Abstract
Egr1, a transcription factor rapidly induced by various stimuli including stress, can elevate transcription of genes for the catecholamine biosynthetic enzymes TH and PNMT. To examine if Egr1 also regulates dopamine β-hydroxylase (DBH) gene expression, PC12 cells were transfected with expression vector for full length or truncated inactive Egr1 and various DBH promoter-driven luciferase constructs. While Egr1 elevated TH promoter activity, DBH promoter activity was reduced. The reduction occurred as early as 4 h and reached maximal inhibition 16–40 h after transfection. Egr1 also reduced the expression of endogenous DBH mRNA and the induction of DBH promoter activity by cAMP. These effects were not observed with truncated Egr1 lacking the DNA binding domain. The first 247, but not 200, nucleotides of DBH promoter are sufficient for this suppression. Several putative Egr1 motifs were identified, and mutagenesis showed that the motif at −227/−224 is required. Binding of Egr1 to this region of the DBH promoter was verified by chromatin immunoprecipitation and electrophoretic mobility shift assays. This study demonstrates that DBH promoter contains at least one functional Egr1 motif; and indicates, for the first time, that Egr1 can play an inhibitory role in regulation of DBH gene transcription.
Keywords: Egr1, Dopamine β-hydroxylase, Transcription, PC12 cells
1. Introductin
Egr1 (Zif268, NGFI-A, TIS8, or Krox24) is a transcription factor with three zinc fingers of the Cys2His2 class [reviewed by [33]]. Egr1 binds to a GC-rich motif (5′-GCG(T/G)GGGCG-3′) through its three zinc finger DNA-binding domains [4], and modulates transcription of a number of genes that participate in various cellular functions [reviewed by [47,53]]. As an immediate early gene, many stimuli such as growth factors, hormones, neurotransmitters, stress, and cytokines induce Egr1 expression [5,7,17,23,36]. This response is presumed to have a key role in orchestrating a second wave of gene expression that underlies long-term effects of these factors on cell growth and differentiation.
Egr1, together with catecholamine biosynthetic enzymes, such as tyrosine hydroxylase (TH), dopamine β-hydroxylase (DBH), and phenylethanolamine N-methyltransferase (PNMT), are induced in adrenal medulla by different types of stress [23,27,30,34,35,38].
A functional Egr1 binding site on proximal TH promoter was previously identified and shown to interact with an AP1 motif in the induction of TH promoter activity [29,35]. Egr1 alone, or together with other factors such as glucocorticoids, also regulates PNMT, the final enzyme in epinephrine biosynthesis [8,28,52]. The role of Egr1 in transcriptional activation of the DBH gene has not been previously studied.
DBH (EC 1.14.17.1) catalyzes the synthesis of norepinephrine (NE) from dopamine (DA). Modulation of DBH activity or expression is implicated in several neuropsychiatric disorders [reviewed by [6]]. For example, an elevated DA/NE ratio resulting from decreased DBH expression or activity is associated with increased vulnerability to psychotic depression [26,41]. Polymorphisms of the human DBH gene have been linked to reactive schizophrenia, psychotic depression, alcoholism, attention deficit hyperactivity disorders, modulation of smoking and other reward-seeking behavior, and susceptibility to Parkinson’s disease [reviewed by [2,6,14]].
DBH gene expression is regulated in response to several pharmacological and physiological stimuli [1,16,18,25,45]. On the DBH promoter, there are several functional motifs that are homologous to known cis-acting regulatory elements including Sp1, three homeodomains, an overlapping cAMP/AP1 response element [(CRE)/AP1], and AP2 motifs [19,20,50]. The three homeobox motifs can bind Phox2a/Arix, which is important for cell-specific DBH gene expression. The overlapping CRE/AP1 motif can bind CREB and also AP1 family members [42,49,51].
In this study, we examined whether Egr1 can regulate DBH gene expression. Our results show that Egr1 suppressed DBH promoter activity, and that the region −227/−224 of the promoter is responsible for this effect. Chromatin immunoprecipitation (ChIP) and electrophoretic mobility shift assays (EMSA) indicated that this involves specific binding of Egr1 to the DBH promoter. Our results suggest, for the first time, that Egr1 may play a role in the physiological regulation of transcription of the DBH gene.
2. Results
2.1 Egr1 reduced the DBH promoter-driven luciferase activity
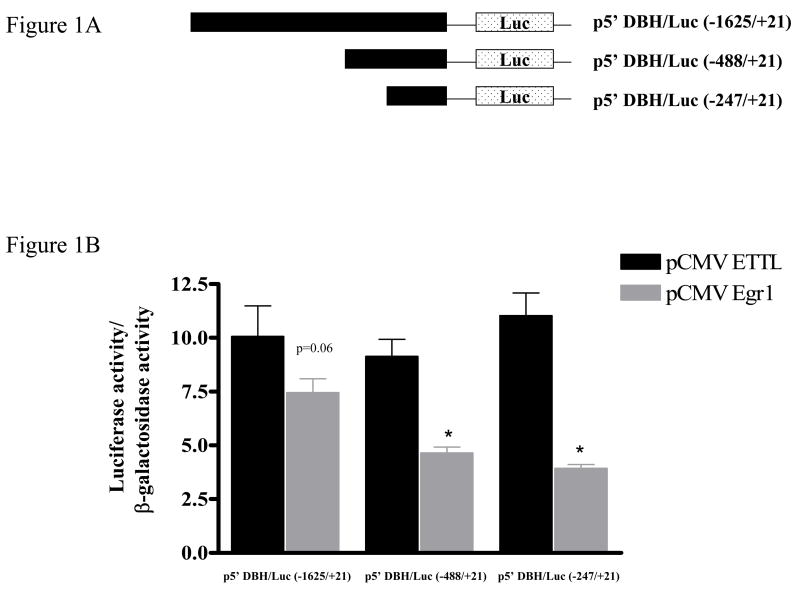
To determine whether DBH promoter-driven transcription can respond to Egr1, constructs with various lengths of rat DBH promoter directing the luciferase reporter gene were used. PC12 cells were cotransfected with either an expression vector for Egr1 (pCMV Egr1); or for truncated inactive Egr1 without the DNA binding domain (pCMV ETTL), and reporter constructs with luciferase activity under control of the first 1625, 488, or 247 bp upstream of the cap site (Fig. 1A). As shown in Figure 1B, the luciferase activity was significantly suppressed (more than 50%) in cells transfected with Egr1 expression vector and with p5′DBH/Luc (−488/+21) or p5′DBH/Luc (−247/+21). With p5′DBH/Luc (−1625/+21), there was about a 25% reduction in luciferase activity. Thus, the first 247 nucleotides of DBH promoter are sufficient for the Egr1-elicited reduction of promoter activity.
Figure 1.
Effect of Egr1 on DBH and TH promoter-driven luciferase activity. (A) Rat DBH promoter constructs used in this study are shown schematically. (B) PC12 cells were transfected with DBH promoter construct and either pCMV Egr1 or pCMV ETTL (truncated Egr1 without DNA binding domain) expression vector and pSV β-galactosidase, as a transfection efficiency control. Cells were harvested at 16 h after transfection. Luciferase activity was determined and normalized to β-galactosidase activity. * p < 0.001 versus respective pCMV ETTL group. Two-way ANOVA revealed analysis significant effect of Egr1 (p < 0.0001), and interaction with DBH promoter length (p = 0.0196). One way ANOVA analysis showed significant inter-group differences between various lengths of DBH promoter in the cells transfected with pCMV Egr1 (F2,11 = 19.65, p < 0.05).
As a positive control, we also cotransfected PC12 cells with a TH promoter construct [p5′TH-Luc (−272/+27)] and with either expression vector pCMV Egr1 or pCMV ETTL. Egr1 was found to increase the luciferase activity about threefold at 24 h post-transfection (data not shown). This is consistent with our previous finding showing that Egr1 can upregulate TH promoter activity [29,35].
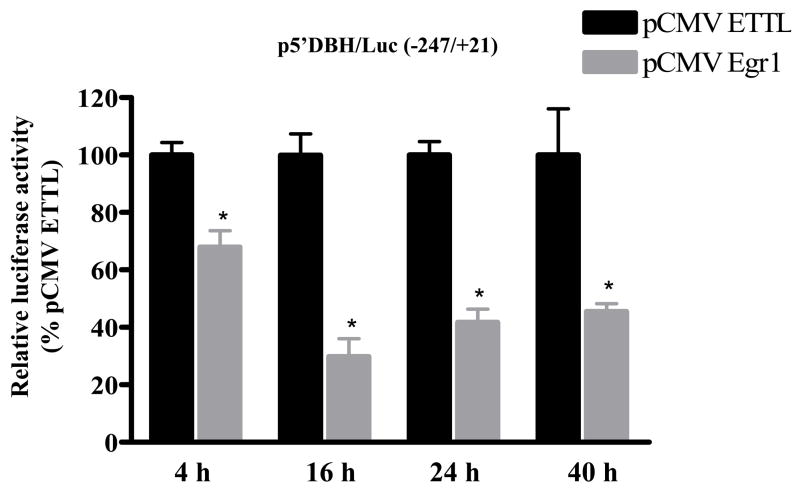
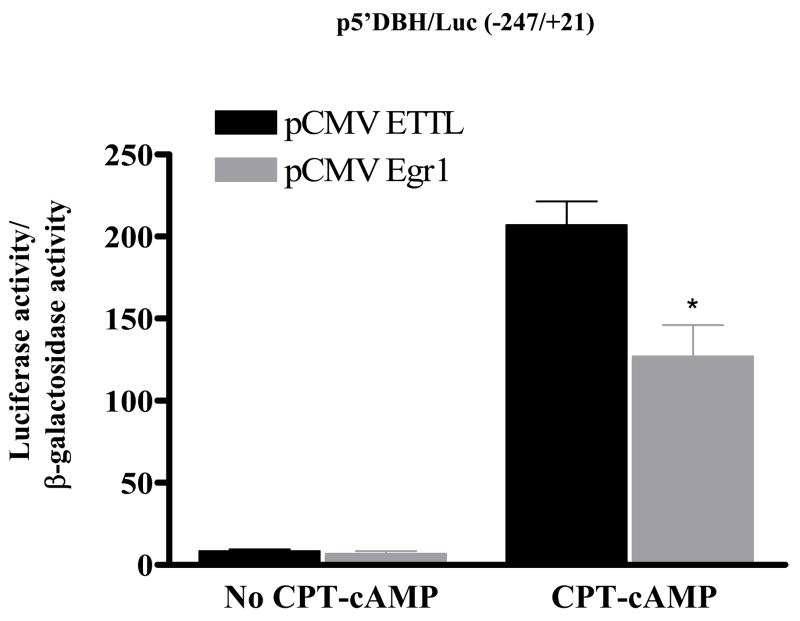
To analyze the time frame required for the effect of Egr1, the responses of DBH promoter activity were examined at various times post-transfection. As shown in Figure 2, a suppression of luciferase activity was observed as early as 4 h after transfection. A greater than 50% reduction was observed at 16–40 h post-transfection. Next, we analyzed whether Egr1 would also have an effect on stimulated DBH promoter activity. Cells transfected with pCMV Egr1 and p5′DBH/Luc (−247/+21) were treated with the non-hydrolysable cAMP analog (CPT-cAMP), which led to stimulation of DBH promoter activity. However, CPT-cAMP induced DBH promoter activity was reduced compared with the control cells transfected with pCMV ETTL (Fig. 3). Thus, Egr1 also reduced the cAMP-triggered stimulation of DBH promoter activity.
Figure 2.
Time course of inhibition of DBH promoter activity by Egr1. PC12 cells were transfected with p5′DBH/Luc (−247/+21) and either pCMV Egr1 or pCMV ETTL expression vector and pSV β-galactosidase. Cells were harvested at different time points after transfection as indicated. Luciferase activity was determined and normalized to β-galactosidase activity. * p < 0.001 versus respective pCMV ETTL group.
Figure 3.
Effect of Egr1 on induction of DBH promoter activity by cAMP. PC12 cells were transfected with p5′DBH/Luc (−247/+21) and either pCMV Egr1 or pCMV ETTL expression vector and pSV β-galactosidase. At 16 h after transfection, cells were treated with 200 μM CPT-cAMP and harvested 24 h later. Luciferase activity was determined and normalized to β-galactosidase activity. * p < 0.001 versus respective pCMV ETTL group.
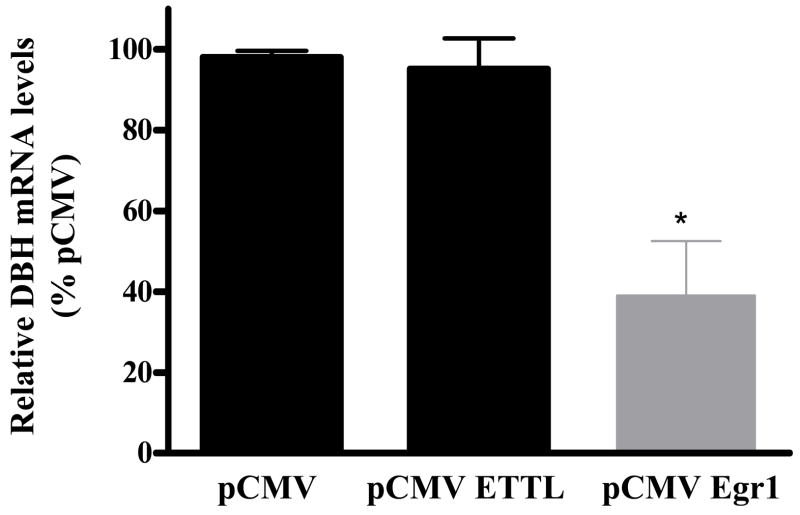
To examine whether Egr1 had a similar effect on endogenous DBH, the mRNA levels were determined by real-time RT-PCR from cells transfected with pCMV, pCMV ETTL, or pCMV Egr1. The results revealed that expression of Egr1 also significantly reduced the level of DBH mRNA (Fig. 4).
Figure 4.
Egr1 reduced endogenous DBH mRNA levels. PC12 cells were transfected with either pCMV, pCMV ETTL, or pCMV Egr1 expression vector. Total RNA was isolated and analyzed by real-time RT-PCR. Relative DBH mRNA levels are shown with the means ± S.E.M. relative to the pCMV group taken as 100. * p < 0.0001 versus the pCMV and pCMV ETTL groups.
2.2 Mapping the functional Egr1 site
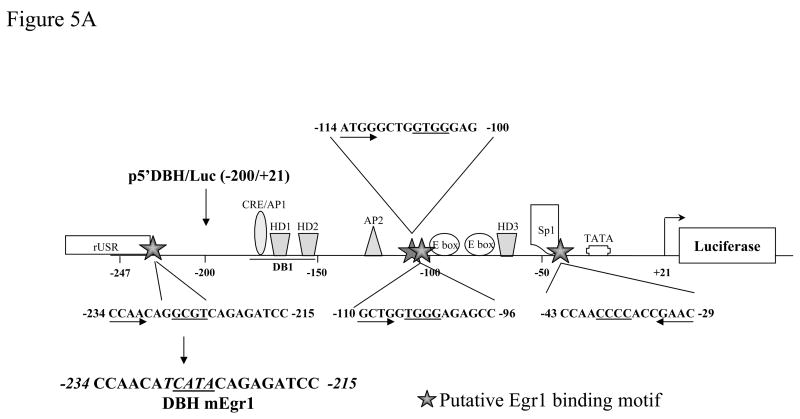
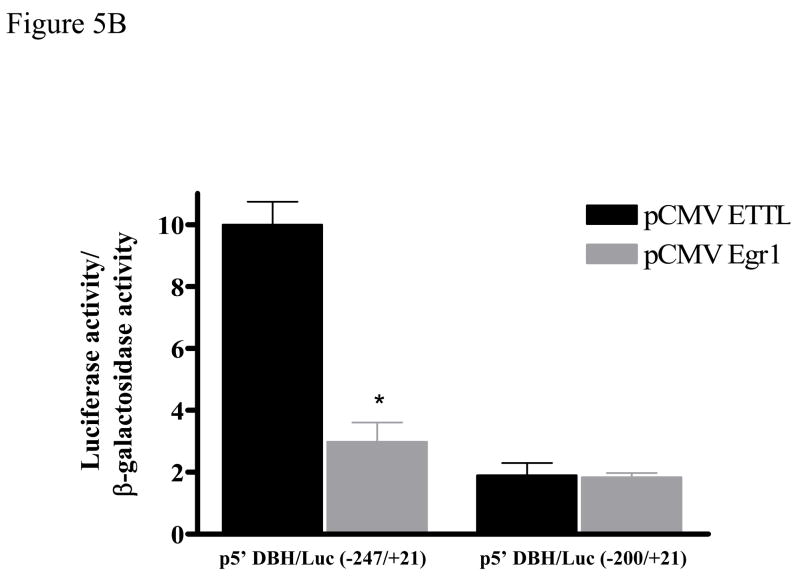
To identify whether the DBH promoter contains an Egr1 motif, which may be mediating the negative regulation, we used MatInspector Professional Software [37] to analyze the sequence between −247 to +21. This revealed four potential binding sites for Egr1 at positions −39/−36, −105/−102, −107/−104, and −227/−224 (Fig. 5A) with the core similarity/matrix similarity above 0.70/0.70 as compared to consensus Egr1 (Table 1). First we examined whether the Egr1 motif at position −227/−224 is important for regulation of DBH gene transcription. The truncated DBH promoter construct [p5′DBH/Luc (−200/+21)] was prepared and failed to elicit any changes in response to Egr1 (Fig. 5B). Thus, the element needed for Egr1-mediated inhibition is localized at the region between −247 to −200 bp of the DBH promoter.
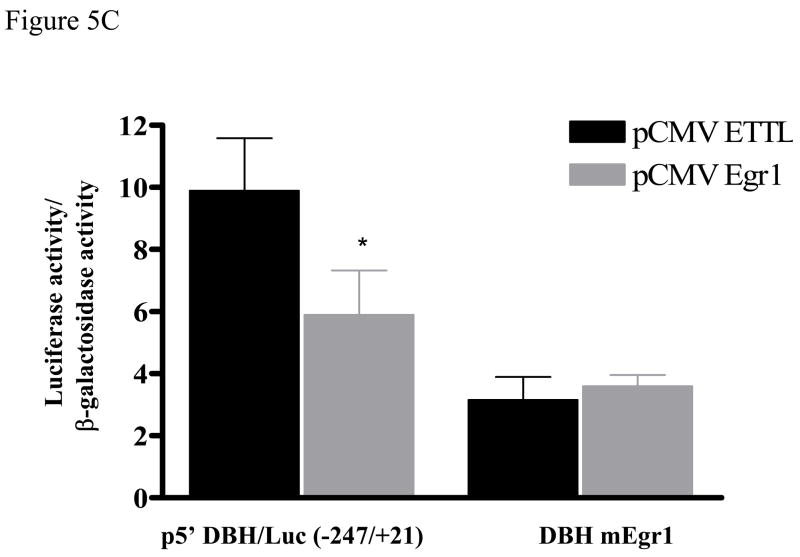
Figure 5.
Mapping the region of DBH promoter responsible for Egr1-mediated effect. A diagram of DBH (−247/+21) promoter regulatory elements is shown schematically (A). It includes the sequence of four potential Egr1 binding sites: −39/−36, −105/−102, −107/−104, and −227/−224. PC12 cells were transfected with various DBH promoter constructs: p5′DBH/Luc (−247/+21) and p5′DBH/Luc (−200/+21) (B), or the mutant DBH promoters construct (DBH mEgr1, −228 GGCGT −224 mutated to −228 TCATA −224) (C), and either pCMV Egr1or pCMV ETTL expression vector and pSV β-galactosidase. Luciferase activity was measured at 16 h after transfection. Data (mean ± S.E.M) are normalized to β-galactosidase activity. * p < 0.001 versus the respective pCMV ETTL group.
Table 1.
The sequences and the core similarity/matrix similarity of the four putative Egr1 binding sites in DBH promoter.
| Egr1 site | Sequence | Core similarity/matrix similarity |
|---|---|---|
| Consensus Egr1 | GCG(G/T)GGGCG | |
| DBH promoter −39/−36 |
|
0.80/0.73 |
| DBH promoter −105/−102 |
|
1.00/0.83 |
| DBH promoter −107/−104 |
|
0.83/0.71 |
| DBH promoter −227/−224 |
|
1.00/0.71 |
The data of the four putative DBH Egr1 binding sites were obtained by using MatInspector Software.
We further determined whether the putative Egr1-binding element in this region is required for the Egr1-mediated inhibition of DBH promoter-driven transcription. The mutated construct DBH mEgr1 with the GGCGT motif replaced with TCATA was generated (Fig. 5A). Mutation of this site abolished the Egr1 responsiveness of the promoter construct. Luciferase activity was similar with pCMV Egr1 and pCMV ETTL (Fig. 5C). These results indicate that the putative Egr1 motif at −227/−224 is needed for Egr1-mediated reduction of DBH promoter-driven transcription.
2.3 Egr1 interacts with the DBH promoter encompassing the region of −227 to −224
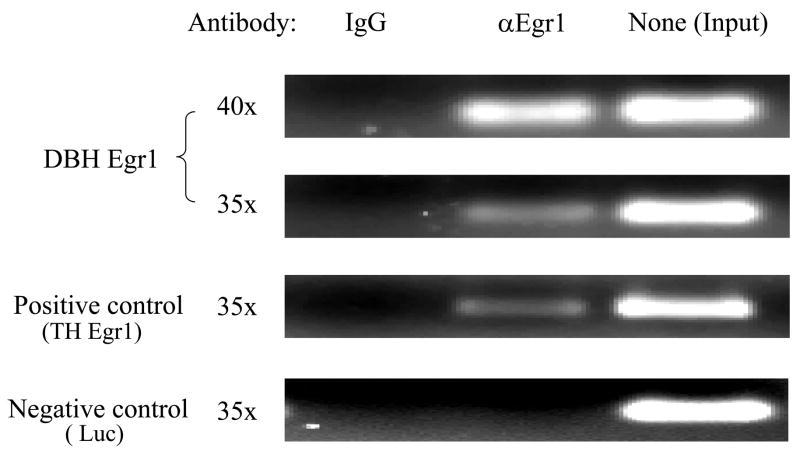
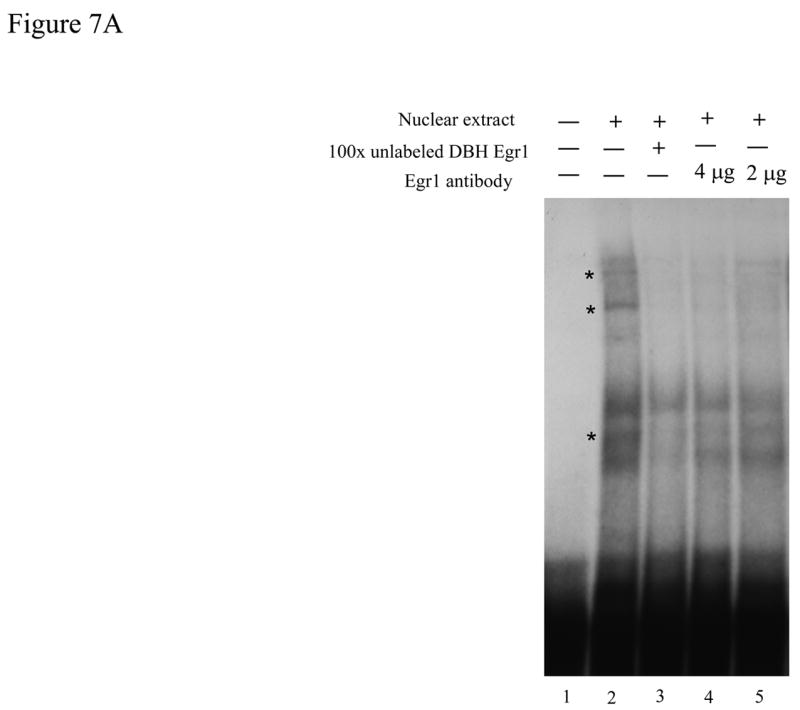
To examine whether Egr1 is associated physically with the putative binding site at the position −227/−224 in vivo, chromatin immunoprecipitation (ChIP) analysis was used. ChIP assay was performed on PC12 cells co-transfected with pCMV Egr1 expression vector and p5′DBH/Luc (−247/+21) reporter vector. Protein-DNA complexes were cross-linked with formaldehyde, and immunoprecipitated with antisera to Egr1 or normal rabbit IgG, as a negative control for non-specific antibody binding. PCR was performed to amplify the specific DNA fragments pulled down by the anti-Egr1 antibody or normal IgG using primers spanning the putative Egr1 binding site (−247/−204). Agarose gel electrophoresis showed amplification of DBH DNA fragments (−247/−204) in the input sample and in the sample immunoprecipitated with the Egr1 antibody, but not in the negative control (normal rabbit IgG) (Fig. 6).
Figure 6.
Egr1 binds to DBH promoter in vivo at the region spanning nucleotides −247 to −204. ChIP assay was carried out as described in the methods. PCR was performed to amplify the DNA fragment bound with Egr1 antibody (αEgr1) or normal IgG (as a negative control for the non-specific antibody binding), and the samples not immunoprecipitated (Input) by using specific primers for the putative DBH Egr1 binding site (−247/−204), the positive control (TH Egr1 motif at −192/−68) and the negative control (Luc at 1231/1275). A representative gel from 3 separate experiments is shown with PCR for either 35 (35×) or 40 (40×) cycles.
Amplifications for the positive control (TH −192/−68), which contains the known Egr1 binding site [34,35], was also seen in Egr1 immunoprecipitated samples. As an additional negative control, we used a luciferase coding region (Luc 1231/1275) that would also be present in the p5′DBH/Luc (−247/+21) plasmid. These data showed that in the cellular environment, Egr1 selectively binds the DBH promoter in a region spanning nucleotides −247 to −204.
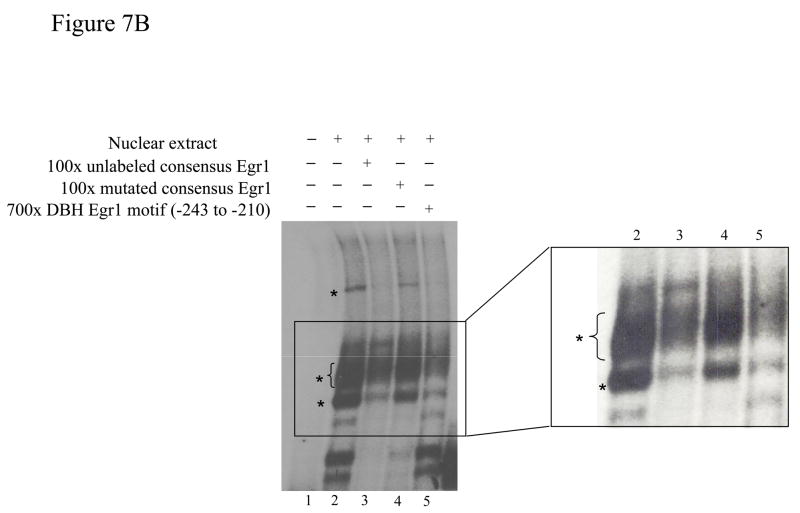
The DNA-protein interaction was further studied in vitro by electrophorectic mobility shift assays (EMSA). The nuclear extracts from pCMV Egr1 transfected cells formed several specific complexes with the 32P-labeled double-stranded DBH Egr1 oligonucleotides spanning −243 to −210 (Fig. 7A, lane 2). The specific complexes were competed with excess unlabeled oligonucleotides (lane 3). Preincubation with antibody against Egr1 (lanes 4 and 5) reduced formation of several complexes (shown by *). These may represent complexes with various phosphorylated forms of Egr1.
Figure 7.
Egr1 in vitro interacts with the putative Egr1 binding motif on the DBH promoter. (A) 32P-labeled DBH Egr1 double-stranded oligonucleotide (−243/−210) was incubated with nuclear extract from pCMV Egr1-transfected cells alone (lane 2), with 100 × excess unlabeled double-stranded oligonucleotide (lane 3), or with 4- or 2 μg specific Egr1 antibody (lanes 4 and 5). Lane 1 is the labeled DBH Egr1 oligonucleotide alone. Bands are indicated by * to show the displacement of the bands by Egr1 antibody. (B) 32P-labeled double-stranded consensus Egr1 oligonucleotide was incubated with nuclear extracts from pCMV Egr1-transfected cells alone (lane 2), with 100 × excess unlabeled double-stranded consensus oligonucleotide (lane 3), with 100 × excess unlabeled double-stranded mutated consensus Egr1 oligonucleotide (lane 4), or with 700 × excess unlabeled double-stranded DBH Egr1 oligonucleotide (−243/−210) (lane 5). Lane 1 is the labeled consensus Egr1 oligonucleotide alone. Bands are indicated by * to show the displacement by consensus Egr1 and DBH Egr1 oligonucleotides (−243/−210), but not by the oligonucleotide with mutated Egr1. Right panel is the amplified image of the representative gel.
To further characterize the specificity of the binding, this oligonucleotide (−243/−210 of DBH promoter) was used for competition with 32P-labeled consensus Egr1 oligonucleotide (Fig. 7B). This consensus Egr1 probe formed several bands with the nuclear extracts from pCMV Egr1 transfected cells (lane 2). The Egr1 specific complexes were identified. Excess mutated consensus Egr1 oligonucleotide (lane 4) did not compete for formation of several complexes (shown by *), compared with the excess consensus Egr1 oligonucleotide (lane 3). Preincubation of the consensus Egr1 oligonucleotide with excess DBH Egr1 oligonucleotide (−243/−210) also competed for these same complexes (lane 5), indicating that it is able to bind Egr1 and thus compete for specific Egr1 motifs.
3. Discussion
This is the first study to characterize the effect of Egr1 on DBH gene expression. We found that Egr1 can play an inhibitory role on DBH promoter activity; it also can reduce endogenous DBH mRNA. Mutation of the promoter indicated that the site at −227/−224 is involved in this inhibition. Both in vitro and in vivo binding studies confirmed the presence of an Egr1 binding site in this region of the proximal DBH promoter.
The Egr1 gene is one of the early response genes dramatically and rapidly induced upon stimulation with many environmental signals including growth factors, hormones, and neurotransmitters [13,17,23,40]. Egr1 functions as a convergence point for many signaling cascades and is thought to couple extracellular signals with long-term responses by altering gene expression of Egr1 target genes [47].
Under the same conditions whereby TH promoter activity was elevated by Egr1, DBH promoter activity and mRNA levels were reduced. The Egr1-mediated inhibition of DBH promoter-driven luciferase activity was evident as early as 4 h post-transfection (earliest time examined), and achieved the maximal reduction at 16–40 h post-transfection. The cAMP-triggered stimulation of DBH promoter activity was also lower in the presence of Egr1.
The suppression of DBH promoter activity by Egr1 was evident with several lengths of the promoter, with the proximal 247 nucleotides being sufficient. With the longest construct tested [p5′DBH/Luc (−1625/+21)], Egr1 also reduced DBH promoter activity, but not as much as with shorter constructs. In this regard, Afar et al. [1] also observed that elevation of reporter activity with 1 kb DBH promoter in response to cAMP was not as great as the responses with the shorter promoter constructs (−210, −190, and −173). It is possible that there is a more dominant regulatory element in the distal region of the first kb of the promoter. Alternatively, the conformation of the promoter may restrict access to Egr1 and other regulatory elements in the longer promoter, leading to an attenuated response.
We initially identified four putative Egr1 binding sites (−39/−36, −105/−102, −107/−104, and −227/−224) on the DBH proximal promoter. The deletion and mutagenesis experiments indicate that the motif at −227/−224 is required for Egr1-elicited reduction in DBH promoter activity. It remains to be determined whether the other motifs also contribute to the regulation. Since pCMV ETTL had no effect on DBH promoter-driven transcription or on endogenous mRNA levels, the Egr1 binding domain must be required for the suppression of DBH transcription. Both ChIP and EMSA experiments revealed that Egr1 could bind to a region of the DBH promoter within −247 to −204. An oligonucleotide containing this sequence was able to compete for binding of a consensus Egr1 oligonucleotide.
How might Egr1 suppress DBH transcription? There are several mechanisms whereby Egr1 may be mediating the decreased DBH promoter activity. It could be due to Egr1 directly binding to DNA or Egr1 indirectly interacting with other transcriptional coactivators, such as the cAMP-response-element-binding protein (CREB)-binding protein (CBP). Egr1 has been shown to interact with CBP/p300 physically and functionally [48]. CBP/p300 are large multifunctional nuclear proteins and their levels are limited [reviewed by [11]]. Egr1, when over-expressed, might compete with other transcriptional factors for a limited amount of CBP/p300. However, this does not appear to be the case for TH, which is elevated by over-expression of Egr1 under the same conditions.
It is also possible that Egr1 might be inhibiting the activation of DBH promoter-driven transcription by decreasing the availability of other transcription factors, such as Sp1. Egr1 has been implicated in inhibition of c-met expression by sequestering Sp1 as a transcriptional activator of c-met. The Sp1/Egr1 complex that was formed presumably resulted in a decreased availability of unbound Sp1 as a transcriptional activator [56]. Therefore, over-expressed Egr1 could repress DBH promoter-driven transcription by decreasing free Sp1.
DBH gene transcription is also regulated by AP1 proteins, especially c-Jun and JunD [51]. Over-expressed Egr1 has been shown to suppress the c-Jun mRNA expression induced by interleukin-17 [9]. Levkovitz and Baraban [22] showed that the zinc finger domain of Egr1 interacts with c-Jun, and then reduces transcriptional activation by c-Jun in response to nerve growth factor. Therefore, sequestration of c-Jun protein may mediate the Egr1-elicited reduction of the basal level of DBH gene expression.
The newly identified Egr1 binding site at −227/−224 is very close to the DBH silencer site [46]. Even though the DBH silencer site is only functional in non-catecholaminergic cells, it remains to be determined if an interaction between Egr1 and the DBH silencer site plays a role in the repression of DBH promoter activity observed here. In this regard, the DNA sequences near the Egr1 binding site were also shown to be important in determining whether Egr1 regulates gene transcription positively or negatively [10].
What is the impact of the ability of Egr1 to reduce DBH gene expression? It was surprising that Egr1 attenuates DBH gene transcription, while it was previously found to elevate transcription of TH and PNMT genes. A reduction of DBH gene expression might increase the dopamine levels in noradrenergic cells. In this regard, an elevated DA/NE ratio resulting from decreased DBH expression or activity is associated with increased vulnerability to psychotic depression [26,41]. Interestingly, with a single exposure of rats to immobilization stress, TH and PNMT mRNA in the adrenal medulla are elevated (transiently) as high as with repeated exposures to this stress [31,54,55], while induction of DBH mRNA is not as high as with repeated stress [21,24]. Both single and repeated stress induce Egr1 expression in the adrenal medulla [34], but there are additional transcriptional changes with repeated stress. These include prolonged phosphorylation of CREB and induction of Fra-2 [32,39], which might overcome any inhibition of DBH by Egr1. Moreover, in the locus coeruleus, where Egr1 is not induced by immobilization stress [15], DBH gene transcription is raised to a similar extent with both single and repeated exposures [44].
In conclusion, we have found that Egr1 can play an inhibitory role on DBH promoter-driven transcription. This inhibition requires a newly identified Egr1 response element at the −227/−224 region of DBH promoter. This inhibition appears to result from Egr1 directly bound to the DBH promoter. We speculate that Egr1 may reduce DBH gene transcription in vivo, thereby leading to altered DA/NE ratio. Further studies should determine the functional role of Egr1 in conjunction with other transcription factors in the regulation of DBH gene expression in catecholaminergic neuronal systems.
4. Experimental procedure
4.1 Cell culture
Rat pheochromocytoma (PC12) cells were grown as described previously [12,25]. For experiments, cells were plated at a density of 3 × 105 per well in 6-well tissue culture dishes (Falcon) on the day prior to transfection.
4.2 Plasmids and transfections
The p5′DBH/Luc (−1625/+21) and p5′DBH/Luc (−247/+21) constructs were generated from p5′DBH/Luc (−2236/+21) [43] by digestion with Kpn1 restriction enzyme (New England Biolabs, Ipswich, MA, USA) for DBH/Luc (−1625/+21) or with Kpn1 and Pst1 (New England Biolabs) for DBH/Luc (−247/+21) followed by religation.
Mutant reporter constructs p5′DBH/Luc (−200/+21) and DBH mEgr1 (−228 GGCGT −224 to −228 TCATA −224) with mutated Egr1 at this position were prepared from wild type p5′DBH/Luc (−247/+21) as the template for PCR using the QuikChange® site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) according to the supplier’s protocol. The following primers were used in the mutagenesis procedure. For p5′DBH/Luc (−200/+21): Sense 5′-CGAGGTCGACGGTATCATGGCCATTCTGCTTC-3′; Antisense 5′-GAAGCAGAATGGCCATGATACCGTCGACCTCG-3′; for DBH mEgr1: Sense 5′-AGAGAGTAGCTGTTTCCAACATCATACAGAGATCCATTGGAGGACATG-3′; Antisense 5′-CATGTCCTCCAATGGATCTCTGTATGATGTTGGAAACAGCTACTCTCT-3′.
The plasmids pCMV Egr1 and pCMV ETTL, which express a full-length and a truncated inactive Egr1 protein respectively, were kind gifts from Dr. Dona Wong (Harvard Medical School). In pCMV ETTL, the translation is terminated after serine 170, which removes the DNA binding domain [13].
The β-galactosidase (pSV β-gal) plasmid used as a control for transfection efficiency was purchased from Promega (Madison, WI, USA). The TH promoter construct was previously described [29].
All plasmids were isolated using EndoFree Plasmid Maxi Kit (Qiagen, Valencia, CA, USA) and dissolved in TRIS/Ethylenediaminetetraacetic acid (TE) buffer. The mutations were confirmed by sequencing (Davis Sequencing, Davis, CA, USA).
For transfection, 1 μg of each plasmid DNA was used for each culture well. The plasmid DNAs were mixed with SuperFect™ (Qiagen) according to the manufacturer’s instructions. At 16 h after transfection, some cells were treated with 200 μM 8-(4-chlorophenyl-thio) adenosine 3′,5′-cyclic monophosphate (CPT-cAMP) (Sigma-Aldrich, St. Louis, MO, USA) for 24 h. Cells were then harvested into 1 ml of PBS and collected by centrifugation at the indicated time points.
The PC12 cells were lysed with 100 μl of 1x Reporter Lysis Buffer (Promega). Firefly luciferase activity was determined using Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Luminescence was measured immediately with Luminometer, model TD-20/20 (Turner, Sunnyvale, CA, U.S.A.). As a control for transfection efficiency, β-galactosidase activity was performed as directed by the manufacture’s protocol (Promega). Briefly, 50 μl of the serially diluted standards and the lysates were added to 50 μl Assay 2x Buffer and the mixtures were incubated at 37°C for 2 h. Then, after adding the stop solution (150 μl of 1 M sodium carbonate), the absorbance of the samples and standards were measured at 420 nm in a microplate reader (Model Synergy HT, Bio-Tek Instruments, Inc., Winooski, VT, USA) and analyzed by KC4™ Signature software (Bio-Tek Instruments, Inc.). Luciferase activity was normalized to β-galactosidase activity.
At least four cell culture replicates were used for each treatment. All experiments were performed at least twice with similar results.
4. 3 RNA isolation and real time quantitative RT-PCR
Total RNA was isolated by using RNA-STAT 60 (Tel-Test, Friendswoods, TX, USA) and quantified by using Quan-it™ Ribo-Green® RNA assay kit (Molecular Probes, Eugene, OR, USA). RNA (300 ng) was reverse-transcribed with AMV reverse transcriptase (Roche Applied Science, Indianapolis, IN, USA) and 1 μM specific reverse transcription primer (1285/1268) for rat DBH gene (5′-GCACAGTAATCACCTTCC-3′) in 5 μl of reverse transcription mixture. Reverse transcription was carried out at 42°C for 1 h followed by 10 min at 85°C.
Real-time quantitative PCR was performed as described previously [3] by using LightCycler® 1.2 System (Roche Applied Science) with LightCycler® FastStart DNA Master SYBR Green I (Roche Applied Science) with the following primers: forward primer, 5′-CACCACATCATCATGTATGAGG-3′ (745/766); reverse primer, 5′-CCTGTCTGTGCAGTAGCCAG-3′ (1194/1175). For quantification assays, a standard curve was used and produced by amplification of several 10-fold dilutions of linearized plasmids containing DBH cDNA.
4.4. Chromatin immunoprecipitation (ChIP) assay
ChIP analysis was performed on PC12 cells transfected with pCMV-Egr1 expression vector and p5′DBH/Luc (−247/+21) reporter vector according to the manufacturer’s EZ-ChIP immunoprecipitation kit protocol (Millipore, Billerica, MA, USA). In brief, cells were cross-linked with 1% formaldehyde for 10 min, and the reaction was terminated by the addition of 0.25 M glycine. Cells were washed twice in ice-cold PBS, harvested with 1 ml cold PBS containing 1 x Protease inhibitor cocktail II (Millipore), then spun at 700 × g at 4°C for 5 min to pellet cells. Pellets were resuspended in ChIP SDS lysis buffer containing 1 x Protease inhibitor cocktail II and then sonicated to shear chromatin into 200–1000 bp fragments. The samples were pre-cleared with 60 μl of Protein G Agarose and subsequently incubated overnight with rotation either with 5 μg of rabbit anti-Egr1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) or with 1 μg of normal rabbit IgG (Santa Cruz Biotechnology) as a negative control for non-specific antibody binding at 4°C. Immunocomplexes were recovered with Protein G Agarose and washed consecutively with low salt immune complex wash buffer, high salt immune complex wash buffer, LiCl immune complex wash buffer, and TE buffer. Protein-DNA complexes were then eluted with ChIP elution buffer. The complexes (200 μl) were uncross-linked at 65°C overnight in 0.2 M NaCl, followed by 1 μl RNase A for 30 min at 37°C and 4 μl 0.5 M EDTA, 8 μl 1 M Tris-HCl, and 1 μl proteinase K for 2 h at 45°C. DNA fragments were then captured by an activated silica membrane filter column to separate from proteins and eluted to isolate the purified DNA. The ChIP-purified DNA fragments were screened by PCR for the rat DBH promoter region (−247/−204) encompassing the putative Egr1 binding sequence, the positive control (TH Egr1 −192/−68) with known Egr1 binding element, and negative control (Luc) lacking Egr1 binding element (pGL3/Luciferase basic vector 1231/1275). PCR reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA). For the DBH Egr1, the PCR conditions were an initial denaturation cycle (2 min, 94°C), 35–40 cycles of denaturation (20 sec, 94°C), annealing (20 sec, 52.5°C), and extension (25 sec, 72°C), followed by a final extension cycle (2 min, 72°C) terminating at 4°C. For the positive control (TH Egr1), the PCR conditions were an initial denaturation cycle (2 min, 94°C), 35 cycles of denaturation (20 sec, 94°C), annealing (20 sec, 57°C), and extension (25 sec, 72°C), followed by a final extension cycle (2 min, 72°C) terminating at 4°C. For the negative control (Luc), the PCR conditions were an initial denaturation cycle (2 min, 94°C), 35 cycles of denaturation (20 sec, 94°C), annealing (20 sec, 53.5°C), and extension (25 sec, 72°C), followed by a final extension cycle (2 min, 72°C) terminating at 4°CThe sequence for oligonucleotide primers were:
| DBH Egr1: | sense 5′-GAGAGTAGCTGTTTCCAAC-3′;
antisense 5′-TGTCCTCCAATGGATC-3′ |
| Positive control (TH Egr1): | sense 5′-GGTGCCTGTGACAGTGGAT-3′;
antisense 5′-TCCTACCTCCTGCGCATC-3′ |
| Negative control (Luc): | sense 5′-CTGGGCGTTAATCAAAGAGG-3′;
antisense 5′-CATAGGACCTCTCACACACA-3′ |
The PCR products were visualized on 2% agarose gels in 1 × TAE buffer stained with ethidium bromide.
4.5 Preparation of nuclear extracts and electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared as described previously [35] from PC12 cells transfected either with pCMV Egr1 or with pCMV empty vectors. Protein concentrations were determined with the Bio-Rad Bradford assay kit (Hercules, CA, USA).
EMSA was performed with the following oligonucleotide and its complementary strand synthesized by Integrated DNA technologies, Inc. (Coralville, IA, USA):
DBH Egr1, 34 bp 5′-AGCTGTTTCCAACAGGCGTCAGAGATCCATTGGA-3′ (−243 to −210). Egr1 consensus (5′-GGATCCAGCGGGGGCGAGCGGGGGCGA-3′) and mutated consensus oligonucleotides (5′-GGATCCAGCTAGGGCGAGCGGGGGCGA-3′) from Santa Cruz Biotechnology.
The experiments were conducted as described before [35]. PC12 nuclear extracts (4 μg) were incubated in 15 μl of reaction buffer containing 10 mM HEPES, pH 7.5, 2.5 mM MgCl2, 50 mM NaCl, 0.5 mM DTT, 4% glycerol, 1 μg of double-stranded poly(dI-dC), 1 μg BSA, and radiolabeled double-stranded oligonucleotide probes (40,000 cpm/reaction) for 30 min at room temperature. In the competition experiments, the unlabeled double-stranded oligonucleotide probes were added prior to the addition of nuclear extracts. For the supershift experiments, the nuclear extracts were incubated with anti-Egr1 antibody for 30 min at room temperature first, and then mixed with reaction buffer containing the radiolabeled double-stranded oligonucleotide probes. Protein-DNA complexes were analyzed on 6% nondenaturing polyacrylamide gels.
4. 6 Data analysis
Statistical significance was determined by Student’s t-test for experiments with two groups or by performing an analysis of variance (ANOVA) followed by Tukey post-test examination for experiments with more than two groups. A level of p < 0.05 was considered statistically significant. Statistical analyses were performed using GraphPad Prism and InStat Programs (GraphPad Software, Inc., San Diego, CA, USA).
Acknowledgments
We gratefully acknowledge support by NIH grants NS28869 and NS44218. We thank Dr. Bistra Nankova (New York Medical College) for guidance with the mobility shift assays and Dr. Dona Wong (Harvard Medical School) for providing the pCMV Egr1 and pCMV ETTL vectors. We thank Dr. V Gueorguiev for help and suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Afar R, Silverman R, Aguanno A, Albert VR. Positive and negative elements contribute to the cell-specific expression of the rat dopamine beta-hydroxylase gene. Brain Res Mol Brain Res. 1996;36:79–92. doi: 10.1016/0169-328x(95)00247-p. [DOI] [PubMed] [Google Scholar]
- 2.Anney RJ, Olsson CA, Lotfi-Miri M, Patton GC, Williamson R. Nicotine dependence in a prospective population-based study of adolescents: the protective role of a functional tyrosine hydroxylase polymorphism. Pharmacogenetics. 2004;14:73–81. doi: 10.1097/00008571-200402000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Cheng SY, Glazkova D, Serova L, Sabban EL. Effect of prolonged nicotine infusion on response of rat catecholamine biosynthetic enzymes to restraint and cold stress. Pharmacol Biochem Behav. 2005;82:559–68. doi: 10.1016/j.pbb.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Christy B, Nathans D. DNA binding site of the growth factor-inducible protein Zif268. Proc Natl Acad Sci U S A. 1989;86:8737–41. doi: 10.1073/pnas.86.22.8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen DM. Urea-inducible Egr-1 transcription in renal inner medullary collecting duct (mIMCD3) cells is mediated by extracellular signal-regulated kinase activation. Proc Natl Acad Sci U S A. 1996;93:11242–7. doi: 10.1073/pnas.93.20.11242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cubells JF, Zabetian CP. Human genetics of plasma dopamine beta-hydroxylase activity: applications to research in psychiatry and neurology. Psychopharmacology (Berl) 2004;174:463–76. doi: 10.1007/s00213-004-1840-8. [DOI] [PubMed] [Google Scholar]
- 7.Dieckgraefe BK, Weems DM. Epithelial injury induces egr-1 and fos expression by a pathway involving protein kinase C and ERK. Am J Physiol. 1999;276:G322–30. doi: 10.1152/ajpgi.1999.276.2.G322. [DOI] [PubMed] [Google Scholar]
- 8.Ebert SN, Balt SL, Hunter JP, Gashler A, Sukhatme V, Wong DL. Egr-1 activation of rat adrenal phenylethanolamine N-methyltransferase gene. J Biol Chem. 1994;269:20885–98. [PubMed] [Google Scholar]
- 9.Faour WH, Alaaeddine N, Mancini A, He QW, Jovanovic D, Di Battista JA. Early growth response factor-1 mediates prostaglandin E2-dependent transcriptional suppression of cytokine-induced tumor necrosis factor-alpha gene expression in human macrophages and rheumatoid arthritis-affected synovial fibroblasts. J Biol Chem. 2005;280:9536–46. doi: 10.1074/jbc.M414067200. [DOI] [PubMed] [Google Scholar]
- 10.Gashler AL, Swaminathan S, Sukhatme VP. A novel repression module, an extensive activation domain, and a bipartite nuclear localization signal defined in the immediate-early transcription factor Egr-1. Mol Cell Biol. 1993;13:4556–71. doi: 10.1128/mcb.13.8.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldman PS, Tran VK, Goodman RH. The multifunctional role of the co-activator CBP in transcriptional regulation. Recent Prog Horm Res. 1997;52:103–19. discussion 119–20. [PubMed] [Google Scholar]
- 12.Gueorguiev VD, Cheng SY, Sabban EL. Prolonged Activation of cAMP-response Element-binding Protein and ATF-2 Needed for Nicotine-triggered Elevation of Tyrosine Hydroxylase Gene Transcription in PC12 Cells. J Biol Chem. 2006;281:10188–95. doi: 10.1074/jbc.M513806200. [DOI] [PubMed] [Google Scholar]
- 13.Gupta MP, Gupta M, Zak R, Sukhatme VP. Egr-1, a serum-inducible zinc finger protein, regulates transcription of the rat cardiac alpha-myosin heavy chain gene. J Biol Chem. 1991;266:12813–6. [PubMed] [Google Scholar]
- 14.Healy DG, Abou-Sleiman PM, Ozawa T, Lees AJ, Bhatia K, Ahmadi KR, Wullner U, Berciano J, Moller JC, Kamm C, Burk K, Barone P, Tolosa E, Quinn N, Goldstein DB, Wood NW. A functional polymorphism regulating dopamine beta-hydroxylase influences against Parkinson’s disease. Ann Neurol. 2004;55:443–6. doi: 10.1002/ana.20063. [DOI] [PubMed] [Google Scholar]
- 15.Hebert MA, Serova LI, Sabban EL. Single and repeated immobilization stress differentially trigger induction and phosphorylation of several transcription factors and mitogen-activated protein kinases in the rat locus coeruleus. J Neurochem. 2005;95:484–98. doi: 10.1111/j.1471-4159.2005.03386.x. [DOI] [PubMed] [Google Scholar]
- 16.Ishiguro H, Yamada K, Ichino N, Nagatsu T. Identification and characterization of a novel phorbol ester-responsive DNA sequence in the 5′-flanking region of the human dopamine beta-hydroxylase gene. J Biol Chem. 1998;273:21941–9. doi: 10.1074/jbc.273.34.21941. [DOI] [PubMed] [Google Scholar]
- 17.Kaufmann K, Bach K, Thiel G. The extracellular signal-regulated protein kinases Erk1/Erk2 stimulate expression and biological activity of the transcriptional regulator Egr-1. Biol Chem. 2001;382:1077–81. doi: 10.1515/BC.2001.135. [DOI] [PubMed] [Google Scholar]
- 18.Kilbourne EJ, Nankova BB, Lewis EJ, McMahon A, Osaka H, Sabban DB, Sabban EL. Regulated expression of the tyrosine hydroxylase gene by membrane depolarization. Identification of the responsive element and possible second messengers. J Biol Chem. 1992;267:7563–9. [PubMed] [Google Scholar]
- 19.Kim HS, Hong SJ, LeDoux MS, Kim KS. Regulation of the tyrosine hydroxylase and dopamine beta-hydroxylase genes by the transcription factor AP-2. J Neurochem. 2001;76:280–94. doi: 10.1046/j.1471-4159.2001.00044.x. [DOI] [PubMed] [Google Scholar]
- 20.Kim HS, Seo H, Yang C, Brunet JF, Kim KS. Noradrenergic-specific transcription of the dopamine beta-hydroxylase gene requires synergy of multiple cis-acting elements including at least two Phox2a-binding sites. J Neurosci. 1998;18:8247–60. doi: 10.1523/JNEUROSCI.18-20-08247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kvetnansky R, Sabban EL. Stress and molecular biology of neurotransmitter-related enzymes. Ann N Y Acad Sci. 1998;851:342–56. doi: 10.1111/j.1749-6632.1998.tb09008.x. [DOI] [PubMed] [Google Scholar]
- 22.Levkovitz Y, Baraban JM. A dominant negative Egr inhibitor blocks nerve growth factor-induced neurite outgrowth by suppressing c-Jun activation: role of an Egr/c-Jun complex. J Neurosci. 2002;22:3845–54. doi: 10.1523/JNEUROSCI.22-10-03845.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Kvetnansky R, Serova L, Sollas A, Sabban EL. Increased susceptibility to transcriptional changes with novel stressor in adrenal medulla of rats exposed to prolonged cold stress. Brain Res Mol Brain Res. 2005;141:19–29. doi: 10.1016/j.molbrainres.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 24.McMahon A, Kvetnansky R, Fukuhara K, Weise VK, Kopin IJ, Sabban EL. Regulation of tyrosine hydroxylase and dopamine beta-hydroxylase mRNA levels in rat adrenals by a single and repeated immobilization stress. J Neurochem. 1992;58:2124–30. doi: 10.1111/j.1471-4159.1992.tb10954.x. [DOI] [PubMed] [Google Scholar]
- 25.McMahon A, Sabban EL. Regulation of expression of dopamine beta-hydroxylase in PC12 cells by glucocorticoids and cyclic AMP analogues. J Neurochem. 1992;59:2040–7. doi: 10.1111/j.1471-4159.1992.tb10092.x. [DOI] [PubMed] [Google Scholar]
- 26.Meyers BS, Alexopoulos GS, Kakuma T, Tirumalasetti F, Gabriele M, Alpert S, Bowden C, Meltzer HY. Decreased dopamine beta-hydroxylase activity in unipolar geriatric delusional depression. Biol Psychiatry. 1999;45:448–52. doi: 10.1016/s0006-3223(98)00085-7. [DOI] [PubMed] [Google Scholar]
- 27.Morita K, Bell RA, Siddall BJ, Wong DL. Neural stimulation of Egr-1 messenger RNA expression in rat adrenal gland: possible relation to phenylethanolamine N-methyltransferase gene regulation. J Pharmacol Exp Ther. 1996;279:379–85. [PubMed] [Google Scholar]
- 28.Morita K, Ebert SN, Wong DL. Role of transcription factor Egr-1 in phorbol ester-induced phenylethanolamine N-methyltransferase gene expression. J Biol Chem. 1995;270:11161–7. doi: 10.1074/jbc.270.19.11161. [DOI] [PubMed] [Google Scholar]
- 29.Nakashima A, Ota A, Sabban EL. Interactions between Egr1 and AP1 factors in regulation of tyrosine hydroxylase transcription. Brain Res Mol Brain Res. 2003;112:61–9. doi: 10.1016/s0169-328x(03)00047-0. [DOI] [PubMed] [Google Scholar]
- 30.Nankova B, Devlin D, Kvetnansky R, Kopin IJ, Sabban EL. Repeated immobilization stress increases the binding of c-Fos-like proteins to a rat dopamine beta-hydroxylase promoter enhancer sequence. J Neurochem. 1993;61:776–9. doi: 10.1111/j.1471-4159.1993.tb02188.x. [DOI] [PubMed] [Google Scholar]
- 31.Nankova B, Kvetnansky R, McMahon A, Viskupic E, Hiremagalur B, Frankle G, Fukuhara K, Kopin IJ, Sabban EL. Induction of tyrosine hydroxylase gene expression by a nonneuronal nonpituitary-mediated mechanism in immobilization stress. Proc Natl Acad Sci U S A. 1994;91:5937–41. doi: 10.1073/pnas.91.13.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nankova BB, Rivkin M, Kelz M, Nestler EJ, Sabban EL. Fos-related antigen 2: potential mediator of the transcriptional activation in rat adrenal medulla evoked by repeated immobilization stress. J Neurosci. 2000;20:5647–53. doi: 10.1523/JNEUROSCI.20-15-05647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Donovan KJ, Tourtellotte WG, Millbrandt J, Baraban JM. The EGR family of transcription-regulatory factors: progress at the interface of molecular and systems neuroscience. Trends Neurosci. 1999;22:167–73. doi: 10.1016/s0166-2236(98)01343-5. [DOI] [PubMed] [Google Scholar]
- 34.Papanikolaou NA, Sabban EL. Sp1/Egr1 motif: a new candidate in the regulation of rat tyrosine hydroxylase gene transcription by immobilization stress. J Neurochem. 1999;73:433–6. doi: 10.1046/j.1471-4159.1999.0730433.x. [DOI] [PubMed] [Google Scholar]
- 35.Papanikolaou NA, Sabban EL. Ability of Egr1 to activate tyrosine hydroxylase transcription in PC12 cells. Cross-talk with AP-1 factors. J Biol Chem. 2000;275:26683–9. doi: 10.1074/jbc.M000049200. [DOI] [PubMed] [Google Scholar]
- 36.Park JA, Koh JY. Induction of an immediate early gene egr-1 by zinc through extracellular signal-regulated kinase activation in cortical culture: its role in zinc-induced neuronal death. J Neurochem. 1999;73:450–6. doi: 10.1046/j.1471-4159.1999.0730450.x. [DOI] [PubMed] [Google Scholar]
- 37.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–84. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabban EL, Kvetnansky R. Stress-triggered activation of gene expression in catecholaminergic systems: dynamics of transcriptional events. Trends Neurosci. 2001;24:91–8. doi: 10.1016/s0166-2236(00)01687-8. [DOI] [PubMed] [Google Scholar]
- 39.Sabban EL, Liu X, Serova L, Gueorguiev V, Kvetnansky R. Stress triggered changes in gene expression in adrenal medulla: transcriptional responses to acute and chronic stress. Cell Mol Neurobiol. 2006;26:843–54. doi: 10.1007/s10571-006-9069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato K, Ishikawa K, Ui M, Okajima F. Sphingosine 1-phosphate induces expression of early growth response-1 and fibroblast growth factor-2 through mechanism involving extracellular signal-regulated kinase in astroglial cells. Brain Res Mol Brain Res. 1999;74:182–9. doi: 10.1016/s0169-328x(99)00279-x. [DOI] [PubMed] [Google Scholar]
- 41.Schatzberg AF, Rothschild AJ. Psychotic (delusional) major depression: should it be included as a distinct syndrome in DSM-IV? Am J Psychiatry. 1992;149:733–45. doi: 10.1176/ajp.149.6.733. [DOI] [PubMed] [Google Scholar]
- 42.Seo H, Yang C, Kim HS, Kim KS. Multiple protein factors interact with the cis-regulatory elements of the proximal promoter in a cell-specific manner and regulate transcription of the dopamine beta-hydroxylase gene. J Neurosci. 1996;16:4102–12. doi: 10.1523/JNEUROSCI.16-13-04102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Serova L, Rivkin M, Nakashima A, Sabban EL. Estradiol stimulates gene expression of norepinephrine biosynthetic enzymes in rat locus coeruleus. Neuroendocrinology. 2002;75:193–200. doi: 10.1159/000048237. [DOI] [PubMed] [Google Scholar]
- 44.Serova LI, Nankova BB, Feng Z, Hong JS, Hutt M, Sabban EL. Heightened transcription for enzymes involved in norepinephrine biosynthesis in the rat locus coeruleus by immobilization stress. Biol Psychiatry. 1999;45:853–62. doi: 10.1016/s0006-3223(98)90360-2. [DOI] [PubMed] [Google Scholar]
- 45.Shaskus J, Greco D, Asnani LP, Lewis EJ. A bifunctional genetic regulatory element of the rat dopamine beta-hydroxylase gene influences cell type specificity and second messenger-mediated transcription. J Biol Chem. 1992;267:18821–30. [PubMed] [Google Scholar]
- 46.Shaskus J, Zellmer E, Lewis EJ. A negative regulatory element in the rat dopamine beta-hydroxylase gene contributes to the cell type specificity of expression. J Neurochem. 1995;64:52–60. doi: 10.1046/j.1471-4159.1995.64010052.x. [DOI] [PubMed] [Google Scholar]
- 47.Silverman ES, Collins T. Pathways of Egr-1-mediated gene transcription in vascular biology. Am J Pathol. 1999;154:665–70. doi: 10.1016/S0002-9440(10)65312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silverman ES, Du J, Williams AJ, Wadgaonkar R, Drazen JM, Collins T. cAMP-response-element-binding-protein-binding protein (CBP) and p300 are transcriptional co-activators of early growth response factor-1 (Egr-1) Biochem J. 1998;336(Pt 1):183–9. doi: 10.1042/bj3360183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swanson DJ, Adachi M, Lewis EJ. The homeodomain protein Arix promotes protein kinase A-dependent activation of the dopamine beta-hydroxylase promoter through multiple elements and interaction with the coactivator cAMP-response element-binding protein-binding protein. J Biol Chem. 2000;275:2911–23. doi: 10.1074/jbc.275.4.2911. [DOI] [PubMed] [Google Scholar]
- 50.Swanson DJ, Zellmer E, Lewis EJ. The homeodomain protein Arix interacts synergistically with cyclic AMP to regulate expression of neurotransmitter biosynthetic genes. J Biol Chem. 1997;272:27382–92. doi: 10.1074/jbc.272.43.27382. [DOI] [PubMed] [Google Scholar]
- 51.Swanson DJ, Zellmer E, Lewis EJ. AP1 proteins mediate the cAMP response of the dopamine beta-hydroxylase gene. J Biol Chem. 1998;273:24065–74. doi: 10.1074/jbc.273.37.24065. [DOI] [PubMed] [Google Scholar]
- 52.Tai TC, Morita K, Wong DL. Role of Egr-1 in cAMP-dependent protein kinase regulation of the phenylethanolamine N-methyltransferase gene. J Neurochem. 2001;76:1851–9. doi: 10.1046/j.1471-4159.2001.00189.x. [DOI] [PubMed] [Google Scholar]
- 53.Thiel G, Cibelli G. Regulation of life and death by the zinc finger transcription factor Egr-1. J Cell Physiol. 2002;193:287–92. doi: 10.1002/jcp.10178. [DOI] [PubMed] [Google Scholar]
- 54.Viskupic E, Kvetnansky R, Sabban EL, Fukuhara K, Weise VK, Kopin IJ, Schwartz JP. Increase in rat adrenal phenylethanolamine N-methyltransferase mRNA level caused by immobilization stress depends on intact pituitary-adrenocortical axis. J Neurochem. 1994;63:808–14. doi: 10.1046/j.1471-4159.1994.63030808.x. [DOI] [PubMed] [Google Scholar]
- 55.Wong DL, Her S, Tai TC, Bell RA, Rusnak M, Farkas R, Kvetnansky R, Shih JC. Stress-Induced Expression of Phenylethanolamine N-Methytransferase: Normal and Knock out Animals. In: Kvetnansky R, editor. Stress: Neural, Endocrine and Molecular Studies. Taylor and Francis; London: 2002. pp. 129–135. [Google Scholar]
- 56.Zhang X, Liu Y. Suppression of HGF receptor gene expression by oxidative stress is mediated through the interplay between Sp1 and Egr-1. Am J Physiol Renal Physiol. 2003;284:F1216–25. doi: 10.1152/ajprenal.00426.2002. [DOI] [PubMed] [Google Scholar]