Abstract
Background
In humans, chromosome fragile sites are regions that are especially prone to forming non-staining gaps, constrictions or breaks in one or both of the chromatids on metaphase chromosomes either spontaneously or following partial inhibition of DNA synthesis and have been well identified. So far, no plant chromosome fragile sites similar to those in human chromosomes have been reported.
Methods and Results
During the course of cytological mapping of rDNA on ryegrass chromosomes, we found that the number of chromosomes plus chromosome fragments was often more than the expected 14 in most cells for Lolium perenne L. cv. Player by close cytological examination using a routine chromosome preparation procedure. Further fluorescent in situ hybridization (FISH) using 45S rDNA as a probe indicated that the root-tip cells having more than a 14-chromosome plus chromosome fragment count were a result of chromosome breakage or gap formation in vitro (referred to as chromosome lesions) at 45S rDNA sites, and 86% of the cells exhibited chromosome breaks or gaps and all occurred at the sites of 45S rDNA in Lolium perenne L. cv. Player, as well as in L. multiflorum Lam. cv. Top One. Chromatin depletion or decondensation occurred at various locations within the 45S rDNA regions, suggesting heterogeneity of lesions of 45S rDNA sites with respect to their position within the rDNA region.
Conclusions
The chromosome lesions observed in this study are very similar cytologically to that of fragile sites observed in human chromosomes, and thus we conclude that the high frequency of chromosome lesions in vitro in Lolium species is the result of the expression of 45S rDNA fragile sites. Possible causes for the spontaneous expression of fragile sites and their potential biological significance are discussed.
Introduction
The number of ribosomal DNA (rDNA) sites in a genome differs considerably among species. 45S rDNA sites on the chromosomes are referred to as secondary constriction regions. They occasionally show a lightly stained chromatin structure during metaphase in some plant species when stained with propidium iodide (PI) or 4, 6-diamidino-2-phenylindole (DAPI) [1]. However the gap or constriction at 45S rDNA sites on metaphase chromosomes has not been well identified and studied. During the course of mapping the rDNA in ryegrasses, we serendipitously discovered many chromosome breakages or gap formations, and all occurred exclusively in the 45S rDNA sites in root-tip meristematic cells in Lolium spp. This unusual and interesting phenomenon led us to the effort of characterizing these chromosome breaks or gaps associated with rDNAs and establishing possible links with fragile-site expression frequently reported in humans [2], [3].
In humans, chromosome fragile sites are regions that are especially prone to forming non-staining gaps, constrictions or breaks in one or both of the chromatids on metaphase chromosomes either spontaneously or following partial inhibition of DNA synthesis [2], [3]. Since their discovery more than three decades ago [4], more than 120 chromosome fragile sites have been identified in human [5]. They are classified as either rare or common based on their frequency and mode of induction. Rare fragile sites are archetypal dynamic mutations and can be sensitive to folate or induced by replication inhibitors [6]. They are present in fewer than 2.5% of the human populations but they have not been implicated in cancer. In contrast, common fragile sites are seen in all humans and are regions of normal chromosome structure that are typically stable in somatic cells [3], [7]. However, under DNA replication stresses, such as treatment with aphidicolin, an inhibitor of DNA polymerase alpha, these sites are prone to breakage [8]. Such breakages are frequently involved in chromosomal rearrangements in cancer cells and have been associated with other human diseases [9], [10]. Fragile sites are known to extend over large regions on a chromosome and have been associated with genes [11]. It is generally agreed that fragile sites comprise regions of high DNA flexibility and display delayed replication [12], [13]. Chromatin modifications such as DNA methylation and histone methylation and acetylation are involved in the expression of fragile sites [14], [15]. A heterochromatin-like compact chromatin structure contributes to the expression of fragile sites and chromosomal fragility may be indicative of altered higher-order DNA organization or stalled replication [16], [17]. Fragile sites were found to be preferred sites of DNA recombination, gene amplification and plasmid integration [18]–[20].
So far, no plant chromosome fragile sites similar to those in human chromosomes have been reported, although changes in chromosome number and structure often occur in natural plant populations as well as in tissue-cultured cells [21], [22]. In this study, we investigated the cause of the varied number of metaphase chromosomes (should actually be chromosomes plus chromosome fragments) among root-tip cells, which is often more than the expected 14 in most cells for Lolium perenne L. cv. Player by close cytological examination using a routine chromosome preparation procedure. Further fluorescent in situ hybridization (FISH) using 45S rDNA as a probe showed that most of the root-tip cells having more than a 14-chromosome plus chromosome fragment count are a result of chromosome breakage or gap formation (referred to as chromosome lesions) and these chromosome lesions occurred exclusively in the 45S rDNA sites. FISH also revealed that some gaps of 45S rDNA segments showed a depleted chromatin structure during metaphase, which looked like one or a few thin threads and some showed no DNA fibers between the two separated parts. Based on cytological observations and prior knowledge of fragile sites on human chromosomes, we conclude that the high frequency of chromosome lesions is the result of the expression of fragile sites in 45S rDNA. Interestingly, the 45S rDNA fragile sites we observed here are highly expressed under normal growth conditions without an addition of any DNA replication stress agents, possibly suggesting that the fragility is indicative of inherent unique chromosomal structures of the 45S rDNA site.
Results
1. Chromosome gaps and breaks occur at a high frequency on mitotic chromosomes in vitro in Lolium perenne cv. Player
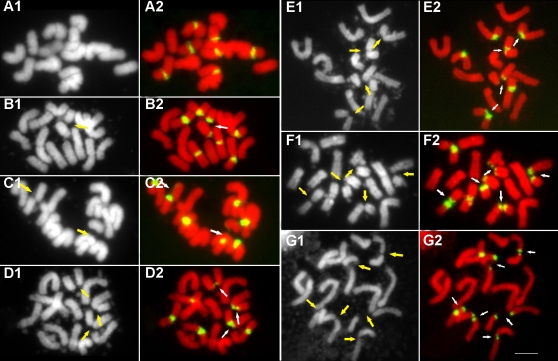
During the course of cytological mapping of rDNA on ryegrass chromosomes, we found that the number of chromosomes plus chromosome fragments was often more than the expected 14 in most cells for Lolium perenne L. cv. Player by close cytological examination using a routine chromosome preparation procedure (Figure 1a). However, it is difficult to distinguish whether one “chromosome” is a complete chromosome or merely a chromosome fragment before mapping 45S rDNA by FISH. The frequency of chromosome lesions in vitro was quite high. In the 119 metaphase cells analyzed, there were only 18 cells with the normal 14 chromosomes (15%), while the number of cells with 15, 16, 17, 18, 19 or 20 chromosomes plus chromosome fragments accounted for the majority of cells (85%, Figure 1b).
Figure 1. a: Varied chromosome plus chromosome fragment count of mitotic chromosomes in vitro in the diploid Lolium perenne cv. Player (2n = 14) due to chromosome lesions.
Figs A–F show the variation of chromosome plus chromosome fragment numbers from 15 to 20 in different metaphase cells. A: 15, B: 16, C: 17, D: 18, E: 19, F: 20. b: The pie chart represents the percentages of cells with different chromosome plus chromosome fragment counts. Bar = 5 µm.
2. Chromosome gaps and breaks were exclusively associated with the 45S rDNA and resembled fragile sites in human chromosomes
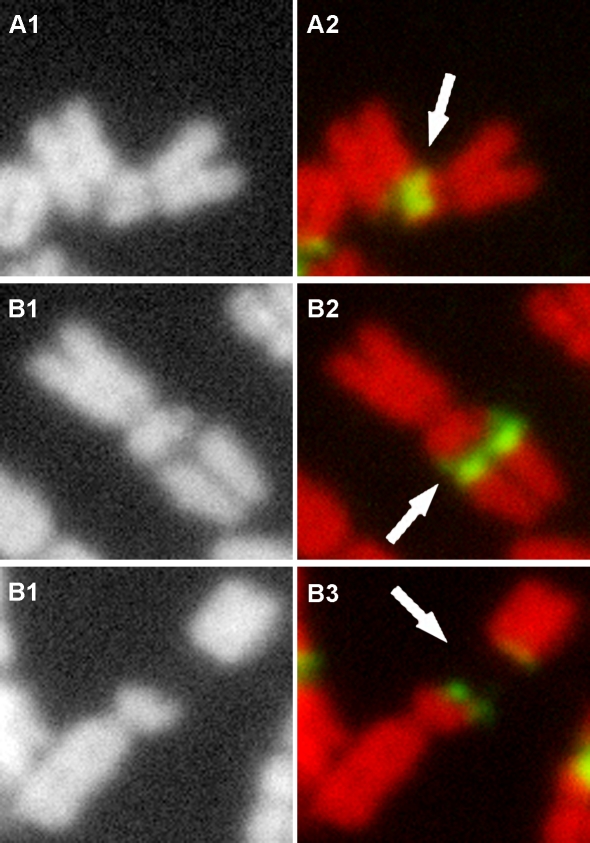
Following FISH with 45S rDNA as a probe, we detected seven 45S rDNA hybridization sites in the diploid L. perenne cv. Player and our FISH results also revealed all of 45S rDNA signals occurred in the middle of the chromosomes when no lesions happened (e.g. 14 chromosomes) (Figure 2). These are agreement with the results reported in ryegrasses [23]. Chromosome preparations were examined for the presence or absence of lesions and the pattern of the lesions. A cell was considered to contain chromosomes with lesion sites in vitro if one or both of the chromatids of one or more chromosome were broken or had gaps on one or more chromosomes. If there is only one chromosome break at a single site or only one gap appears on one chromosome, there will be 14 chromosomes plus 1 chromosome fragment in a cell, resulting in a chromosome plus chromosome fragment count of 15. Accordingly cells with 2, 3, 4, 5 or 6 lesions would result in cells with chromosome plus chromosome fragment counts of 16, 17, 18, 19 and 20, respectively. In the 100 cells analyzed, 86 cells showed at least one chromosome lesion. FISH revealed that chromosome lesions occurred exclusively at the 45S rDNA sites and each one of the seven sites could be involved with a lesion, although the number of lesions varied among different cells in vitro. However, we are not sure that whether each of the seven 45S regions is as frequently involved in chromosome breaks as the other, e.g. whether some fragile sites are more prone to breakage than others because the identification of specific chromosomes is difficult. We also did not determine whether the broken chromosomes are homologous, however based on the fact that the number of breaks varies from cells to cells, we guess that breaks could occur randomly and could occur on either or both of a pair of homologous chromosomes when multiple breaks occur in a cell. In cells without any chromosome lesions at the 45S rDNA regions in vitro, metaphase chromosome number is always 14, confirming that lesions occurred exclusively at the 45S rDNA regions and there were no lesions at any other parts of the chromosomes. In L. multiflorum cv. Top One (2n = 28), chromosome lesions were also frequent at 45S rDNA sites (Figure 3). Furthermore, FISH analysis on the chromosome preparations in the other two cultivars of L. perenne L. and L. multiflorum Lam. showed that 45S rDNAs regions were the sites of chromosome lesions (data not shown). These results lead us to believe that 45S rDNA is a region of chromosome fragility in Lolium. Cytological appearance of lesions at 45S rDNA fragile sites in Lolium appears to be analogous to that of fragile sites observed in human chromosomes. Three different cytological appearances of lesions were observed at the 45S rDNA sites in Lolium: first, breakage or constriction occurred to a single chromatid within the 45S rDNA region (Figure 4, A1 and A2); second, one formed a gap within the rDNA between the two chromosome ends, with the chromosome still connected through one or a few thin DNA fibers (local despiralizations of the chromatid) (Figure 4, B1 and B2); and third, breakage occurred to both chromatids of a chromosome with no detectable DNA hybridization signals between the broken ends of the two chromosome fragments (Figure 4, C1 and C2).
Figure 2. Fluorescence in situ hybridization with 45S rDNA as the probe shows that 45S rDNAs (green) are the sites of chromosome lesions in meristematic cells of root tips in diploid Lolium perenne cv. Player.
The number of lesion sites varys in different cells from 0 to 6 due to the existence of multiple 45S rDNA sites. The left panel A1–G1: black layers and the right panel A2–G2: color images by merging red layers and green layers. Arrows indicate lesion sites. Bar = 5 µm.
Figure 3. Fluorescence in situ hybridization with 45S rDNA as a probe shows that 45S (green) rDNAs are the sites of chromosome lesions in meristematic cells of root tips in tetraploid Lolium multiflorum cv. Top One.
A: black layer; B: color image by merging red layers and green layers. Arrows indicate sites. Bar = 5 µm.
Figure 4. Different cytological appearances of lesions at the 45S rDNA fragile sites.
A: breakage or constriction occurs to a single chromatid within the 45S rDNA region. B: a gap forms within the rDNA between the two chromosome ends, but is still connected through one or a few thin DNA fibers (local despiralizations of the chromatid). C: A chromosome is broken and completely separated into two parts without any DNA hybridization signals detected within the gap. A1–C1: black layer; A2–C2: color image by merging red layers and green layers. Arrows indicate lesion sites.
3. Chromatin depletion or decondensation occurred at various sites within the 45S rDNA repeat unit
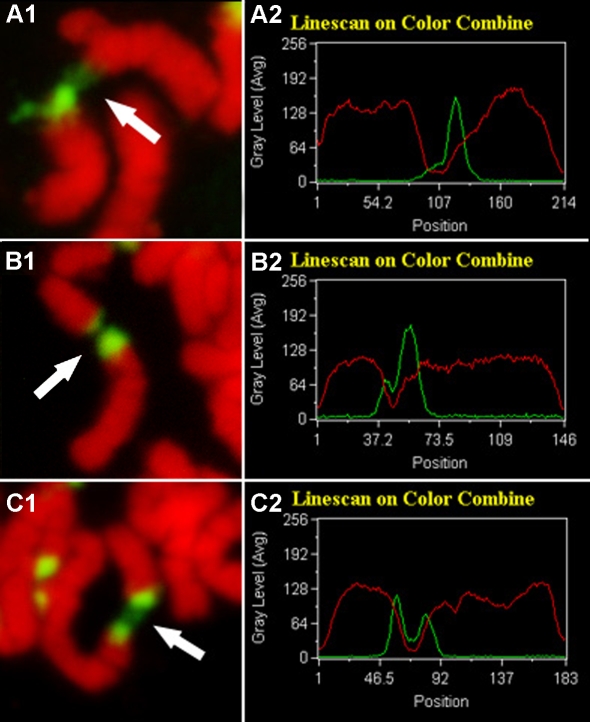
Linescan curve analysis of fluorescence signals for chromosomes and the 45S rDNAs indicated that chromatin depletion or decondensation occurred at various sites within a 45S rDNA region (Figure 5). In figure 5, A2, the occurrence of a single wave crest in the green linescan curve indicated that a strong fluorescence signal was present only at one end of a lesioned chromosome while the green line to the right of the crest is almost flat, suggesting that few or no 45S rDNAs remain on this part of the chromosome. This result indicated that chromatin depletion or decondensation took place at a 45S rDNA terminus. Figure 5, B2 showed that both chromosome lesion ends had concentrated fluorescence signals, but the two signals had different fluorescence intensity with one having a much stronger signal than the other. In the linescan curve, this difference in signal intensity was reflected by the height of the wave crests. This meant that chromatin depletion or decondensation was close to a 45S rDNA terminus, but remained within the 45S rDNA repeats. In figure 5, C2, the two signal wave crests in the linescan curve had similar height, indicating that both lesion ends of the chromosome have similar 45S rDNA signal intensity, therefore the chromatin must have been depleted or decondensed in the middle of a 45S rDNA repeat unit. The linescan curve data are in good agreement with the cytological observations provided in figures 5, A1, B1 and C1.
Figure 5. Chromatin depletion or decondensation occurs at various positions of the 45S rDNA repeat unit.
Pictures in the left panel represent three different lesions at various positions in the 45S rDNA region. Arrows indicate lesion sites. Pictures in the right panel are kymograms showing the intensity of the signals for the hybridization sites. The horizontal axis is the length of one chromosome; the vertical axis is the gray level which measures the intensity of the fluorescent dye and signals. The green line represents the hybridization site and signal intensity, and the red line shows PI-stained chromosomes.
Discussion
Fragile sites are expressed either as gaps, constrictions or breaks on human metaphase chromosomes [24]. In this study, we report, for the first time in a plant species, the finding of 45S rDNA as the chromosome fragile sites spontaneously expressed in vitro on metaphase chromosomes from root-tip meristematic cells in Lolium species. The percentage of cells with chromosome lesions in vitro was very high (∼85%). Examination of a large number of cytological chromosome preparations suggested that chromosome lesions in Lolium spp. occurred exclusively at the sites of the 45S rDNA repeat unit, although the exact location of a particular lesion varies with the chromosome. The lesions observed in this study resembled the appearance of fragile sites in human chromosomes. In humans, these lesions or gaps are either complete breaks or decondensed chromatins or fibers that can't be seen through routine cytological observations. In our study, some gaps or lesions are not really DNA breaks. FISH using 45S rDNA as a probe revealed that some gaps of 45S rDNA segments showed a depleted or decondensed chromatin structure during metaphase, which looked like a or a few thin threads joined together and some showed no any DNA fibers between the two separated parts and should be complete breaks. It was speculated that a failure of the complex folding of the chromatin fibers occurred at fragile sites, resulting in gap formation or break of fragile sites. From these results, we conclude that the 45S rDNA regions are chromosome fragile sites in Lolium spp..
Thomas et al. reported the existence of extensive chromosome rearrangements based on the variation in the number and positions of rDNA sites in Lolium rigidum [25]. One possibility is that the subsequent rejoining of the broken chromosome ends in vivo following chromosome breakage caused the varied number and locations of 45S rDNA in Lolium rigidum. 45S rDNA is also involved in the chromosome breakage-fusion-bridge cycle and rearrangements in late generation telomerase-deficient Arabidopsis [26]. In Allium, varied numbers and positions of the nucleolus organizer regions (NORs) which contain the rDNA gene have also been observed in clones of a genotype [27]. This led the authors to believe that the NORs of some Allium chromosomes are free to jump from one locus to another. In addition, in partial diploid strains of Neurospora, chromosome breakage in NORs results in large terminal deletions [28], [29]. Furthermore, the presence of ectopic rDNA created a new chromosome breakage site in the partial diploid genome of Neurospora [28]. These results possibly suggested that rDNA might break as the fragile sites under the in vivo conditions in some plants.
The presence of unique DNA sequences in fragile sites may result in chromosome fragile site expressions. Sequence analyses of the human common fragile sites, however, revealed no cis-acting sequences that can explain their instability [30]–[33]. Nevertheless all rare fragile site sequences identified so far contain expanded repeating sequences and the expression of these fragile sites is directly linked to the increased length of the triplet CGG repeats or AT rich repeats [34], [35]. In a ciliated protozoon Tetrahymena in which chromosome breakage is a normal and highly regulated event in the development of the new macronucleus following conjugation, a highly conserved 15 bp cis-acting sequence has been identified as necessary and sufficient to induce chromosome breakage [36]. It has been demonstrated that chromosome breakage between rDNA and its flanking sequence leads to the excision of the rDNA gene during macronucleus development in Tetrahymena [37]. Because the sequence of the 45S rDNA repeating unit is highly conservative among different plants and no obvious lesions in 45S rDNA regions are reported in the other plants, therefore the sequence of 45S rDNA repeating unit could not be main cause for the fragility. Nevertheless, it is possible that the copy number of the rDNA repeat unit or the intergenic space between these repeat units is associated with the expression of rDNA fragile sites in Lolium spp. because the number of repeat units and the intergenic space between these repeat units is variable among species.
It is also possible that other factors are involved in regulating expression of fragile sites. Recent findings have shown that key cell cycle checkpoint functions are associated with fragile site stability. For example, ataxia-telangiectasia and Rad3-related protein (ATR), a DNA replication checkpoint kinase that is essential for cellular response to DNA damage and replication stresses has been shown to be critical for genome stability at fragile sites in humans [17]. The authors found that chromosome fragile sites were expressed in cells that lack the replication checkpoint protein ATR and that had not been exposed to replication inhibitors. ATR was also shown to regulate a G2-phase cell-cycle checkpoint in Arabidopsis [38]. Furthermore, the breast cancer 1 (BRCA1) protein, one of the downstream targets of ATR in response to DNA damage, has been shown to be required for fragile site stability [33], [39]–[41]. These results represent the first characterized major molecular pathway that regulates fragile site expression.
DNA repair or epigenetic modification may also be involved in expression of chromosome fragile sites. There is clear evidence in humans that a low level of DNA repair may account for the extreme fragility of constitutive heterochromatin and epigenetic marks such as DNA and histone modifications, which alter chromatin structures, and therefore are related to chromosome fragility [42]. Hypermethylation of the DNA in the fragile sites led to transcriptional silencing of genes at the site [43]. Hyperacetylation of histone proteins also reduces the expression of fragile site FRAXA in humans [44]. 45S rDNA genes are highly conserved among plant species and are typically organized in tandem repeats with several hundreds or thousands of copies, but only a few of the rRNA genes are transcriptionally active at a particular time [45], therefore, the number of active rDNA genes must be strictly regulated in the form of dosage control which also operates in nucleolar dominance and in which DNA methylation and histone modification have been shown to play a role [46], [47]. Recent studies demonstrated that the transcriptionally inactive rDNA genes are correlated with DNA hypermethylation and histone hypoacetylation [48]. In both S. cerevisiae and S. pombe, transcriptional silencing at rDNA repeats involves the assembly of large regions of DNA into a specialized chromatin structure by modification of chromatin [49]. Sirtuin Hst2 in S. pombe was proved to play a similar role as the other Sirtuins in transcriptional silencing of the rDNA regions [50]. Synthetic interactions between hst2 and sir2 were also reported in the silencing of budding yeast rDNA [51]. Considering that DNA and histone modification is involved in both gene silencing and the expression of the fragile sites, we speculate that 45S rDNA fragility may be indicative of inactive 45S rRNA genes due to DNA and histone modifications, and of unique chromatin structures. Spontaneous fragile site expression in the form of chromosome breakage is rare in cultured human cells, but can be triggered and enhanced by treatment of cells with agents such as aphidicolin that slightly delay DNA replication fork progression [3], [52]. It was suggested that the expression of fragile sites in humans might be an indicator of changed chromatin structure and stalled replication [16], [17].
Very little is known about the biological cause of 45S rDNA fragility, but the potential mobility of the 45S rDNA caused by breaking and subsequent rejoining and the fact that the fragile sites are preferred sites for foreign gene integration and gene recombination in humans [18]–[20] may have practical applications in agricultural biotechnology. The molecular mechanism that regulates spontaneous expression of the 45S rDNA fragility remains to be elucidated.
Materials and Methods
Plant material
Naturally occurring Lolium are diploid with 2n = 14. Plants from a diploid turf type cultivar, Player of perennial ryegrass (Lolium perenne L.) were used for the current research. Seeds were kindly provided by Turf Seed (Hubbard, OR, USA).
Chromosome preparation and analysis
Metaphase chromosome preparation was performed using the outline protoplast technique as described by Song and Gustafson in 1995 [53]. Root tips were harvested when the primary roots were 0.5–1.0 cm long from seedlings grown on moist filter papers in a culture tank. The excised roots were treated in freezing deionized water overnight. After being fixed in ethanol-glacial acetic acid (3:1 v/v) at 4°C overnight, roots were treated with an enzyme mixture of 2% cellulase and 2% pectolyase for 50–70 min at 28°C. More than 100 cells from different genotypes of each cultivar were analyzed.
Digoxigenin labeling DNA and fluorescence in situ hybridization
Plasmid 45S rDNAs were digoxigenin-labeled by nick translation using Dig-Nick Translation Mix purchased from Boehringer Mannheim Corporation (IN, USA). In situ hybridization was performed using the procedure described by Li et al. [54]. The hybridization mixture contained 50% deionized formamide, 10% dextran sulphate, 2×SSC, 1 mg/mL of sheared salmon sperm DNA and 1–2 µg/mL probes. Hybridization was performed at 37°C overnight.
Detection
Digoxigenin-labeled probes were detected with sheep-anti-digoxigenin-FITC (Roche Molecular Biochemical) and amplified with rabbit-anti-sheep-FITC (Vector Laboratories, Burlingame, CA, USA). In both steps of the immune reactions, slides were placed in a wet chamber at 37°C for 1 h and then washed with 1×PBS three times, each for 5 min, at room temperature. Chromosomes were counterstained with 1 µg/mL PI in Vectashield (Vector Laboratories, Burlingame, CA, USA).
Image capture
Chromosome preparations were examined with an Olympus BX-60 fluorescence microscope with filter blocks for PI and FITC. The filter blocks on this microscope have been coaligned so that no image would be shifted with filter changes. Images were captured with a CCD monochrome camera Sensys 1401E and a computer using the software MetaMorph 4.6.3 (Universal Imaging Corp., Downingtown, PA, USA). Separate monochrome images were captured for chromosomes (PI) or 45S rDNA (FITC), and then converted into red and green images, respectively. Kymograms were recorded by using the “linescan” command in the software MetaMorph 4.6.3 with the PI and 45S rDNA fluorescence signal intensity as key parameters.
Acknowledgments
We are grateful to Dr. Antonio Musio (Istituto di Tecnologie Biomediche C.N.R. Via Fratelli Cervi, Italy) and Dr. Loren Stephens (Department of Horticulture, Iowa State University, Ames, IA, USA) for critical reading of this manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by grants from the National Natural Science Foundation of China (No. 30771204), Program for New Century Excellent Talents in University (NCET-07-0634) and the Natural Science Foundation of Hubei Province (No. 2005ABA097).
References
- 1.Robert-Fortel I, Junéra HR, Géraud G, Hernadez-Verdun D. Three-dimensional organization of the ribosomal genes and Ag-NOR proteins during interphase and mitosis in PtK 1 cells studied by confocal microscopy. Chromosoma. 1993;102:146–157. doi: 10.1007/BF00387729. [DOI] [PubMed] [Google Scholar]
- 2.Richards RI. Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet. 2001;17:339–345. doi: 10.1016/s0168-9525(01)02303-4. [DOI] [PubMed] [Google Scholar]
- 3.Glover TW. Common fragile sites. Cancer Lett. 2006;232:4–12. doi: 10.1016/j.canlet.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 4.Magenis RE, Hecht F, Lovrien EW. Heritable fragile site on chromosome 16: probable localization of haptoglobin locus in man. Science. 1970;170:85–87. doi: 10.1126/science.170.3953.85. [DOI] [PubMed] [Google Scholar]
- 5.Buttel I, Fechter A, Schwab M. Common fragile sites and cancer: targeted cloning by insertional mutagenesis. Ann NY Acad Sci. 2004;1028:14–27. doi: 10.1196/annals.1322.002. [DOI] [PubMed] [Google Scholar]
- 6.Sutherland GR, Baker E, Richards RI. Fragile sites still breaking. Trends Genet. 1998;14:501–506. doi: 10.1016/s0168-9525(98)01628-x. [DOI] [PubMed] [Google Scholar]
- 7.Huebner K, Croce CM. FRA3B and other common fragile sites: the weakest links. Nat Rev Cancer. 2001;1:214–221. doi: 10.1038/35106058. [DOI] [PubMed] [Google Scholar]
- 8.Glover TW, Berger C, Coyle J, Echo B. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum Genet. 1984;67:136–142. doi: 10.1007/BF00272988. [DOI] [PubMed] [Google Scholar]
- 9.Nelson DL. The fragile X syndromes. Semin Cell Biol. 1995;6:5–11. doi: 10.1016/1043-4682(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 10.Popescu NC. Genetic alterations in cancer as a result of breakage at fragile sites. Cancer Lett. 2003;192:1–17. doi: 10.1016/s0304-3835(02)00596-7. [DOI] [PubMed] [Google Scholar]
- 11.Boldog FL, Waggoner B, Glover TW, Chumakov I, Le Paslier D, et al. Integrated YAC contig containing the 3p14. 2 hereditary renal carcinoma 3; 8 translocation breakpoint and the fragile site FRA3B. Genes Chromosomes Cancer. 1994;11:216–221. doi: 10.1002/gcc.2870110403. [DOI] [PubMed] [Google Scholar]
- 12.Laird C, Jaffe E, Karpen G, Lamb M, Nelson R. Fragile sites in human chromosomes as regions of late-replicating DNA. Trends Genet. 1987;3:274–281. [Google Scholar]
- 13.Handt O, Baker E, Dayan S, Gartler SM, Woollatt E, et al. Analysis of replication timing at the FRA10B and FRA16B fragile site loci. Chromosome Res. 2000;8:677–688. doi: 10.1023/a:1026737203447. [DOI] [PubMed] [Google Scholar]
- 14.Coffee B, Zhang F, Ceman S, Warren ST, Reines D. Histone modifications depict an aberrantly heterochromatinized FMR1 gene in fragile X syndrome. Am J Hum Genet. 2002;71:923–932. doi: 10.1086/342931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y. Chromatin structure of human chromosomal fragile sites. Cancer Lett. 2006;232:70–78. doi: 10.1016/j.canlet.2005.07.040. [DOI] [PubMed] [Google Scholar]
- 16.Gericke GS. Chromosomal fragility may be indicative of altered higher-order DNA organization as the underlying genetic diathesis in complex neurobehavioural disorders. Med Hypotheses. 1999;52:201–208. doi: 10.1054/mehy.1997.0643. [DOI] [PubMed] [Google Scholar]
- 17.Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 18.Rassool FV, McKeithan TW, Neilly ME, Melle EV, Espinosa R, III, et al. Preferential integration of marker DNA into the chromosomal fragile site at 3p14: an approach to cloning fragile sites. Proc Natl Acad Sci USA. 1991;88:6657–6661. doi: 10.1073/pnas.88.15.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coquelle A, Pipiras E, Toledo F, Buttin G, Debatisse M. Expression of fragile sites triggers intrachromosomal mammalian gene amplification and sets boundaries to early amplicons. Cell. 1997;89:215–225. doi: 10.1016/s0092-8674(00)80201-9. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz M, Zlotorynski E, Kerem B. The molecular basis of common and rare fragile sites. Cancer Lett. 2006;232:13–26. doi: 10.1016/j.canlet.2005.07.039. [DOI] [PubMed] [Google Scholar]
- 21.Winfield MO, Schmitt M, Lorz H, Davey MR, Karp A. Nonrandom chromosome variation and morphogenic potential in cell lines of bread wheat. Genome. 1995;38:869–878. doi: 10.1139/g95-115. [DOI] [PubMed] [Google Scholar]
- 22.Morgan WG, King IP, Harper JA, Thomas HM. The cytogenetic analysis of a Robertsonian rearrangement in Lolium multiflorum. Cytologia. 2000;65:173–177. [Google Scholar]
- 23.Thomas HM, Harper JA, Meredith MR, Morgan WG, Thomas ID, et al. Comparisons of ribosomal DNA sites in Lolium species by fluorescent in situ hybridization. Chromosome Res. 1996;4:486–490. doi: 10.1007/BF02261775. [DOI] [PubMed] [Google Scholar]
- 24.Cimprich KA. Fragile Sites: breaking up over a slowdown. Curr Biol. 2003;13:231–233. doi: 10.1016/s0960-9822(03)00158-1. [DOI] [PubMed] [Google Scholar]
- 25.Thomas HM, Harper JA, Morgan WG. Gross chromosome rearrangements are occurring in an accession of the grass Lolium rigidum. Chromosome Res. 2001;9:585–590. doi: 10.1023/a:1012499303514. [DOI] [PubMed] [Google Scholar]
- 26.Siroky J, Zluvova J, Riha K, Shippen DE, Vyskot B. Rearrangements of ribosomal DNA clusters in late generation telomerase-deficient Arabidopsis. Chromosoma. 2003;112:116–123. doi: 10.1007/s00412-003-0251-7. [DOI] [PubMed] [Google Scholar]
- 27.Schubert I, Wobus U. In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma. 1985;92:143–148. [Google Scholar]
- 28.Butler DK. Ribosomal DNA is a site of chromosome breakage in aneuploid strains of Neurospora. Genetics. 1992;131:581–592. doi: 10.1093/genetics/131.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perkins DD, Raju NB, Barry EG, Butler DK. Chromosome rearrangements that involve the nucleolus organizer region in Neurospora. Genetics. 1995;141:909–923. doi: 10.1093/genetics/141.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boldog F, Gemmill RM, West J, Robinson M, Robinson L, et al. Chromosome 3p14 homozygous deletions and sequence analysis of FRA3B. Hum Mol Genet. 1997;6:193–203. doi: 10.1093/hmg/6.2.193. [DOI] [PubMed] [Google Scholar]
- 31.Mishmar D, Rahat A, Scherer SW, Nyakatura G, Hinzmann B, et al. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc Natl Acad Sci USA. 1998;95:8141–8146. doi: 10.1073/pnas.95.14.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ried K, Finnis M, Hobson L, Mangelsdorf M, Dayan S, et al. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum Mol Genet. 2000;9:1651–1663. doi: 10.1093/hmg/9.11.1651. [DOI] [PubMed] [Google Scholar]
- 33.Arlt MF, Xu B, Durkin SG, Casper AM, Kastan MB, et al. BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Mol Cell Biol. 2004;24:6701–6709. doi: 10.1128/MCB.24.15.6701-6709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang YH, Griffith J. Methylation of expanded CCG triplet repeat DNA from fragile X syndrome patients enhances nucleosome exclusion. J Biol Chem. 1996;271:22937–22940. [PubMed] [Google Scholar]
- 35.Yu S, Mangelsdorf M, Hewett D, Hobson L, Baker E, et al. Human chromosomal fragile site FRA16B is an amplified AT-rich minisatellite repeat. Cell. 1997;88:367–374. doi: 10.1016/s0092-8674(00)81875-9. [DOI] [PubMed] [Google Scholar]
- 36.Fan Q, Yao MC, Journals O. A long stringent sequence signal for programmed chromosome breakage in Tetrahymena thermophila. Nucleic Acids Res. 2000;28:895–900. doi: 10.1093/nar/28.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao MC. Ribosomal RNA gene amplification in Tetrahymena may be associated with chromosome breakage and DNA elimination. Cell. 1981;24:765–774. doi: 10.1016/0092-8674(81)90102-1. [DOI] [PubMed] [Google Scholar]
- 38.Culligan K, Tissier A, Britt A. ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell. 2004;16:1091–1104. doi: 10.1105/tpc.018903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, et al. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Gene Dev. 2000;14:2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gatei M, Shkedy D, Khanna KK, Uziel T, Shiloh Y, et al. Ataxia-telangiectasia: chronic activation of damage-responsive functions is reduced by alpha-lipoic acid. Oncogene. 2001;20:289–294. doi: 10.1038/sj.onc.1204111. [DOI] [PubMed] [Google Scholar]
- 41.Yarden RI, Pardo-Reoyo S, Sgagias M, Cowan KH, Brody LC. BRCA1 regulates the G 2/M checkpoint by activating Chk 1 kinase upon DNA damage. Nat Genet. 2002;30:285–289. doi: 10.1038/ng837. [DOI] [PubMed] [Google Scholar]
- 42.Ramanathan B, Smerdon MJ. Enhanced DNA repair synthesis in hyperacetylated nucleosomes. J Biol Chem. 1989;264:11026–11034. [PubMed] [Google Scholar]
- 43.Gu Y, Shen Y, Gibbs RA, Nelson DL. Identification of FMR2, a novel gene associated with the FRAXE CCG repeat and CpG island. Nat Genet. 1996;13:109–113. doi: 10.1038/ng0596-109. [DOI] [PubMed] [Google Scholar]
- 44.Pomponi MG, Neri G. Butyrate and acetyl-carnitine inhibit the cytogenetic expression of the fragile X in vitro. Am J Med Genet. 1994;51:447–450. doi: 10.1002/ajmg.1320510428. [DOI] [PubMed] [Google Scholar]
- 45.Conconi A, Widmer RM, Koller T, Sogo JM. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- 46.Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, et al. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell. 2004;13:599–609. doi: 10.1016/s1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- 47.McStay B. Nucleolar dominance: a model for rRNA gene silencing. Gene Dev. 2006;20:1207–1214. doi: 10.1101/gad.1436906. [DOI] [PubMed] [Google Scholar]
- 48.Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, et al. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Gene Dev. 2006;20:1283–1293. doi: 10.1101/gad.1417706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Y. Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Nucleic Acids Res. 2002;30:1465–1482. doi: 10.1093/nar/30.7.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durand-Dubief M, Sinha I, Fagerström-Billai F, Bonilla C, Wright A, et al. Specific functions for the fission yeast Sirtuins Hst2 and Hst4 in gene regulation and retrotransposon silencing. EMBO J. 2007;26:2477–2488. doi: 10.1038/sj.emboj.7601690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, et al. Response to comment on ‘HST2 mediates SIR2-independent life-span extension by calorie restriction’. Science. 2006;312:1312. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- 52.Sutherland G, Jacky P, Baker E. Heritable fragile sites on human chromosomes. XI. Factors affecting expression of fragile sites at 10q25, 16q22, and 17p12. Am J Hum Genet. 1984;36:110–122. [PMC free article] [PubMed] [Google Scholar]
- 53.Song YC, Gustafson JP. The physical location of fourteen RFLP markers in rice (Oryza sativa L.). Theor Appl Genet. 1995;90:113–119. doi: 10.1007/BF00221003. [DOI] [PubMed] [Google Scholar]
- 54.Li LJ, Arumuganathan K, Rines HW, Phillips RL, Riera-Lizarazu O, et al. Flow cytometric sorting of maize chromosome 9 from an oat-maize chromosome addition line. Theor Appl Genet. 2001;102:658–663. [Google Scholar]