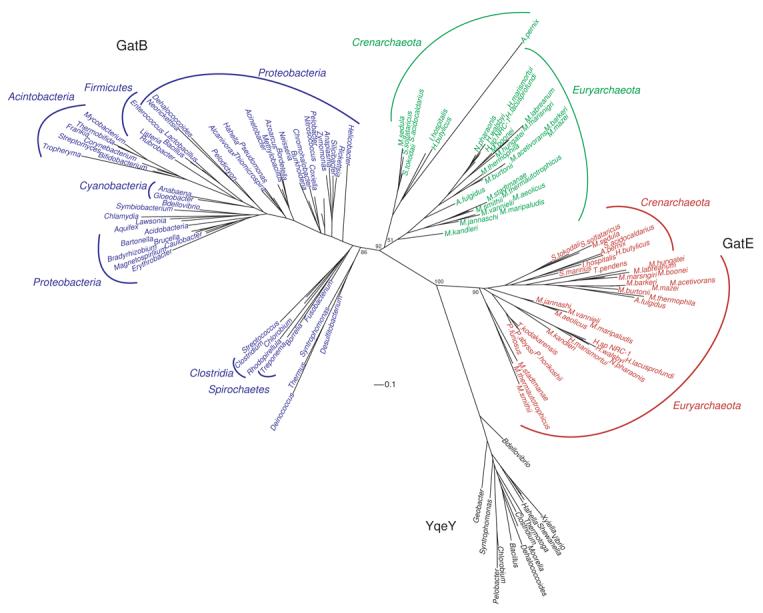
Representative bacterial enzymes are shown. Scale bar represents 0.1 changes/site. Only bootstrap values for the branch points discussed in the text are shown for clarity. A tree with the additional bootstrap values can be found in the supplementary material. YqeY sequences (black) served as the outgroup. See Materials and Methods for details.
The Thermoplasmata and Thermoproteales have a discriminating AspRS and use AsnRS and not GatCAB to form Asn-tRNAAsn. The tail domain of GatE has been proposed as being important for that discrimination by recognizing bases in the D-loop of tRNAGln.22 The tail domains of Thermoplasmata and Thermoproteales GatE enzymes lack a GXXAXGX motif found in other archaeal GatE sequences suggesting the motif may play a role in distinguishing Glu-tRNAGln from Asp-tRNAAsn by the enzyme (Supplemental Fig. 2). Therefore, the GatDE enzymes in these organisms do not and likely cannot distinguish Glu-tRNAGln from Asp-tRNAAsn.
The YqeY sequences placed the root between GatB and GatE (Fig. 3). This suggests that GatE diverged from GatB in LUCA given the GatB tail domains further split into bacterial and archaeal lineages. The bacterial branches are not well resolved in the tail domain, and the same is true of the GatE sequences. On the other hand, standard archaeal phylogeny is observed in the history of the archaeal GatB tail domain. This may be an indication of the relative importance the tail domain plays in substrate specificity between the archaeal GatCAB and the bacterial GatCAB and GatDE. Bacterial GatB and GatE use the cradle domain to recognize the first base-pair in the acceptor stem. This allows the enzymes to distinguish their mischarged tRNA substrates from other aa-tRNA species.17, 21, 22 Thus, the bacterial GatB and GatE use both the tail and cradle domains for tRNA recognition. In the case of the archaeal GatCAB, the enzyme does not use the first-base pair of the acceptor stem to recognize Asp-tRNAAsn,17, 43 so the archaeal GatB may rely more on its tail domain for aa-tRNA specificity. The fact the bacterial GatB tail domain accommodates tRNAGln as well as tRNAAsn may have allowed for greater drift in this region of bacterial enzymes than is found in their archaeal counterparts, partially obscuring the phylogenetic signal of this short segment of the enzyme.
Phylogeny of GatD follows that of GatE
In Archaea, GatE associates with the AnsA-like GatD. To determine the evolutionary origin of GatD we examined the phylogenetic relationship between GatD and related AnsA sequences from bacteria and archaea. Type II L-aspraginases (AnsB) sequences served to root the tree (Fig. 4). From the root, two branches diverge, the AnsA subfamily and the GatD subfamily. The results indicate that GatD has co-evolved with GatE, and GatDE holoenzyme originated before the phylogentic split of the archaeal and bacterial domains.
Figure 4. Rooted phylogenetic tree of the bacterial (cyan) and archaeal (green) type I L-asparaginases (AnsA), and archaeal GatD (red) sequences.
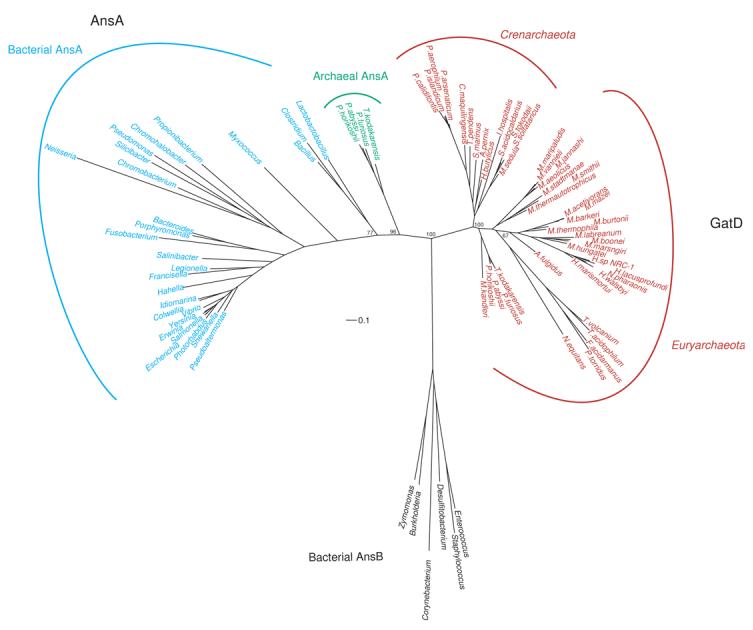
Representative bacterial enzymes are shown. Scale bar represents 0.1 changes/site. Only bootstrap values for the branch points discussed in the text are shown for clarity. A tree with the additional bootstrap values can be found in the supplementary material. Representative bacterial type II L-asparaginases (AnsB) were used as the outgroup (black) in this reconstruction. See Materials and Methods for details.
For the most part, the phylogeny of the D-subunit tracks with the E-subunit. The main incongruence between the phylogenies is found in the Thermoplasmata. Unlike the GatE phylogeny, the D-subunit from Thermoplasmata grouped as expected with the other Euryarchaea. We theorize that their GatE no longer needs to distinguish Glu-tRNAGln from Asp-tRNAAsn as these organisms have a discriminating AspRS (discussed above) and thus their GatE sequences are freer to drift than the other euryarchaeal GatE sequences. No such constraints have been lifted for the GatD from the Thermoplasmata as the subunit is the glutaminase domain of the AdT and does not interact with the tRNA.22
Phylogenies of GatA and GatB are congruent
We next examined the evolutionary relationship between the A-subunits of GatCAB. While GatA is homologous to other amidases in its catalytic core,10, 21, 44 no amidase is similar enough to GatA across the entire enzyme to root the GatA tree (Fig. 5). The phylogenetic distribution of GatAs is in general agreement with that of the core of the GatB enzymes (Fig. 2), supporting the expected co-evolution of the subunits of this AdT. The sequences divided into bacterial and archaeal lineages. There were some differences in the branch order of the bacterial GatAs versus the GatBs. The most noticeable were the GatAs from pathogenetic bacteria such as Helicobacter pylori and Chlamydia trachomatis, which grouped together. The long branches of these sequences suggest this is a treeing artifact33 and that the A-subunits in these organisms have evolved at a different rate than their GatB counterparts. Though less likely, we cannot rule out the possibility of HGT of the A-subunit into these organisms.
Figure 5. Unrooted phylogenetic tree of the bacterial GatA (blue) and archaeal GatA (green) sequences.
Representative bacterial enzymes are shown. Scale bar represents 0.1 changes/site. Only bootstrap values for the branch points discussed in the text are shown for clarity. A tree with the additional bootstrap values can be found in the supplementary material. The Methanosaeta thermophila GatA was used as the outgroup for this reconstruction. See Materials and Methods for details.
For the most part the euryarchaeal-crenearchaeal divide is upheld in the GatA tree. A deviation from canonical archaeal phylogeny in the Methanobacteria, Methanocci, and Methanopyrus is likely the result of long branch attraction.33 A similar artifact was present in the core of GatB from these organisms (Fig. 2), but the tail domains displayed typical archaeal phylogeny (Fig. 3). Based on alignment to the Staphylococcus aureus GatCAB structure21 conserved residues in the GatA and GatB from this group differ from those in other archaeal sequences in the interface between GatC, GatB and GatA.
GatAB divides into bacterial and archaeal types
In the phylogenies of GatA (Fig. 5) and the core and tail domains of GatB (Fig. 2 and 3), we do not see a deep divide between the bacterial and archaeal versions. For the GatB phylogenies this may be due to the fact we examined just segments of the enzyme. With GatA, approximately 150 amino acids of the alignment is comprised of the Gly/Ser rich amidase signature sequence which is highly conserved within the greater amidase family.45 In order to resolve the divergence of archaeal and bacterial GatCAB, we examined a phylogeny based on concatenation of GatA and GatB sequence (Fig. 6). The tree shows a deep and well-supported divide between the archaeal and bacterial GatAB.
Figure 6. Unrooted phylogenetic tree of concatenated bacterial GatAB (blue) and archaeal GatAB (green) sequences.
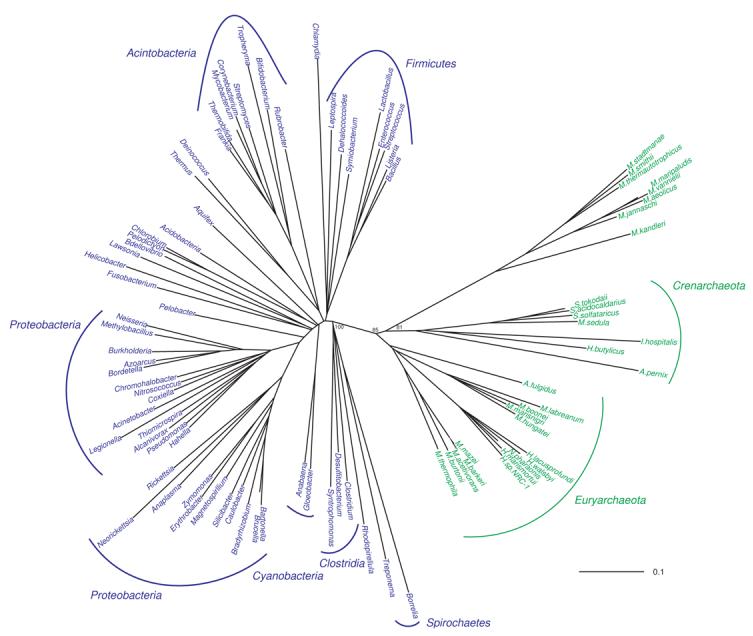
Representative bacterial enzymes are shown. Scale bar represents 0.1 changes/site. Only bootstrap values for the branch points discussed in the text are shown for clarity. A tree with the additional bootstrap values can be found in the supplementary material. The M. thermophila GatAB was used as the outgroup for this reconstruction. See Materials and Methods for details.
Divergence in tRNAGln isoacceptors and convergence in tRNAAsn isoacceptors
In Archaea, GatDE and GatCAB have co-evolved with tRNAGln and tRNAAsn to enable GatDE to serve as a Glu-AdT and GatCAB as an Asp-AdT (see accompanying article). Biochemical and structural results indicate GatDE specifically recognizes the archaeal Glu-tRNAGln.22 The archaeal GatCAB evolution is characterized by rejection of archaeal Glu-tRNAGln and Asp-tRNAAsp rather than adaptation to specifically recognize the archaeal Asp-tRNAAsn.17, 43 The net effect is the bacterial mischarged tRNA serves as a poor substrate for GatDE (see accompanying article) while the archaeal GatCAB can readily use the bacterial Asp-tRNAAsn as a substrate (see accompanying article).17, 43 Given the differences in the recognition of aa-tRNA substrates between the two AdTs in Archaea, we wanted to determine if this had an effect on the phylogenies of their tRNAGln (Fig. 7) and tRNAAsn isoacceptors (Fig. 8).
Figure 7. Unrooted phylogenetic tree of tRNAGln gene sequences from bacterial (blue), archaeal (green) and eukaryotic (gold) genomes.
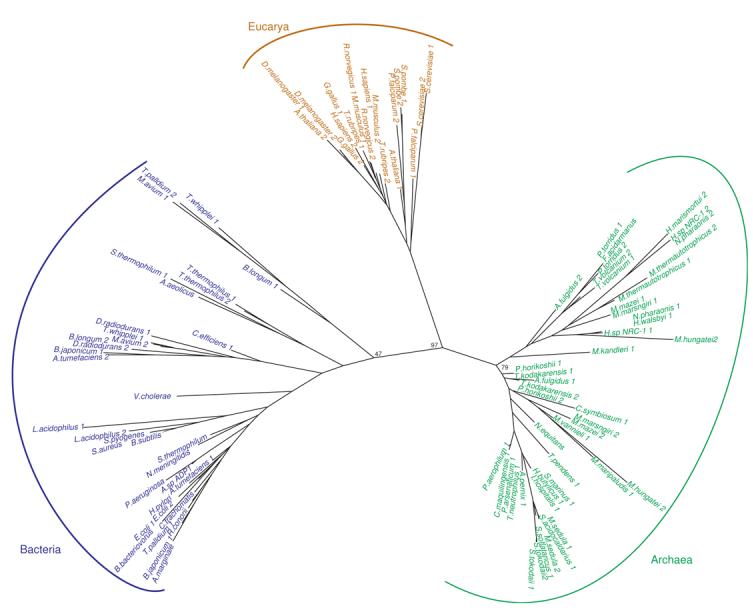
Representative tRNA genes are shown. Only bootstrap values for the branch points discussed in the text are shown for clarity. A tree with the additional bootstrap values can be found in the supplementary material. See Materials and Methods for details.
Figure 8. Unrooted phylogenetic tree of tRNAAsn gene sequences from bacterial (blue), archaeal (green) and eukaryotic (gold) genomes.
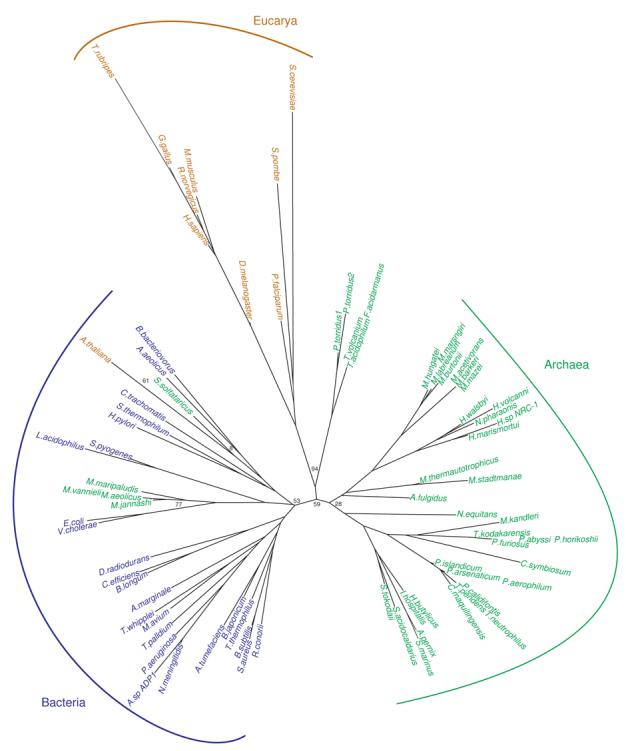
Representative tRNA genes are shown. Only bootstrap values for the branch points discussed in the text are shown for clarity. A tree with the additional bootstrap values can be found in the supplementary material. See Materials and Methods for details.
As expected, given the unique elements recognized by GatDE, the archaeal tRNAGln was distinct from the isoacceptors from the other two domains of life (Fig. 7). The phylogeny displayed a three-domain pattern with the eukaryotic tRNAGln isoacceptors grouping closer to the archaeal sequences than the bacterial ones. Since each of three domains of life for the most part use distinct enzymes for Gln-tRNAGln formation, the tRNAGln phylogeny is untouched by HGT. GatDE is used exclusively in all known archaea3 while all eukaryotes use GlnRS. In most bacteria GatCAB is used to synthesize Gln-tRNAGln though in a few bacteria, in particular the γ- and β-Proteobacteria, GlnRS is used instead.2
The tRNAAsn sequences do appear to group as distinct bacterial, archaeal and eukaryotic types (Fig. 8), yet the divisions are not as clear as in the tRNAGln tree. For example, the Methanococci tRNAAsn isoacceptors group with the bacterial sequences from Escherichia coli and Vibrio cholrea. Thermoplasmata tRNAAsn sequences are more similar to the eukaryotic tRNAAsn and like eukaryotes these organisms use AsnRS and not GatCAB to form Asn-tRNAAsn. Finally, S. solfataricus and A. thaliana tRNAAsn are of the bacterial type.
Since tRNA genes are prime targets for integration events46-48 which include HGT, we examined the genomic context to determine if there is additional evidence of HGT from bacteria into these organisms. These tRNAAsn genes are not adjacent to any detectable bacterial genes, integrases or recombinases.
The phylogenetic discrepancies in the tRNAAsn tree therefore maybe due to convergence rather than gene transfer events. With regards to Asn-tRNAAsn formation the three domains of life to do not use domain specific enzymes. GatCAB is used in a number of bacteria and archaea while AsnRS is found in all three domains of life.2, 3, 6 Therefore, there might not be the same selective pressure to maintain distinct tRNAAsn sequences as there is with tRNAGln.
The archaeal GatCAB can use archaeal or bacterial Asp-tRNAAsn equally well.17, 43 In addition, the archaeal ND-AspRS can aminoacylate bacterial tRNAAsn.2, 3, 6 In fact, Thermus thermophilus and D. radiodurans rely on an archaeal ND-AspRS6, 49, 50 to recognize their bacterial tRNAAsn as the first step of their tRNA-dependent biosynthesis of Asn.51 This would suggest recognition of tRNAAsn by the ND-AspRS and GatCAB is not a barrier to HGT of the bacterial tRNAAsn to archaea or convergence of the archaeal tRNAAsn sequences to the bacterial type.
It is also worth noting the major identity elements the E. coli AsnRS recognized in tRNAAsn,52 G73 and the anticodon, are conserved in tRNAAsn isoacceptors from all three domains of life, suggesting the aaRS should be able to cross-react with heterologous tRNAAsn species. Thus, recognition of the tRNAAsn by AsnRS may also not place a strong evolutionary constraint upon the tRNAAsn sequences enabling greater genetic drift or HGT.
Discussion
Model of the evolution of the two AdTs
The hypothesis that GatDE is an archaeal invention30 is not supported by our phylogenetic results. In the unrooted phylogeny of the GatB and GatE core, GatE sequences did not cluster within one of the GatB branches but rather were on a long branch away from the GatB sequences (Fig. 2). Without rooting the phylogeny, we can not rule out a specific archaeal GatB/GatE relationship with GatE evolving at an accelerated rate as compared to GatB. However, the phylogeny of the tail domain of the kinase subunits (GatB and GatE) rooted with YqeY sequences from diverse bacteria places the root between GatB and GatE (Fig. 3). The divide is well supported (bootstrap value of 100) and the GatB tail domains further split into bacterial and archaeal lineages which is also well supported (bootstrap value of 92). In a similar fashion, the phylogenetic tree of AnsA/GatD places the root between the type I L-asparaginases and GatD (bootstrap value of 100) (Fig. 4). Taken in total, the results strongly suggest that even though GatDE is an archaeal signature protein it emerged prior to the phylogenetic separation of Archaea and Bacteria.
GatA and GatB divide into archaeal and bacterial types. Generally, the phylogeny of both archaeal subunits follows that of established taxonomy, the core of the GatB and the GatA of the Methanobacteria, Methanocci, and Methanopyrus being notable exceptions. This could be an indication of a second archaeal type of GatCAB or due to accelerated co-evolution of the AdT subunits. Canonical archaeal phylogeny derived from the tail domain supports the latter scenario.
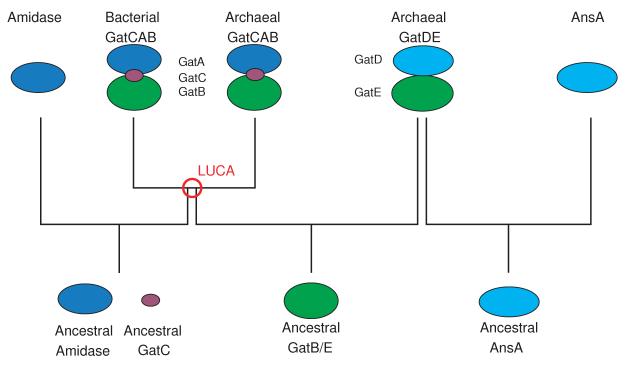
The deep divide between GatB and GatE, the co-evolution of GatA with GatB and of GatD with GatE, and the presence of GatCAB in Bacteria and Archaea while GatDE only being present in Archaea leads us to the following proposal for the evolution of the two AdTs (Fig. 9). In the ancestral community, pre-LUCA, there was a duplication event of the ancient GatB/E. The glutaminase partner of this ancient AdT is unknown. It is possible the ancestral GatB/E did not require a partner. After the duplication, one ancient GatB/E paired with an amidase (ancestral GatA) possibly with the assistance of a helper protein, GatC giving rise to GatCAB. The other paired with an ancestral AnsA (GatD) forming the early heterodimeric AdT. Archaea emerged from LUCA with GatDE and GatCAB. Bacteria emerged with just GatCAB. Why Archaea may have retained two AdTs is discussed further in the accompanying article.
Figure 9. Evolutionary pedigree of the two AdTs.
GatA and amidases are represented in blue and GatC in violet. GatD and AnsA are in cyan. The kinase subunit (GatB or GatE) is green. The figure is discussed in detail in the text. Briefly, we propose that there was a duplication event of ancestral GatB/E prior to the split in LUCA (denoted by the red circle). One copy (GatB) associated or remained associated with an amidase (GatA) and GatC. The other (GatE) paired with an L-asparaginase (GatD). We predict GatDE was only retained in the archaeal progenitors in LUCA.
GatCAB and GatDE both being present in LUCA would not be unusual. For example, both the indirect and direct pathways for Cys-tRNACys were also present in the ancestral community29, 53 as were different versions of glycyl-tRNA synthetase, seryl-tRNA synthetase, and lysyl-tRNA synthetase.27, 28 Likely, LUCA was comprised of a diverse group of organisms, as is the case in modern microbial communities, in which HGT was common.25 Within this community some of the organisms may have only encoded GatCAB while others may have used both GatCAB and GatDE. It is not out of the realm of possibility that a subpopulation in LUCA could have encoded only the ancestral GatDE to function as both a Glu-AdT and an Asp-AdT. If the progenitors of bacteria in LUCA possessed only GatCAB while the archaeal progenitors had both AdTs that would explain the modern distribution of GatCAB and GatDE, and be consistent with our phylogenetic calculations.
Co-evolution of the AdTs with tRNAGln and tRNAAsn
Given the evolutionary relationship of the bacterial GatB and GatE and the fact both enzymes, GatDE and the bacterial GatCAB, recognize the first base-pair of the acceptor stem17, 21, 22 would suggest the ancient AdT also recognized the element in its aa-tRNA substrates, presumably Glu-tRNAGln and Asp-tRNAAsn. The selective pressure to separate Glu-AdT and Asp-AdT activity in Archaea evidently favored the archaeal GatCAB to no longer recognize the first base-pair in Asp-tRNAAsn in order to retain its Asp-AdT function.17, 43
In addition, the selection for a separate Glu-AdT and Asp-AdT in Archaea evidently favored GatDE to co-evolve with tRNAGln such that the enzyme recognizes specific bases in its Glu-tRNAGln.22 Archaeal GatCAB on the other hand seems to have evolved to be an Asp-AdT more by rejecting its own Glu-tRNAGln (see accompanying article) than on recognition of specific elements in Asp-tRNAAsn.17, 43 The differences in the co-evolution of the two archaeal AdTs with their tRNA substrates may be a reason as to why AsnRS is found in all three domains of life while GlnRS is absent in Archaea.
The phylogeny of tRNAGln shows the archaeal tRNAGln to be distinct while the tRNAAsn phylogeny indicates possible cases of convergence between archaeal, bacterial and eukaryotic sequences. Previously, it was shown that neither the E. coli nor yeast GlnRS could aminoacylate the M. thermautotrophicus tRNAGln.3 Transplanting identity elements recognized by the E. coli GlnRS onto the archaeal tRNAGln yielded a tRNA that the bacterial GlnRS could aminoacylate.3 The unique archaeal tRNAGln favored by GatDE may therefore be preventing Archaea from successfully acquiring GlnRS via HGT.
The archaeal GatCAB on the other hand does not seem to have favored a distinct archaeal tRNAAsn that AsnRS could not recognize. In fact, the major identity elements recognized by the E. coli AsnRS, G73 and the anticodon in tRNAAsn,52 are features common in tRNAAsn isoacceptors in all three domains of life. Recognition of the archaeal tRNAAsn by AsnRS therefore is probably not a major barrier preventing the retention of the AsnRS gene in more archaeal genomes. Differences in Asn metabolism between different organisms may play a bigger role in whether AsnRS is acquired or not.2, 6, 51
Alternative function(s) for the GatCAB subunit?
The role of Pet112 in S. cerevisiae remains unresolved but our phylogentic analysis shows it is an evolutionary remnant of mitochondrial GatCAB. In yeast, Pet112 has been shown to localize to both the mitochondria54-56 and the nucleus54, 56 as well as to be essential for mitochondrial function.31 Pet112 in S. cerevisiae has been shown to interact with nuclear protein kinase and a transcription factor of unknown function (RDS1) but not an amidase nor an L-asparaginase, the partners of Pet112 orthologs GatB and GatE, respectively.57, 58 The presence of a mitochondrial AsnRS in S. cerevisiae seemingly negates the need for an Asp-AdT for protein synthesis.59 In addition, the fact the S. cerevisiae mitochondrial GluRS is discriminating in nature (i.e. does not form Glu-tRNAGln) and the cytoplasmic GlnRS is present in the yeast mitochondria suggests a Glu-AdT is also not required in the organelle.60 The conservation of the important residues in Pet112 for the kinase activity of GatB and GatE indicates a catalytic role for the enzyme in the cell.
Pet112 in S. cerevisiae therefore maybe serving an alternative catalytic function in the organism other than as part of an AdT for amide aa-tRNA biosynthesis for translation. It is interesting to note that GatE, the Pet112 archaeal homolog, can form pyroglutamate from glutamate on tRNAGln in the absence of a functional glutaminase subunit.20 Given the conservation of the residues essential for the kinase activity of GatE in the Pet112 subfamily suggests the latter enzymes may also be able to catalyze the same conversion of glutamate to pyroglutame. Interestingly, the latter has been implicated as a stimulant of Na+-dependent amino acid transporters in mammals.61-63 It should be noted, use of aa-tRNA for purposes other than peptide elongation is not that unusual. For example, Glu-tRNAGlu formation is the first step in heme and chlorophyll biosynthesis in plants and many bacteria and archaea.64, 65 Whether S. cerevisiae Pet112 recognizes an aa-tRNA substrate for use other than protein synthesis awaits further investigations.
Such studies may help uncover a second function for bacterial GatB. Mutation of pet112 in S. cerevisae gives rise to a petite phenotype,31 which can be rescued when the B. subtilis gatB gene is provided.32 The result suggests that bacterial GatB can recognize the same cellular substrate as Pet112. In three genera of bacteria, Hahella, Lawsonia, and Myxococcus, it has been noted that the cellular function of their GatCAB is unknown.2 These bacteria are predicted to lack both ND-aaRSs since they encode in their genomes the necessary enzymes for Gln-tRNAGln, Asn-tRNAAsn and free Asn biosynthesis.2 It is possible the AdT in these bacteria were retained for a function besides amide aa-tRNA synthesis for translation. A second function for the AdTs or one of the subunits may be an additional barrier preventing the replacement of the indirect pathways with the direct ones by the aaRSs.
Conclusion
While GlnRS and AsnRS were late inventions, evolving from GluRS and AspRS respectively after the split in LUCA, Gln and Asn coding are ancient.6, 26-29 The primordial route to amide aa-tRNA formation was likely via the indirect pathway.66 The ancestor to GatE and GatB probably recognized both Asp-tRNAAsn and Glu-tRNAGln. While GatDE is an archaeal signature protein, it likely evolved before the phylogenetic split between Archaea and Bacteria and was present in LUCA together with GatCAB.
Materials and Methods
Phylogenetic Analysis
The bacterial and the archaeal GatA, GatB, AnsA, and AnsB along with the archaeal GatD and GatE, the eukaryotic homologs of GatB, and YqeY sequences were taken from the NCBI database. The AspRS-like insertion domain in the GatE sequences was manually removed prior to aligning with GatB and Pet112 using Geneious 2.5.367 based on the structural alignment of the P. abyssi GatE with the S. aureus GatB.21 GatA sequences were aligned with one another. GatD was aligned with AnsA. Alignments were done using ClustalW in Geneious 2.5.3 followed by manual refinement of the alignment in Geneious 2.5.3.67 This manual refinement included removing large insertions found within enzymes sequences in order to minimize artifacts in our phylogenetic analyses. These included insertions specifically found in GatB sequences and not GatE sequences and vice versa.21 The phylogenies were determined by a maximum parsimony/maximum likelihood combined method as previously described in detail.53 Briefly, the thousand most parsimonious trees were generated with PAUP4b10.68 Gaps were counted as the 21st amino acid and the Blosum45 substitution matrix was used as the parsimony cost matrix.69 PHYML v.2.4.4 was used to choose the maximum likelihood topology from the thousand most parsimonious trees.70 PROTML from the Molphy 3.2 package was used to calculate Bootstrap values using the re-estimation of log likelihoods (RELL) method.71
For the tRNA phylogenies sequences were obtained from the Genomic tRNA database72 and additional archaeal tRNA sequences were obtained from the UCSC Archaeal Genome Browser.73 The tRNA sequences were manually aligned within Geneious 2.5.3.67 The gene sequences were used for the alignments with introns and CCA ends manually removed when present. Given the short length of the tRNA sequences, we used a distance-based method to minimize assumptions previously used to examine the phylogeny of tRNACys sequences.74 Distance-matrices were computed by using DNADIST in Phylip 3.66 with Jukes-Cantor set as the model.33 The trees were then built using NEIGHBOR and DRAWTREE also in Phylip 3.66.33 Bootstrap values were calculated as described above.
Supplementary Material
Acknowledgements
We thank Patrick O'Donoghue and Michael Hohn for advice and stimulating discussions. This work was supported by grants from the National Institute of General Medical Sciences (GM22854) and Department of Energy (DE-FG02-98ER20311).
The abbreviations used are
- aaRS
aminoacyl-tRNA synthetase
- GlnRS
glutaminyl-tRNA synthetase
- AsnRS
asparaginyl-tRNA synthetase
- ND
non-discriminating
- AdT
tRNA-dependent amidotransferase
- GluRS
glutamyl-tRNA synthetase
- Glu-AdT
glutamyl-tRNAGln amidotransferase
- AspRS
aspartyl-tRNA synthetase
- Asp-AdT
aspartyl-tRNAAsn amidotransferase
- AnsA
type I L-asparaginase
- AnsB
type II L-asparaginase
- LUCA
last univeral communal ancestor
- HGT
horizontal gene transfer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Ann. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Sheppard K, Akochy PM, Salazar JC, Söll D. The Helicobacter pylori amidotransferase GatCAB is equally efficient in glutamine-dependent transamidation of Asp-tRNAAsn and Glu-tRNAGln. J. Biol. Chem. 2007;282:11866–18873. doi: 10.1074/jbc.M700398200. [DOI] [PubMed] [Google Scholar]
- 3.Tumbula DL, Becker HD, Chang WZ, Söll D. Domain-specific recruitment of amide amino acids for protein synthesis. Nature. 2000;407:106–110. doi: 10.1038/35024120. [DOI] [PubMed] [Google Scholar]
- 4.Lapointe J, Duplain L, Proulx M. A single glutamyl-tRNA synthetase aminoacylates tRNAGlu and tRNAGln in Bacillus subtilis and efficiently misacylates Escherichia coli tRNAGln1in vitro. J. Bacteriol. 1986;165:88–93. doi: 10.1128/jb.165.1.88-93.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilcox M, Nirenberg M. Transfer RNA as a cofactor coupling amino acid synthesis with that of protein. Proc. Natl. Acad. Sci. USA. 1968;61:229–236. doi: 10.1073/pnas.61.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy H, Becker HD, Reinbolt J, Kern D. When contemporary aminoacyl-tRNA synthetases invent their cognate amino acid metabolism. Proc. Natl. Acad. Sci. USA. 2003;100:9837–9842. doi: 10.1073/pnas.1632156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker HD, Reinbolt J, Kreutzer R, Giege R, Kern D. Existence of two distinct aspartyl-tRNA synthetases in Thermus thermophilus. Structural and biochemical properties of the two enzymes. Biochemistry. 1997;36:8785–97. doi: 10.1021/bi970392v. [DOI] [PubMed] [Google Scholar]
- 8.Curnow AW, Ibba M, Söll D. tRNA-dependent asparagine formation. Nature. 1996;382:589–590. doi: 10.1038/382589b0. [DOI] [PubMed] [Google Scholar]
- 9.Becker HD, Kern D. Thermus thermophilus: a link in evolution of the tRNA-dependent amino acid amidation pathways. Proc. Natl. Acad. Sci. USA. 1998;95:12832–12837. doi: 10.1073/pnas.95.22.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curnow AW, Hong K, Yuan R, Kim S, Martins O, Winkler W, Henkin TM, Söll D. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl. Acad. Sci. USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham DE, Overbeek R, Olsen GJ, Woese CR. An archaeal genomic signature. Proc. Natl. Acad. Sci. USA. 2000;97:3304–3308. doi: 10.1073/pnas.050564797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curnow AW, Tumbula DL, Pelaschier JT, Min B, Söll D. Glutamyl-tRNA(Gln) amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis. Proc. Natl. Acad. Sci. USA. 1998;95:12838–12843. doi: 10.1073/pnas.95.22.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker HD, Min B, Jacobi C, Raczniak G, Pelaschier J, Roy H, Klein S, Kern D, Söll D. The heterotrimeric Thermus thermophilus Asp-tRNA(Asn) amidotransferase can also generate Gln-tRNA(Gln) F.E.B.S. Lett. 2000;476:140–144. doi: 10.1016/s0014-5793(00)01697-5. [DOI] [PubMed] [Google Scholar]
- 14.Raczniak G, Becker HD, Min B, Söll D. A single amidotransferase forms asparaginyl-tRNA and glutaminyl-tRNA in Chlamydia trachomatis. J. Biol. Chem. 2001;276:45862–45867. doi: 10.1074/jbc.M109494200. [DOI] [PubMed] [Google Scholar]
- 15.Cathopoulis TJ, Chuawong P, Hendrickson TL. A thin-layer electrophoretic assay for Asp-tRNA(Asn)/Glu-tRNA(Gln) amidotransferase. Anal. Biochem. 2007;360:151–153. doi: 10.1016/j.ab.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akochy PM, Bernard D, Roy PH, Lapointe J. Direct glutaminyl-tRNA biosynthesis and indirect asparaginyl-tRNA biosynthesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2004;186:767–776. doi: 10.1128/JB.186.3.767-776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailly M, Giannouli S, Blaise M, Stathopoulos C, Kern D, Becker HD. A single tRNA base pair mediates bacterial tRNA-dependent biosynthesis of asparagine. Nucleic Acids Res. 2006;34:6083–6094. doi: 10.1093/nar/gkl622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cathopoulis T, Chuawong P, Hendrickson TL. Novel tRNA aminoacylation mechanisms. Mol. Biosyst. 2007;3:408–418. doi: 10.1039/b618899k. [DOI] [PubMed] [Google Scholar]
- 19.Wilcox M. Gamma-phosphoryl ester of Glu-tRNAGln as an intermediate in Bacillus subtilis glutaminyl-tRNA synthesis. Cold Spring Harb. Symp. Quant. Biol. 1969;34:521–528. doi: 10.1101/sqb.1969.034.01.059. [DOI] [PubMed] [Google Scholar]
- 20.Feng L, Sheppard K, Tumbula-Hansen D, Söll D. Gln-tRNAGln formation from Glu-tRNAGln requires cooperation of an asparaginase and a Glu-tRNAGln kinase. J. Biol. Chem. 2005;280:8150–8155. doi: 10.1074/jbc.M411098200. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura A, Yao M, Chimnaronk S, Sakai N, Tanaka I. Ammonia channel couples glutaminase with transamidase reactions in GatCAB. Science. 2006;312:1954–1958. doi: 10.1126/science.1127156. [DOI] [PubMed] [Google Scholar]
- 22.Oshikane H, Sheppard K, Fukai S, Nakamura Y, Ishitani R, Numata T, Sherrer RL, Feng L, Schmitt E, Panvert M, Blanquet S, Mechulam Y, Söll D, Nureki O. Structural basis of RNA-dependent recruitment of glutamine to the genetic code. Science. 2006;312:1950–1954. doi: 10.1126/science.1128470. [DOI] [PubMed] [Google Scholar]
- 23.Schmitt E, Panvert M, Blanquet S, Mechulam Y. Structural basis for tRNA-dependent amidotransferase function. Structure. 2005;13:1421–1433. doi: 10.1016/j.str.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 24.Harpel MR, Horiuchi KY, Luo Y, Shen L, Jiang W, Nelson DJ, Rogers KC, Decicco CP, Copeland RA. Mutagenesis and mechanism-based inhibition of Streptococcus pyogenes Glu-tRNAGln amidotransferase implicate a serine-based glutaminase site. Biochemistry. 2002;41:6398–6407. doi: 10.1021/bi012126u. [DOI] [PubMed] [Google Scholar]
- 25.Vetsigian K, Woese C, Goldenfeld N. Collective evolution and the genetic code. Proc. Natl. Acad. Sci. USA. 2006;103:10696–10701. doi: 10.1073/pnas.0603780103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamour V, Quevillon S, Diriong S, N'Guyen VC, Lipinski M, Mirande M. Evolution of the Glx-tRNA synthetase family: the glutaminyl enzyme as a case of horizontal gene transfer. Proc. Natl. Acad. Sci. USA. 1994;91:8670–8674. doi: 10.1073/pnas.91.18.8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woese CR, Olsen GJ, Ibba M, Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Donoghue P, Luthey-Schulten Z. On the evolution of structure in aminoacyl-tRNA synthetases. Microbiol Mol. Biol. Rev. 2003;67:550–573. doi: 10.1128/MMBR.67.4.550-573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Donoghue P, Luthey-Schulten Z. Evolutionary profiles derived from the QR factorization of multiple structural alignments gives an economy of information. J. Mol. Biol. 2005;346:875–894. doi: 10.1016/j.jmb.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 30.Ibba M, Becker HD, Stathopoulos C, Tumbula DL, Söll D. The adaptor hypothesis revisited. Trends Biochem. Sci. 2000;25:311–316. doi: 10.1016/s0968-0004(00)01600-5. [DOI] [PubMed] [Google Scholar]
- 31.Mulero JJ, Rosenthal JK, Fox TL. PET112, a Saccharomyces cerevisiae nuclear gene required to maintain rho+ mitochondrial DNA. Current Genetics. 1994;25:299–304. doi: 10.1007/BF00351481. [DOI] [PubMed] [Google Scholar]
- 32.Kim SI, Stange-Thomann N, Martins O, Hong KW, Söll D, Fox TD. A nuclear genetic lesion affecting Saccharomyces cerevisiae mitochondrial translation is complemented by a homologous Bacillus gene. J. Bacteriol. 1997;179:5625–5627. doi: 10.1128/jb.179.17.5625-5627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Felsenstein J. PHYLIP -- Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 34.Iwabe N, Kuma K, Hasegawa M, Osawa S, Miyata T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc. Natl. Acad. Sci. USA. 1989;86:9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown JR, Doolittle WF. Gene descent, duplication, and horizontal transfer in the evolution of glutamyl- and glutaminyl-tRNA synthetases. J Mol Evol. 1999;49:485–495. doi: 10.1007/pl00006571. [DOI] [PubMed] [Google Scholar]
- 36.Schön A, Kannangara CG, Gough S, Söll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1988;331:187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- 37.Jahn D, Kim YC, Ishino Y, Chen MW, Söll D. Purification and functional characterization of the Glu-tRNA(Gln) amidotransferase from Chlamydomonas reinhardtii. J. Biol. Chem. 1990;265:8059–8064. [PubMed] [Google Scholar]
- 38.Dyall SD, Brown MT, Johnson PJ. Ancient invasions: from endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- 39.Andersson SG, Karlberg O, Canback B, Kurland CG. On the origin of mitochondria: a genomics perspective. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2003;358:165–77. doi: 10.1098/rstb.2002.1193. discussion 177-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurland CG, Andersson SG. Origin and evolution of the mitochondrial proteome. Microbiol Mol. Biol. Rev. 2000;64:786–820. doi: 10.1128/mmbr.64.4.786-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417:63–67. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- 42.Deniziak M, Sauter C, Becker HD, Paulus CA, Giege R, Kern D. Deinococcus glutaminyl-tRNA synthetase is a chimer between proteins from an ancient and the modern pathways of aminoacyl-tRNA formation. Nucleic Acids Res. 2007;35:1421–1431. doi: 10.1093/nar/gkl1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Namgoong S, Sheppard K, Sherrer RL, Söll D. Co-evolution of the archaeal tRNA-dependent amidotransferase GatCAB with tRNA(Asn) F.E.B.S. Lett. 2007;581:309–314. doi: 10.1016/j.febslet.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin S, Lee TH, Ha NC, Koo HM, Kim SY, Lee HS, Kim YS, Oh BH. Structure of malonamidase E2 reveals a novel Ser-cisSer-Lys catalytic triad in a new serine hydrolase fold that is prevalent in nature. E.M.B.O.J. 2002;21:2509–2516. doi: 10.1093/emboj/21.11.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chebrou H, Bigey F, Arnaud A, Galzy P. Study of the amidase signature group. Biochim. Biophys. Acta. 1996;1298:285–293. doi: 10.1016/s0167-4838(96)00145-8. [DOI] [PubMed] [Google Scholar]
- 46.Reiter WD, Palm P, Yeats S. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 1989;17:1907–1914. doi: 10.1093/nar/17.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reiter WD, Palm P. Identification and characterization of a defective SSV1 genome integrated into a tRNA gene in the archaebacterium Sulfolobus sp. B12. Mol. Gen. Genet. 1990;221:65–71. doi: 10.1007/BF00280369. [DOI] [PubMed] [Google Scholar]
- 48.Williams KP. Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: sublocation preference of integrase subfamilies. Nucleic Acids Res. 2002;30:866–875. doi: 10.1093/nar/30.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Becker HD, Roy H, Moulinier L, Mazauric MH, Keith G, Kern D. Thermus thermophilus contains an eubacterial and an archaebacterial aspartyl-tRNA synthetase. Biochemistry. 2000;39:3216–3230. doi: 10.1021/bi992573y. [DOI] [PubMed] [Google Scholar]
- 50.Tumbula-Hansen D, Feng L, Toogood H, Stetter KO, Söll D. Evolutionary divergence of the archaeal aspartyl-tRNA synthetases into discriminating and nondiscriminating forms. J. Biol. Chem. 2002;277:37184–37190. doi: 10.1074/jbc.M204767200. [DOI] [PubMed] [Google Scholar]
- 51.Min B, Pelaschier JT, Graham DE, Tumbula-Hansen D, Söll D. Transfer RNA-dependent amino acid biosynthesis: an essential route to asparagine formation. Proc. Natl. Acad. Sci. USA. 2002;99:2678–2683. doi: 10.1073/pnas.012027399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Donoghue P, Sethi A, Woese CR, Luthey-Schulten ZA. The evolutionary history of Cys-tRNACys formation. Proc. Natl. Acad. Sci. USA. 2005;102:19003–19008. doi: 10.1073/pnas.0509617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, Piccirillo S, Umansky L, Drawid A, Jansen R, Liu Y, Cheung KH, Miller P, Gerstein M, Roeder GS, Snyder M. Subcellular localization of the yeast proteome. Genes Dev. 2002;16:707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schönfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rinehart J. Glutaminyl-tRNA synthesis in mitochondria. Yale University; 2004. [Google Scholar]
- 57.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 59.Landrieu I, Vandenbol M, Hartlein M, Portetelle D. Mitochondrial asparaginyl-tRNA synthetase is encoded by the yeast nuclear gene YCR24c. Eur. J. Biochem. 1997;243:268–273. doi: 10.1111/j.1432-1033.1997.0268a.x. [DOI] [PubMed] [Google Scholar]
- 60.Rinehart J, Krett B, Rubio MA, Alfonzo JD, Söll D. Saccharomyces cerevisiae imports the cytosolic pathway for Gln-tRNA synthesis into the mitochondrion. Genes Dev. 2005;19:583–592. doi: 10.1101/gad.1269305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hawkins RA, Simpson IA, Mokashi A, Vina JR. Pyroglutamate stimulates Na+ -dependent glutamate transport across the blood-brain barrier. F.E.B.S. Lett. 2006;580:4382–4286. doi: 10.1016/j.febslet.2006.06.097. [DOI] [PubMed] [Google Scholar]
- 62.Vina JR, Puertes IR, Montoro JB, Saez GT, Vina J. Gamma-glutamyl-amino acids as signals for the hormonal regulation of amino acid uptake by the mammary gland of the lactating rat. Biol. Neonate. 1985;48:250–256. doi: 10.1159/000242178. [DOI] [PubMed] [Google Scholar]
- 63.Vina JR, Palacin M, Puertes IR, Hernandez R, Vina J. Role of the gamma-glutamyl cycle in the regulation of amino acid translocation. Am. J. Physiol. 1989;257:E916–E922. doi: 10.1152/ajpendo.1989.257.6.E916. [DOI] [PubMed] [Google Scholar]
- 64.Schön A, Krupp G, Gough S, Berry-Lowe S, Kannangara CG, Söll D. The RNA required in the first step of chlorophyll biosynthesis is a chloroplast glutamate tRNA. Nature. 1986;322:281–284. doi: 10.1038/322281a0. [DOI] [PubMed] [Google Scholar]
- 65.Huang DD, Wang WY, Gough SP, Kannangara CG. delta-Aminolevulinic acid-synthesizing enzymes need an RNA moiety for activity. Science. 1984;225:1482–1494. doi: 10.1126/science.6206568. [DOI] [PubMed] [Google Scholar]
- 66.Di Giulio M. Genetic code origin: are the pathways of type GlutRNA(Gln) -> Gln-tRNA(Gln) molecular fossils or not? J. Mol. Evol. 2002;55:616–622. doi: 10.1007/s00239-002-2357-6. [DOI] [PubMed] [Google Scholar]
- 67.Drummond AJ, Kearse M, Heled J, Moir R, Thierer T, Ashton B, Wilson A, Stones-Havas S. Geneious v2.5. 2006 http://www.geneious.com/
- 68.Swofford DL. PAUP* - Phylogenetic Analysis Using Parsimony (*and other Methods), Version 4.0b 10 4.0b10 edit. Sinauer Associates; Sunderland, Massachusetts: 2003. [Google Scholar]
- 69.Marsh TL, Reich CI, Whitelock RB, Olsen GJ. Transcription factor IID in the Archaea: sequences in the Thermococcus celer genome would encode a product closely related to the TATA-binding protein of eukaryotes. Proc. Natl. Acad. Sci. USA. 1994;91:4180–4184. doi: 10.1073/pnas.91.10.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systems Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 71.Adachi J, Hasegawa M. MOLPHY version 2.3: programs for molecular phylogenetics based on maximum likelihood. Comput. Sci. Monogr. 1996;28:1–150. [Google Scholar]
- 72.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schneider KL, Pollard KS, Baertsch R, Pohl A, Lowe TM. The UCSC Archaeal Genome Browser. Nucleic Acids Res. 2006;34:D407–D410. doi: 10.1093/nar/gkj134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hohn MJ, Park HS, O'Donoghue P, Schnitzbauer M, Söll D. Emergence of the universal genetic code imprinted in an RNA record. Proc. Natl. Acad. Sci. USA. 2006;103:18095–18100. doi: 10.1073/pnas.0608762103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.