Abstract
Purpose
Lens fiber cell differentiation is marked by the onset of βB1-crystallin expression and is controlled by the cooperative action of a set of transcription factors including Prox1, an atypical homeodomain protein. Previously, the authors reported that Prox1 directly interacts with the OL2 element found in the chicken βB1-crystallin basal promoter to activate the expression of this gene. Here they mapped the location of activating and repressing sequences of the full-length chicken βB1-crystallin promoter (−432/+30) in lens epithelial cells, annular pad cells, and intact lens and characterized Prox1-binding sites found in this region.
Methods
Transfection analysis and transgenic mice were used to characterize upstream regions of the chicken βB1-crystallin gene. DNaseI footprinting and chromatin immunoprecipitation was performed to identify Prox1-binding sites, and transfection analyses were used to characterize these sites functionally.
Results
Sequences between −152 and −432 of the chicken βB1-crystallin promoter mediated either promoter activation or repression, depending on the stage of lens differentiation tested. Two new Prox1-binding sites were found in this region that bound Prox1 more avidly than the OL2 element. However, neither binding site conferred Prox1-mediated activation on a heterologous promoter; instead, each allowed Prox1 to repress promoter function.
Conclusions
The function of the upstream region of the chicken βB1-crystallin promoter changes depending on cellular context. These data suggest that Prox1 function as a transcriptional activator could be regulated at the DNA level based on the characteristics of the responsive elements.
The ocular lens is a transparent tissue consisting of a monolayer of anterior epithelial cells, with the remainder of the lens consisting of terminally differentiated fiber cells.1 During embryonic development, the lens is first apparent as a placodal thickening of the head ectoderm, which invaginates to form the lens vesicle. The cells in the anterior portion of the vesicle remain proliferative and form the lens epithelial cells, whereas the cells of the posterior lens vesicle leave the cell cycle and elongate to become the primary lens fibers.2,3 The lens increases in size as epithelial cells found along the lens equator leave the cell cycle and elongate into additional fiber cells.4 The fiber cell differentiation program is marked by dramatic changes in gene expression regulated by the transcription factors responsible for the initial development of the lens and additional ones whose expression initiates later.5
The clarity and refractive properties of the lens are largely conferred by the presence of high levels of water-soluble protein, the crystallins, in lens fiber cells.6 The expression of βB1-crystallin is highly lens fiber cell preferred,7,8 and its expression levels reach 2.8% of total mRNA in 1-day-old chicken lens fibers9 and 8.5% of the water-soluble protein of the newborn mouse lens.10 Previously, we determined that the −432/+30 fragment of the chicken βB1-crystallin promoter was sufficient to drive reporter gene expression, specifically in the lens fiber cells of transgenic mice,8 at levels approaching those of the endogenous mouse βB1-crystallin gene,11 though the overall sequence similarity between this fragment and similar sequences from mammalian βB1-crystallin genes is not high.12 Deletion analysis revealed that the −432/+30 fragment was 10-fold more active than a −152/+30 fragment and 200-fold more active than a −126/+30 fragment in transgenic lenses.8,13 In contrast, transfection analysis of these promoter fragments into chicken patched lens epithelial cells, which only express low levels of βB1-crystallin, revealed that the −432/+30 promoter was less active than the minimal promoter (−126/+30),14 suggesting that lens epithelial cells repress βB1-crystallin promoter activity through elements located between −126 and −432.
Prox1, the vertebrate homolog of Drosophila Prospero, is a divergent homeodomain protein with a highly conserved C terminus containing the homeodomain and a novel Prospero domain that folds into a single structural unit capable of sequence-specific DNA binding.15–19 Prox1 is expressed in the lens epithelium; however, its expression level upregulates sharply as lens epithelial cells differentiate into lens fiber cells.15,16,20 Prox1 is essential for fiber cell differentiation because primary lens fibers do not properly exit the cell cycle and never elongate in Prox1 null lenses.21 Prox1 is also essential for many other developmental processes, including cell fate determination in the retina and morphogenesis of the liver and lymphatic system.22–24 Most recently, Prox1 gene inactivation has been observed in a variety of tumors and tumor cell lines, implicating roles for this protein in tumor suppression.25–27 The biochemical functions of Prox1 are likely to be complex because this protein has been shown to function as both a sequence-specific transcriptional activator19,28,29 and a transcriptional corepressor of at least some orphan nuclear receptors.30,31
We have previously determined that Prox1 binds to the OL2 element (−75 to −68) of the chicken βB1-crystallin promoter and activates promoter activity in lens and nonlens cells.19 However, deletion of the OL2 element within the −432/+30 construct reduced Prox1 responsiveness by only 50%,19 suggesting that other Prox1 responsive elements exist in this promoter. Here we further investigated the function of sequences between −126 and −432 of the chicken βB1-crystallin promoter in different cellular contexts, identified two additional high-affinity Prox1-binding sites in this region, and discovered that Prox1, like its homolog Drosophila Prospero, can also function as a direct transcriptional repressor.
MATERIALS AND METHODS
Constructs
The chicken βB1-crystallin promoter plasmid 432/+30/CAT (p432) was described previously.13 The −402/+30/CAT plasmid was made by digesting the βB1 −432/+30 pBasic CAT plasmid with PstI and religating the remaining plasmid. The chicken βB1 −245/+30 pBasic CAT plasmid was made by double digesting the βB1 −432/+30 pBasic CAT plasmid with SalI and SstII and religation of the plasmid. The other chicken βB1-crystallin deletion constructs were made by PCR amplification of −432/+30 using the reverse primer −30 (5′-CCC TCT AGA CAG CTG CTG CTT CTT GTT GGA G-3′) and the forward primers −282 (5′-CGG GTC GAC CCG GTG GGC TCT-3′); −206 (5′-AAT GTA TAC TGT TGT GCC GCT GGA TAA AGG AAA-3′); −152 (5′-CCC TCT AGA GGA GAG GCT TCC AGC TCG CCC A-3′); and −126 (5′-CCC TCT AGA AGC TTT GCA GGA TGT GAT-3′). Restriction sites are noted in bold. The −282/+30 and −206/+30 PCR fragments were digested with AccI and XbaI, whereas the −152/+30 and −126/+30 PCR fragments were digested with XbaI and ligated into the pCAT-Basic vector.
A construct containing −432/+30 pBasic CAT with a 10-bp substitution mutation at −220 (−432 mut220) was created by ligating the regions upstream and downstream of the −220 site into pBasic CAT and replacing the −220 site with an AccI restriction site. Primers used in PCR to amplify the upstream portion were (forward) 5′-TAT GCA TGC CTG GGT ATG CCC AAG GTG-3′ and (reverse) 5′-ATA ATG TAT ACT ACG CAC GCC GAC ACC GCG-3′ on the upstream portion of the promoter (−432/−231). Primers for amplifying the downstream region were (forward) 5′-CGA TAG TAT ACA TGC GGT CAG GTC TTC TGT TGT GC-3′ and (reverse) 5′-CCC TCT AGA CAG CTG CTG CTT CTT GTT GGA G-3′. The construct containing an 11-bp substitution mutation at the −220 site within the −245/+30 promoter (−245 mut220) was made by amplifying the promoter (−432 mut220) with the forward primer 5′-AAT GCA TGC CGG TGT CGG CGT GC-3′ and the reverse primer of −432 mut220. Restriction sites are noted in bold. The mutated constructs were confirmed by sequencing.
The 3XPL2 β-actin–CAT construct has been reported.8 The −220 Prox1-binding site (5′-TCG GCG TGC GGC AAA GTG GCG CGG TCA GGT-3′) was synthesized as a trimer (Celtek Genes, Nashville, TN) with BamHI ends, and the −290 Prox1-binding site (5′-AGC AGG AGT GCT GGA TCC AGG TGC TGG TGG GCT-3′) was synthesized (Celtek Genes) as a trimer with BglII ends. The trimerized elements were ligated into the BamHI site of chicken β-actin–CAT32 or the BglII site of pCAT3-control (Promega, Madison, WI).
The eukaryotic chicken Prox1 expression vector pCMV/Prox1 was previously described.28 The pCMV/βGAL plasmid was purchased from Clontech (Palo Alto, CA). Production and purification of the recombinant Prox1-GST protein was reported previously.33
Generation of Transgenic Mice
The −245/+30 CAT construct was digested away from prokaryotic vector sequences and prepared for transgenic mouse production as described.13 Transgenic mice were created with this construct on the FVB/N34 genetic background by University of Delaware Transgenic Mouse Facility staff, as described.11 The −432/+30 and −152/+30 transgenic mice were reported previously.8,13 Mice were screened for the presence of the transgene by PCR, as previously described.8 Genomic DNA of adult transgenic animals was then digested with EcoRI and subjected to Southern blot analysis to determine the number of integration sites and the integrity of the transgenes. Lenses and other tissues were isolated from eight independent lines of transgenic mice and assayed for CAT activity, as described previously.13 The mean CAT activity in the lens for each independently derived −245 line was determined by assaying two or three different animals by pooling both lenses from a single animal into one determination. Mean ± SD of the activity for the −245 construct was calculated by combining the data obtained from all eight independent lines. All experiments using animals were approved by University of Delaware institutional review boards and conformed to the ARVO Statement for the Use of Animals in Ophthalmic Research.
Cell Culture and Transfection Analysis
N/N 1003A cells, derived from rabbit lens epithelia (a gift of John Reddan, Oakland University)35 and Chinese hamster ovary (CHO) cells were plated at a density of 1.2 × 105 per 60-mm dish the day before transfection. Transfections were performed by lipofection (Lipofectamine Plus; Invitrogen, Carlsbad, CA) with 3.5 μg total plasmid, including 0.25 μg pCMV/β-GAL, 3 μg promoter/chloramphenicol acetyltransferase (CAT) reporter plasmid, and 0.25 μg pcDNA/Prox1 expression plasmid. Cells were harvested 48 hours after transfection, and cellular extracts were prepared by multiple cycles of freeze/thaw. The extracts were assayed for CAT and β-galactosidase activity, as previously described.14 All cotransfection experiments were performed at least twice in triplicate and were analyzed statistically by Student’s t-test.
Chicken lens annular pad (CLAP) cells were prepared, as previously described,36 from the lenses of 18-day-old embryonic chickens and plated in 30-mm dishes. Cultures were transfected 1 day after plating, as described.
Western Blot Analysis
Freshly isolated CLAP cells, CLAPs cultured for various times, whole chicken lenses, and isolated chicken lens fibers were harvested and homogenized in RIPA buffer (1× PBS, 1% igepal, 0.5% sodium deoxycholate, 0.1% SDS) with freshly added protease inhibitors: 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 μg/mL aprotinin, 100 μM sodium orthovanadate, 1 mM EDTA. Proteins were separated by PAGE gel and transferred to nitrocellulose membrane (Invitrogen). The membrane was blocked with blocking buffer (5% powdered milk, 0.1% Tween-20 in TBS) and incubated with a 1:7000 dilution of anti-chicken βB1-crystallin antibody,7 followed by the addition of horseradish per-oxidase (HRP)-conjugated secondary anti-rabbit antibody (1:2000; Cell Signaling Technology, Beverly, MA). After washing, the membrane was incubated in 5 mL of 1× reagent (LumiGLO; Cell Signaling) and 1× peroxide (Cell Signaling) for 1 minute. The blot was exposed to film (BioMax MR; Kodak, Rochester, NY) to obtain an image.
DNaseI Footprinting Analysis
Solid-phase DNaseI footprinting was performed on the −432/−152 fragment of the chicken βB1-crystallin promoter using a modification of the method described by Sandaltzopoulos and Becker.37 Briefly, the probe was created by PCR amplification using the following primers, which were either P32 or biotin labeled, depending on the strand to be studied (forward, 5′-TAT GTC GAC CTG GGT ATG CCC AAG GTG CGG-3′; reverse, 5′-AAT GTC GAC CTC TGG GCG AGC TGG AAG CCT-3′). Bold denotes incorporated SalI restriction sites. The probe DNA was then immobilized on the avidin-coupled magnetic beads (New England Biolabs, Ipswich, MA) and incubated in the presence or absence of recombinant Prox1 protein for 15 minutes at room temperature. Various amounts of DNaseI were incubated with the bead mixture for 3 minutes, and digestion was stopped with the addition of an excess of EDTA. Beads were washed and mixed with 8 μL formamide sample buffer (96% formamide, 0.05% xylene cyanol, 0.05% bromophenol blue, 10 mM EDTA, freshly mixed 3:1 with 100 mM NaOH). Samples were denatured for 5 minutes at 76°C and loaded onto a sequencing gel adjacent to a Maxam-Gilbert sequencing reaction of the same probe.
Electrophoretic Mobility Shift Assay
All electrophoretic mobility shift assays (EMSAs) were performed in a total volume of 12.5 μL containing 2.5 μL of 20% Ficoll, 1.25 μL of 10 × EMSA buffer, 100 ng nonspecific poly(dA-dT), 1 μL recombinant Prox1-GST protein, and 20,000 Cerenkov counts of double-stranded DNA probe, as described previously.38 Nonradioactive competitors were added at 100-fold molar excess. After 15-minute preincubation at room temperature, reactions were electrophoresed through 5% poly-acrylamide gels using 0.5 × Tris borate-EDTA as the buffer, at 4°C. The oligonucleotides used for gel shift analysis are given in Table 1.38
Table 1.
Oligonucleotides Used for Electrophoretic Mobility Shift Analysis (Sense Strands Only Are Shown)
| Oligonucleotides | Sense Strands | Reference |
|---|---|---|
| PL1 | 5′-GGA TGT GAT GAC TGG GCG GCC GCA-3′ | 38 |
| OL2 | 5′-AGA CAC TGA TGA GCT GGC ACT TCC-3′ | 38 |
| Ol2 mut | 5′-AGA CAC TGA TGA GCT GGG CGT TCC-3′ | 38 |
| −220 | 5′-TGC GGC AAA GTG GCG CGG TCA GGT CTT CTG-3′ | — |
| −220 mut | 5′-TGC GGC AAA TGT GCG CGG TCA GGT CTT CTG-3′ | — |
| −290 | 5′-AGG AGT GCT GGA TCC AGG TGC TGG TGG GCT-3′ | — |
| E2F | 5′-GCC CTC GGG CGG GCG CGA GGG CGG GAC GGG-3′ | — |
Bold denotes position of incorporated mutation.
Chromatin Immunoprecipitation Analysis
Lenses and livers were removed from 14-day-old chicken embryos and sheared, and formaldehyde cross-linked chromatin (average size, 600 bp) was generated by sonication, as described in Yang et al.39 Aliquots of chromatin prepared from 10 lenses or 1 to 2 mg liver were incubated with 1 μL well-characterized rabbit polyclonal antibody prepared against the homeo- and Prospero domains of Prox120 or nonimmune rabbit serum (catalog no. 31884; Pierce, Rockford, IL). Antibody–chromatin complexes were captured by the addition of 100 μL protein G–agarose beads preblocked with salmon sperm DNA (Upstate/Millipore, Billerica, MA). The immunoprecipitates were washed three times and resuspended in a buffer containing 10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 25 mM EDTA supplemented with 0.1 mg/mL RNase and 0.2 mg/mL proteinase K. After 2-hour incubation at 55°C, the cross-links were reversed by overnight incubation at 65°C. Genomic DNA was eluted into 200 μL water using an extraction kit (QIAquick Spin Gel Extraction; Qiagen, Valencia, CA).
The amounts of each specific DNA fragment in immunoprecipitates were determined by quantitative real-time PCR using a standard curve generated for each primer set with 0.2%, 1.0%, and 5% input DNA samples. The amount of a specific DNA fragment in each assay was compared to the amount of that fragment before immunoprecipitation (input DNA). Values obtained with control normal rabbit serum were subtracted from corresponding values.
Primers were designed using available software (Primer 3; (http://frodo.wi.mit.edu/cgi-bin/primer3). PCR products measured between 90 and 110 bp. Reactions were analyzed with a Bio-Rad (Hercules, CA) system (iCycler iQ) and a Qiagen PCR kit (QuantiTech SybrGreen). Parameters were 95°C for 15 minutes followed by 45 cycles of 95°C for 30 seconds, 58°C for 45 seconds, and 72°C for 20 seconds. All reactions were performed in triplicate and analyzed statistically by Student’s unpaired t-test or ANOVA. Primers used for these studies were −2-kb CRYBB1 (5′-GCA TTA CGG AAA GCC CTA ACT-3′ and 5′-GGG AGC AGA ACT GTG GTG TC′), −0.9-kb CRYBB1 (5′-GCA TAG CAC GTG TTC AAT GTG-3′ and 5′-TCT CCT CTA AGG ATG CCA CAG3′), −0.15-kb CRYBB1 (5′-TCT GTT GTG CCG CTG GAT A-3′ and 5′-CCA GTC ATC ACA TCC TGC AA-3′), and +4.8-kb CRYBB1 (5′-TGC TGC AGT GTG CAG AAG G-3′ and 5′-GCA GAG CAG CAT GCC TCA-3′).
Computer Modeling of Prox1 Structure and Its Interaction with DNA
The crystal structures of both apo- and DNA-bound Drosophila Pros-pero (accession codes 1mij and 1xpx)17,18 were used as templates to predict the structure of amino acids 580 to 727 of chicken Prox1 (100% identical in this region to the human sequence) using the Swiss-Model homology modeling server.40–42 The predicted model was subjected to 2000 cycles of energy minimization using AMBER43 to relieve unfavorable steric interactions and to optimize the stereochemistry. The B-form double-stranded DNA structures of the OL2 (5′-GCA CTT CCA-3′), −220 (5′-TGC GGC AAA GTG GCG CGG-3′), and −290 (5′-AGT GCT GGA TCC AGG TGC TGG-3′) sites were generated using the biopolymer module of InsightII and subjected to a short 500-cycle minimization using AMBER. Docking of these DNA molecules onto Prox1 was then carried out using ZDOCK for the initial-stage docking to optimize desolvation, grid-based shape complementarity, and electrostatics to generate plausible protein-ligand poses.44–46 Ranking and refinement were carried out with RDOCK to minimize the predicted structures using CHARMM, and each structure was ranked based on its desolvation and electrostatic energy.47 The three most favorable complexes were subjected to a short 1000-femtosecond molecular dynamics simulation (Discover module of Insight II; Accelrys Inc., Burlington, MA) at 300°K, and the ensemble average was taken to be the final orientation of the Prox1–DNA complexes. The solvent-accessible surface area of the complexes generated from the OL2, −220, and −290 sites was calculated using NACCESS (obtained from http://www.bioinf.manchester.ac.uk/naccess/, University of Manchester, Manchester, UK).
RESULTS
Chicken βB1-Crystallin Minimal Promoter Repression by Upstream Sequences in Lens Epithelial Cells but Activation by Upstream Sequences in Lens Fiber Cells
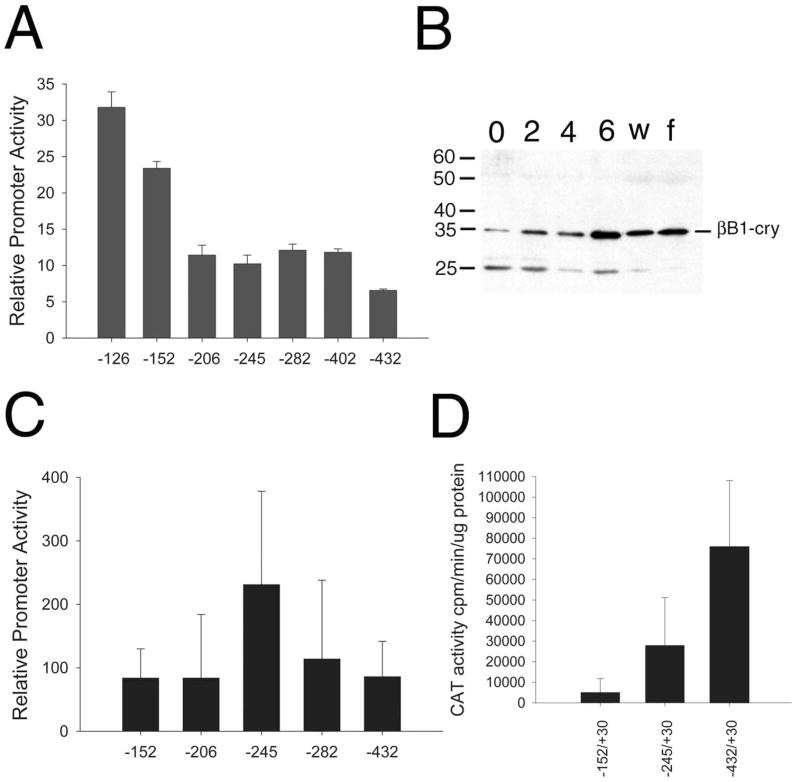
Previously, it was reported that a −126/+30 fragment of the chicken βB1 crystallin promoter was less active than longer promoter fragments in patched chick lens epithelial cell cultures,14 whereas this promoter was more than 200-fold less active than a −432/+30 fragment in the lenses of transgenic mice.8 To further investigate the function of the −126 to −432 region of the chicken βB1-crystallin promoter, we transfected a deletion series encompassing this region into the established lens epithelial cell line N/N1003a (Fig. 1A) and in primary cultures of CLAPs (Fig. 1C). Notably, the −126/+30 minimal promoter was nearly fivefold more active in N/N1003A cells than in the −432/+30 promoter fragment (Fig. 1A) necessary for crystallin level expression in transgenic mice.11 The observed repression appears to be conferred by at least three different negative elements in this region because significant repression was seen between the −126 and −152, −152 and −206, and −402 and −432 fragments (Fig. 1A). In contrast, transfections performed in primary CLAP cells, which express appreciable amounts of endogenous βB1-crystallin (Fig. 1B), revealed that the most active construct was −245/+30, whereas the addition of sequences to −432 repressed promoter activity (Fig. 1C). Thus, transgenic mice were created with the −245/+30 construct; however, these experiments revealed that the activity of this construct in the lens was intermediate between that obtained for −152/+30 and −432/+30 (Fig. 1D). Overall, these data show that the chicken βB1-crystallin promoter has a number of activating and repressing elements between −126 and −432 whose activity is dependent on cell type and/or chromatin context.
FIGURE 1.
Activity of various chicken βB1-promoter fragments in lens cells at different stages of differentiation. (A) Transfection analysis of chicken βB1-promoter fragments in comparison with promoterless vector (=1) in N/N1003a cells, an established lens epithelial cell line. The differences between −126 and −152 (P < 0.01), −125 and −206 (P < 0.001), −245 and −282 (P < 0.049), and −402 and −432 (P < 0.001) are statistically significant, as determined by Student’s t-test. The differences between −206 and −245 (P < 0.15) and −282 and −402 (P < 0.32) are not statistically significant. (B) Western blot analysis of endogenous βB1-crystallin expression in CLAP cells from the time of isolation (0) and days 2, 4, and 6 culture in comparison with whole 18-day embryonic lenses (w) and isolated fiber cells (f). The apparent molecular weight of chicken βB1-crystallin is 35 kDa, as was previously reported.48 (C) Activity of βB1-promoter CAT plasmids in CLAP cells. The differences between −206 and −245 (P = 0.004), −245 and −282 (P = 0.04), −152 and −245 (P = 0.0006), and −245 and −432 (P = 0.0009) are statistically significant, as determined by Student’s t-test. The differences between −152 and −206 (P = 0.5) and −282 and −432 (P = 0.3) are not statistically significant. Results were combined from multiple transfection experiments. The total number of transfected plates for −152 is 17, for −206 is 9, for −245 is 16, for −282 is 6, and for −432 is 11. (D) Activity of βB1-promoter truncations in lenses of transgenic mice. The difference between −152/CAT and −245/CAT was significant (P = 0.01), and the difference between −245/CAT and −432/CAT was significant (P = 0.005). The −432/CAT and −152/CAT expression levels were previously determined to be significantly different (P = 0.0001). Absolute numbers reported here are different from those previously reported for the −152/CAT and −432/CAT transgenic mice8 because of differences in counting efficiency between the previous and current scintillation counter used in the laboratory. All error bars shown in this figure represent SD.
Identification of Two Additional Prox1-Binding Elements at −220 and −290 in the Chicken βB1-Crystallin Promoter
Given that our previous work suggested that the chicken βB1-crystallin promoter has functional Prox1-binding sites upstream of −126,19 we used recombinant protein consisting of the homeo and Prospero domains of Prox133 in solid-phase DNaseI footprinting analysis of the −152/−432 region of the chicken βB1-crystallin promoter. Small regions surrounding nucleotides −290 and −220 (Fig. 2A) were protected from DNaseI digestion, and a DNaseI hypersensitive site was observed around −215 in the presence of Prox1.
FIGURE 2.
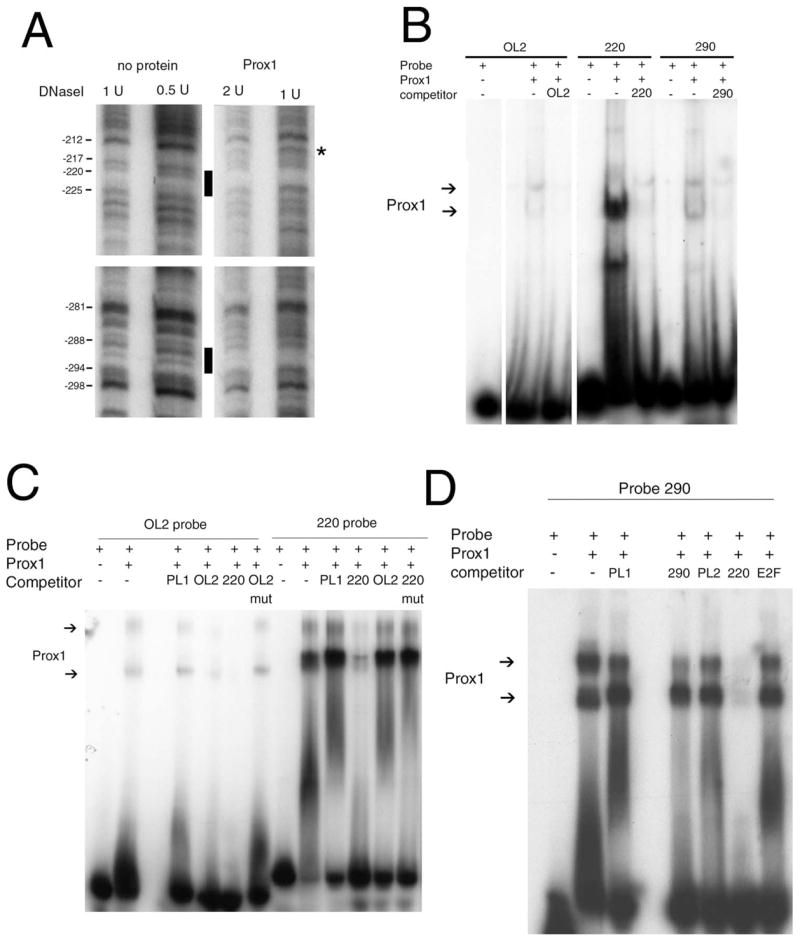
Interaction of Prox1 with three different sites in the chicken βB1-crystallin promoter. (A) Solid-phase DNaseI footprinting analysis of Prox1 interaction with sequences between −126 and −432. Black bars: regions protected from DNaseI digestion by Prox1. Asterisk: hypersensitive site. (B) Gel shift analysis comparing the relative interaction of recombinant Prox1 with the previously identified OL1 site and the newly identified −220 and −290 sites. The experiment was run with the same amount of oligonucleotide and protein on the same gel. All lanes come from the same exposure, and gaps in the figure denote lanes that were deleted. (C) Comparison between Prox1/OL2 and Prox1/−220 interactions by gel shift analysis. PL1 is a MARE site found in the chicken βB1-crystallin promoter shown previously not to interact with Prox1,19 −220 mut contains a substitution that disrupts the G of the AG dinucleotide, and OL2 mut contains a substitution that disrupts the AC dinucleotide. (D) Gel shift analysis of the −290 element of the chicken βB1-crystallin promoter showing the −220 site outcompetes the −290 site for Prox1 binding. Note that this gel was given a longer exposure than that of (B) to make the −220 competition more obvious.
As we previously reported,19 Prox1 interacts specifically, but weakly, with the OL2 element found in the chicken βB1-crystallin basal promoter. In contrast, the newly identified sequences at −220 and −290 interacted with Prox1 more strongly than OL2 (Fig. 2B). Competitions with a 100-fold molar excess of unlabeled oligonucleotides indicate that the −220 site has the highest relative affinity for Prox1 and that an AG dinucleotide found in this site was required for binding (Fig. 2C and data not shown). The −290 site had relatively lower affinity for Prox1 binding, whereas Prox1 interactions with the previously identified OL2 site were far weaker that either the −220 or the −290 site (Figs. 2B–D).
Interaction of Prox1 with the Chicken βB1-Crystallin Promoter in Lens Chromatin
Prox1 is required for lens and liver development.21,23 However, βB1-crystallin promoter sequences (−432/+30) drive gene expression exclusively in the lenses of transgenic mice,8 but βB1-crystallin mRNA is not detectable in the liver by Northern blotting.14 To determine the occupancy of Prox1 at the βB1-crystallin locus in vivo, chromatin immunoprecipitation (ChIP) was performed on chromatin isolated from embryonic chicken lens and liver tissue and was analyzed by Q-PCR using primers amplifying different regions of the βB1-crystallin locus (Fig. 3A). In lens chromatin, the highest Prox1 interaction was detected with primers corresponding to −150 of the promoter, located in the region of the three identified Prox1 sites, whereas Prox1 interactions were fewer, with fragments ampli-fied by primers from regions of the locus both 5′and 3′of the proximal promoter (Fig. 3B). In liver chromatin, the results were more variable between replicates, though Prox1 appeared to interact to some extent with fragments from across the βB1-crystallin locus (Fig. 3C).
FIGURE 3.
ChIP analysis of Prox1 interaction with the βB1-crystallin locus in vivo. (A) Schematic diagram of the chicken βB1-crystallin locus showing the relative location of the tightly linked βA4-crystallin (CRYBA4) and βB1-crystallin (CRYBB1) genes.49 Boxes: entire transcribed unit consisting of six exons and five introns for both genes. Arrowheads: positions of the primer sets used in real-time PCR analysis of chromatin im-munoprecipitated with the anti-Prox1 antibody. (B) ChIP analysis of Prox1 interaction with the chicken βB1-crystallin locus in the 14-day embryonic chicken lens. All data are expressed as relative enrichment (RE) of Prox1 in chromatin compared with nonimmune control. Prox1 was enriched in lens chromatin from −150 of the βB1-crystallin promoter compared with all other tested portions of the locus at a significance of P ≤ 0.0003 calculated by one-way ANOVA. (C) ChIP analysis of Prox1 interaction with the βB1-crystallin locus in the 14-day embryonic chicken liver. All data are expressed as relative enrichment (RE) of Prox1 in chromatin compared with nonimmune control. There was no statistical difference in Prox1 enrichment in any portion of the locus tested.
Transcriptional Repression by Prox1 Sites at −220 and −290 of the Chicken βB1-Crystallin Promoter Confer
We previously demonstrated that multimers of the OL2 element placed in front of the chicken β-actin basal promoter conferred lens-preferred promoter activation,8 whereas Prox1 binding to the OL2 element mediated transcriptional activation of the chicken βB1-crystallin promoter.19 Thus, we tested the function of the newly identified Prox1-binding sites by placing three copies of either the OL2, −220, or −290 elements upstream of the chicken β-actin basal promoter and performing cotransfections with a Prox1 expression vector in CHO cells (Fig. 4A). Cotransfection of Prox1 with the 3XOL2/β-actin–CAT construct resulted in a 10-fold increase in reporter activity (P < 0.001), but the minimal β-actin promoter alone did not respond to the addition of Prox1 (Fig. 4A). In contrast, the expression of β-actin–CAT constructs containing trimers of either the −220 or the −290 Prox1-binding sites was not activated by Prox1 cotransfection (Fig. 4A). Because the −220 element resembles a previously identified Prospero binding site that mediates Prospero-mediated transcriptional repression of the Drosophila R8 rhodopsin promoter50 (see Fig. 6), we tested the ability of the −220 and −290 Prox1 sites to mediate transcriptional repression by cloning trimers of these sites into the pCAT-control vector, which contained SV40-derived promoter and enhancer sequences. The inclusion of either −220 or −290 did not significantly affect the activity of the pCAT-control vector in CHO cells; however, cotransfection of these constructs with a Prox1 expression vector resulted in a 40% to 50% reduction in reporter activity, indicating that −220 and −290 function as Prox1-responsive repressor elements (Fig. 4B). We then tested the function of the −220 Prox1 site in the context of the βB1-crystallin promoter by cotransfection with a series of promoter truncations/mutations in CHO cells. A −206/+30 promoter fragment was activated nearly 10-fold by cotransfection with Prox1, but the inclusion of sequences −207/−245 squelched this activation to less than sixfold (Fig. 4C). Notably, mutation of the core AG dinucleotide of the −220 element in the context of the −245/+30 fragment resulted in a significant increase in Prox1 responsiveness; however, no increase was observed when a similar mutation was made in the context of the −432/+30 promoter, which still contained an intact −290 site.
FIGURE 4.
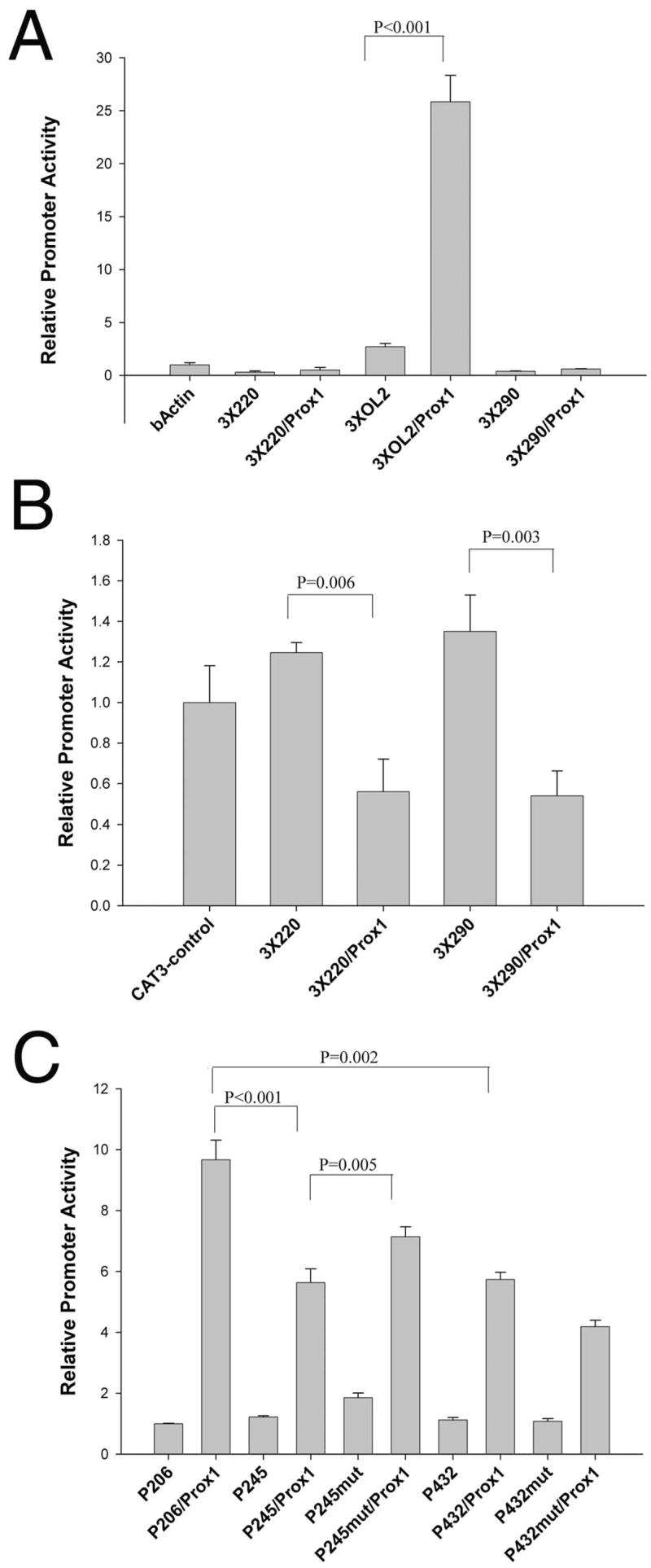
Functional analysis of the −220 and −290 Prox1-binding sites. (A) The ability of the −220, −290, and OL2 elements to confer Prox1 responsiveness to a heterologous promoter. Trimers of each element were placed upstream of the chicken β-actin basal promoter, linked to CAT, and transfected into CHO cells. The OL2 element was able to confer Prox1 responsiveness on this promoter (P ≤ 0.001), but neither −220 nor −290 had a similar effect. (B) The ability of −220 and −290 to confer Prox1-mediated repression on a heterologous promoter. Trimers of the −220 and −290 element were ligated up-stream of the SV40 promoter of the pCAT3-control plasmid, which also contained a strong enhancer. Neither the −220 nor the −290 elements alone significantly affected the activity of the vector in CHO cell transfections (P = 0.06). However, cotransfection with a Prox1 expression vector led to 40% reduction in the activity of 3 × 220 and 3 × 290 pCAT control that was statistically significant (P = 0.006). (C) Prox1 responsiveness of the upstream portions of the βB1-crystallin promoter. The promoter −206/+30 had the most robust response to Prox1 cotransfection, whereas the inclusion of sequences from −207 to −245, which encompassed the −220 Prox1 element, significantly reduced Prox1 responsiveness of the promoter. Mutation of the −220 element in the context of the −245/+30 fragment significantly increased Prox1 responsiveness of this fragment (P < 0.005). The −432/+30 fragment containing all three known Prox1-binding sites was also significantly less Prox1 responsive than −206/+30, which only contained the OL2 site (P = 0.002); however, its Prox1 responsiveness was not significantly different from −245/+30. Notably, mutation of the −220 element in the context of the −432/+30 promoter reduced Prox1 responsiveness of this fragment.
FIGURE 6.

Alignment of the Prox1-binding sites of the chicken βB1-crystallin promoter with the previously identified Prox1 and Prospero sites. (A) Alignment of the Prox1- and Prospero-binding sites containing an AG dinucleotide. The PRORE site is a Prox1-responsive element identified in the mouse γF-crystallin promoter,28 sequence 56 is a Prospero site found in the Drosophila R8 rhodopsin promoter,50 and the consensus Prospero-binding site was used in conjunction with in vivo binding site profiling to identify putative Prospero-responsive genes.62 (B) Alignment of the known Prox1- and Prospero-binding sites containing a CA dinucleotide. The OL2 site is the previously identified Prox1 site found in the chicken βB1-crystallin promoter.19 The −190 site of the FGFR3 promoter was previously described,29 and TDA-pros is the Prospero consensus sequence previously identified by a target detection assay.61
Modeling of the Prox1 Homeo-Prospero Domain and Its Interaction with DNA
Prox1 shares 66% sequence identity with Drosophila Prospero in the homeo-Prospero domain (HPD), located at the C-terminal end of the protein.15,16,51 Given that the crystal structure of the HPD has been solved in the presence and absence of DNA,17,18 we were able to use knowledge-based homology modeling to predict the structure of the Prox1 HPD and its interactions with the three Prox1-binding sites found in the βB1-crystallin promoter. More than 99% of the residues in the model had backbone torsional angles in the allowed region of a Ramachandran plot generated by RAMPAGE (obtained from http://mordred.bioc.cam.ac.uk/~rapper/rampage.php, University of Cambridge, Cambridge, UK; data not shown), and almost all residues modeled were in an energetically favorable local environment. The predicted Prox1 structure was then superimposed on the Prospero template; these two structures are highly superimposable and have an overall root-mean-squared deviation of 1.1Å (Fig. 5A). These results indicate that the predicted model is a good workable structure for use in further analysis.
FIGURE 5.
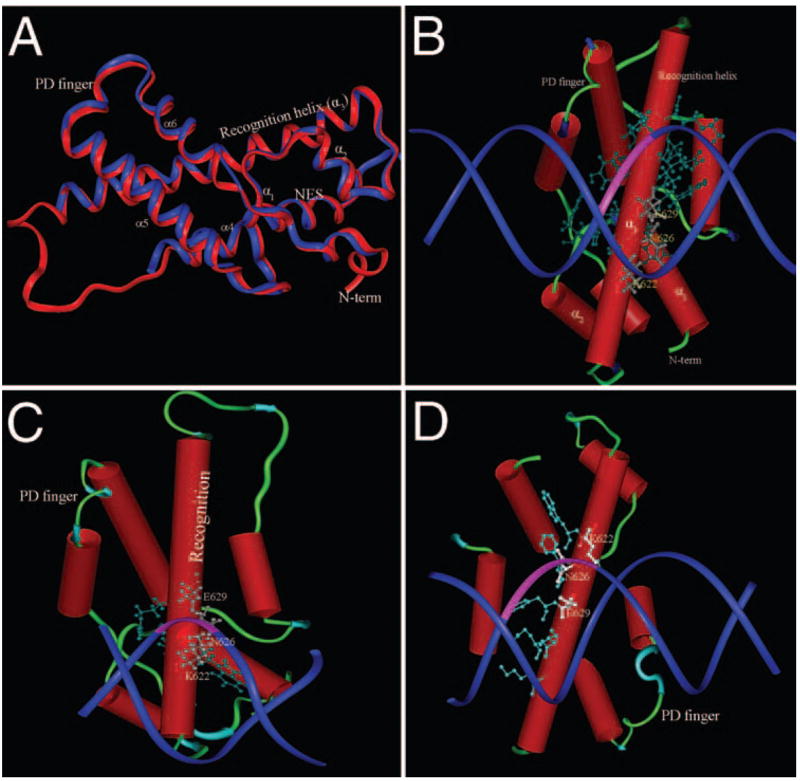
Computer modeling of Prox1 and its interaction with the Prox1-binding sites of the chicken βB1-crystallin promoter. (A) Superposition of the ribbon structure of the homeo-Prospero domain from Prospero determined from crystallography data (blue) over the structure of the homeo-Prospero domain of Prox1 (red) calculated by homology modeling. The region between helix 3 (the homeo-Prospero domain recognition helix) and helix 4 (part of the Prospero domain) is not solved in the Prospero crystal structure and is portrayed as a random coil for Prox1. (B) Model of Prox1 interaction with the −220 site in the chicken βB1-crystallin promoter. Amino acids of the Prox1 recognition helix making contact with the DNA are shown as ball-and-stick representations, and the three main amino acids involved in AG dinucleotide (pink) interaction are numbered. (C) Model of Prox1 interaction with the OL2 site of the chicken βB1-crystallin promoter. Amino acids of the Prox1 recognition helix making contact with the DNA are shown as ball-and-stick representations, and the three main amino acids involved in CA dinucleotide (pink) interaction are numbered. (D) Model of Prox1 interaction with the −290 site in the chicken βB1-crystallin promoter. Amino acids of the Prox1 recognition helix making contact with the DNA are shown as ball-and-stick representations, and the three main amino acids involved in CA dinucleotide (pink) interaction are numbered. Note that in all three cases, the same amino acids of Prox1 are involved in the major groove contact with DNA.
We then used the previously reported OL2 site19 and the newly discovered −220 and −290 sites to determine whether there was good geometric and chemical compatibility with the Prox1 recognition helix (Figs. 5B–D). Our docking model suggested that the −220 site bound to Prox1 through a critical AG dinucleotide (Fig. 5B), which is consistent with the results of our EMSA analysis shown in Figures 2B and 2C. This AG dinucleotide resides in the major groove of DNA, and three conserved residues of Prox1, Lys622, Asn626, and Glu629 make sequence-specific hydrogen bond contacts. Flanking nucleotides make van der Walls or hydrophobic contacts with the recognition helix and further stabilize the complex. The three residues responsible for making hydrogen bond contacts with AG dinucleotide are also conserved in Prospero, where they carry out a similar function in DNA binding.17 The −290 and OL2 sites, however, do not bind through the AG nucleotide; rather, CA is predicted to be important for recognition. Notably, there are more contacts between Prox1 and the −220 site than the other two sites. Further, though the −290 site is similar in sequence to the OL2 site (Fig. 6), −290 makes more contacts with Prox1 than OL2 but fewer than −220. These observations result in the calculated buried surface area of the interface region between Prox1 and −220 to be 1800Å2 (20% of the total surface area), whereas it is calculated to be 1705Å2 (17.5% of the total surface area) for the Prox1/−290 complex and only 1620Å2 (14% of the total surface are) for the Prox1/OL2 complex. Taken together, these data suggest that Prox1 binds most tightly at the −220 site, the next most tightly at −290, and least tightly at the OL2 site.
DISCUSSION
The accumulation of crystallins to concentrations up to 450 mg/mL in the lens fiber cell cytoplasm52 is necessary for the transparency and the high refractive index of the ocular lens.6 The high-level transcription of most crystallin genes is lens fiber cell preferred,5 and individual crystallin mRNAs can represent 1% to 40% of total lens fiber cell mRNA.9,53 Previously, we demonstrated that sequences from −432/+30 of the chicken βB1-crystallin promoter are sufficient to drive both the developmental expression pattern and the high level accumulation of mRNA in the lens, whereas sequences from −152/+30 are less active though still lens specific.8,11,13 Here we dissected the function of the region between −152 and −432 in cells of three different stages of lens fiber cell differentiation; the lens epithelium, the annular pad which represents the early stages of lens fiber cell differentiation,54 and the intact lens.
Transfection analyses in the lens epithelial cell line N/1003a (used to model the lens epithelium in vitro because of its endogenous expression of Pax655 and Prox128) and previous work in primary chicken lens epithelial cells14 suggest that the βB1-crystallin promoter sequences upstream of −152 contain two or more elements that repress its promoter function in lens fiber cell precursors. As lens epithelial cells begin to differentiate into fibers (represented by annular pad cells), an element between −206 and −245 becomes active, and, in the lens fiber cells, at least one additional element between −245 and −432 becomes active to confer appropriate levels of crystallin expression in the lens. Although it is possible that these results may be confounded by the use of lens cell models from multiple species, the data in aggregate suggest that the function of the promoter region between −152 and −432 changes as lens epithelial cells differentiate into lens fiber cells.
Because the mutation of a known Prox1 site in the chicken βB1-crystallin basal promoter did not abolish all Prox1 responsiveness of the full length promoter,19 we hypothesized that the chicken βB1-crystallin promoter has Prox1-responsive elements upstream of −152 and analyzed whether Prox1 regulates the upstream sequences of the chicken βB1-crystallin promoter.
Prox1 is a divergent homeodomain protein important for many developmental processes. Prox1 expression in endothe-lial cells leads to a reduction in the expression of genes controlling blood endothelial fate and the induction of genes important for the formation of the lymphatic endothelium,56 including FGF receptor 3,29 whereas Prox1 deletion results in the persistence of vascular endothelial cell markers.22,57 In the retina, the loss of Prox1 results in the lack of horizontal cell development associated with the increased expression of genes involved in cell proliferation and the downregulation of retinal differentiation markers.24 In the lens, Prox1 is necessary for the exit of lens epithelial cells from the cell cycle, their elongation into fiber cells, and the onset of γ-crystallin expression.21,28 Furthermore, the loss of Prox1 by a variety of mechanisms is associated with cancer progression; thus, this molecule has been proposed to be a tumor suppressor.25,27,58
In contrast to its clear importance in vertebrate development, the biochemical function of Prox1 is less well characterized though it can serve as a transcriptional corepressor30,59 in addition to working as a direct transcriptional activator.19,28,29,60 Here we studied the transcriptional functions of Prox1 using the chicken βB1-crystallin promoter as the model system. We identified two additional Prox1-binding sites at −290 and −220 in this promoter and found that both bound Prox1 more avidly than the previously identified OL2 element19 in gel shift assays. Mutation of the AG dinucleotides found in the −220 site led to a loss of Prox1 binding, suggesting that the AG dinucleotides of this site were required for Prox1–DNA interaction.
The DNA-binding activity of Prox1 resides in the C-terminal homeo-Prospero domain, which shares 66% sequence identity to its Drosophila homolog, Prospero.15,16 To further understand the molecular basis of Prox1 interaction with the OL2, −220, and −290 elements, we predicted the structure of Prox1 bound to these sites using the previously solved Prospero homeo-Prospero domain crystal structure17,18 as a template for knowledge-based homology modeling. All the secondary structural elements found in Prospero were well preserved in the model structure of Prox1. Prox1, like Prospero, has a well-defined homeo domain composed of three β-helices and an N-terminal extension. Helices I and II lie parallel to each other and across from Helix III. The third homeodomain helix continues well into the Prospero domain and is essential for DNA recognition and binding. Thus, the homeo and Prospero domains of Prox1, like those of Prospero,17,18 are most properly defined as a single integrated domain.
We then docked short DNA fragments encompassing the previously reported OL2 site19 and the newly discovered −220 and −290 sites to this Prox1 structure to determine whether there is a good geometric and chemical compatibility between these biochemically determined binding targets and the Prox1 recognition helix. Our docking model shows that the −220 site likely binds to Prox1 through a critical AG dinucleotide that resides in the major groove of DNA, and three conserved residues of Prox1—Lys622, Asn626, and Glu629—make sequence-specific hydrogen bond contacts with this sequence. This is consistent with our data demonstrating that the AG dinucleotide in the −220 site was required for Prox1 interaction. Flanking nucleotides make van der Walls or hydrophobic contacts with the recognition helix and further stabilize the complex. It is interesting to note that the three residues responsible for making hydrogen bond contacts with the AG dinucleotide of the −220 site are also conserved in Prospero, where they carry out a similar function in binding to the previously described TDA-pros Prospero binding sites.17 The OL2 and −290 sites, however, do not bind through the AG nucleotide; rather, a cytosine nucleotide is predicted to be important for recognition. In comparing the three structures, there are noticeably more contacts between Prox1 and the −220 site than the other two sites, but −290 makes more contacts with Prox1 than OL2. This is consistent with the relative binding strengths between these sites, as assayed by gel shift analysis.
At this time, it is unclear which mechanistic differences between Prox1 binding at OL2 confer transcriptional activation and which at −220 and −290 confer Prox1-mediated transcriptional repression. Notably though, TDA-pros, the low-affinity Prospero-binding site identified by target site selection, confers Prospero-mediated transcriptional activation to a heterologous promoter61 and shares the CA dinucleotide found in the activating OL2-binding site19 and the −190 element, which mediates Prox1 activation of the FGFR3 promoter29 (Fig. 6). In contrast, Seq 56, the higher affinity Prospero-responsive element reported to be important to repress the Drosophila R8 rhodopsin promoter in R7 photoreceptors,50 is similar to the −220 element of βB1-crystallin. However, it is apparent that the differences between Prox1/Prospero function at various sites is not only mediated by whether the prime specific contacts are with AG or CA. For example, the PRORE element of the γF crystallin promoter, which is activated by Prox1,28 contains a prominent AG dinucleotide while our computer modeling suggests that the −290 element of the chicken βB1-crystallin promoter which confers transcriptional repression makes these specific contacts via a CA dinucleotide, not the neighboring AG. These data highly suggest that Prox1 and Prospero have two sets of responsive elements: those containing AG dinucleotides and those containing CA dinucleotides.
It was recently reported that target genes containing a Prospero consensus-binding site, defined as T A/t A G A/c/g C/t G/a/t, could be repressed or activated by Prospero.62 In fact, our data showing that the region of the chicken βB1-crystallin promoter containing the −220 and −290 sites could mediate transcriptional repression or activation, depending on cellular context, suggests that a changing balance of transcriptional coactivator/corepressor expression or Prox1 posttranslational modifications could alter the function of these elements. Notably, it was recently shown that Prox1 is phosphorylated at multiple serine residues in the 21-day-old mouse liver,63 though the role of these modifications is unknown. A possible model for Prox1 function in the lens is that the low concentrations of Prox1 found in the lens epithelium are sufficient to bind the high-affinity Prox1 sites in the βB1-crystallin promoter and to repress its function. As lens epithelial cells transition to lens fiber cells, Prox1 levels increase and can engage the lower affinity OL2 site, activating gene expression. The high-affinity Prox1 repressive sites could change to sites activated by Prox1 by alterations in Prox1-binding partners/posttranslational modifications in fiber cells. Alternatively, a combination of chromatin remodeling and recruitment of other activating proteins to the −126/−432 region could abolish the ability of Prox1 to repress gene function at the −220 and −290 sites. Future work will be focused on testing this model and the mechanisms underlying the diverse roles of Prox1 as a transcription factor.
Overall, we have shown that the distal region of the chicken βB1-crystallin promoter has multiple repressive and activating elements that function differently, depending on cellular context. We have shown that this region binds Prox1 in the lens in vivo and contains at least one relatively low-affinity Prox1-binding site that mediates Prox1 activation and two higher affinity Prox1 sites that confer Prox1-mediated repression. Computer models have suggested the structural basis of the difference in Prox1 affinity for these sites. Future work will be required to understand the mechanistic basis for Prox1 function with regard to these sites.
Acknowledgments
The authors thank the Department of Chemistry and Biochemistry at the University of Delaware for providing computational support and Li Liao (Department of Computer and Information Sciences, University of Delaware) for helpful discussions.
Supported by National Eye Institute Grant EY012221 (MKD), SigmaXi-NAS grants-in-aid (XC, JRT), and a Beckman Young Scholars award (TPP).
Footnotes
Disclosure: X. Chen, None; J.R. Taube, None; V.I. Simirskii, None; T.P. Patel, None; M.K. Duncan, P
References
- 1.Kuszak JR. The ultrastructure of epithelial and fiber cells in the crystalline lens. Int Rev Cytol. 1995;163:305–350. doi: 10.1016/s0074-7696(08)62213-5. [DOI] [PubMed] [Google Scholar]
- 2.Wride MA. Cellular and molecular features of lens differentiation: a review of recent advances. Differentiation. 1996;61:77–93. doi: 10.1046/j.1432-0436.1996.6120077.x. [DOI] [PubMed] [Google Scholar]
- 3.Chow RL, Lang RA. Early eye development in vertebrates. Annu Rev Cell Dev Biol. 2001;17:255–296. doi: 10.1146/annurev.cellbio.17.1.255. [DOI] [PubMed] [Google Scholar]
- 4.Zelenka PS, Gao C-Y, Rampalli A, Arora J, Chauthaiwale V, He H-Y. Cell cycle regulation in the lens: proliferation, quiescence, apoptosis and differentiation. Prog Ret Eye Res. 1997;16:303–322. [Google Scholar]
- 5.Cvekl A, Duncan MK. Genetic and epigenetic mechanisms of gene regulation during lens development. Prog Retin Eye Res. 2007;26:555–597. doi: 10.1016/j.preteyeres.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piatigorsky J. Lens crystallins and their genes: diversity and tissue-specific expression. FASEB J. 1989;3:1933–1940. doi: 10.1096/fasebj.3.8.2656357. [DOI] [PubMed] [Google Scholar]
- 7.Brahma SK. Ontogeny of beta B1-crystallin polypeptide during chicken lens development. Exp Eye Res. 1988;47:507–510. doi: 10.1016/0014-4835(88)90060-7. [DOI] [PubMed] [Google Scholar]
- 8.Duncan MK, Li X, Ogino H, Yasuda K, Piatigorsky J. Developmental regulation of the chicken beta B1-crystallin promoter in transgenic mice. Mech Dev. 1996;57:79–89. doi: 10.1016/0925-4773(96)00533-3. [DOI] [PubMed] [Google Scholar]
- 9.Sawada K, Agata K, Eguchi G. Characterization of terminally differentiated cell state by categorizing cDNA clones derived from chicken lens fibers. Int J Dev Biol. 1996;40:531–535. [PubMed] [Google Scholar]
- 10.Ueda Y, Duncan MK, David LL. Lens proteomics: the accumulation of crystallin modifications in the mouse lens with age. Invest Ophthalmol Vis Sci. 2002;43:205–215. [PubMed] [Google Scholar]
- 11.Taube JR, Gao CY, Ueda Y, Zelenka PS, David LL, Duncan MK. General utility of the chicken betaB1-crystallin promoter to drive protein expression in lens fiber cells of transgenic mice. Transgenic Res. 2002;11:397–410. doi: 10.1023/a:1016364001095. [DOI] [PubMed] [Google Scholar]
- 12.Chen WV, Fielding Hejtmancik J, Piatigorsky J, Duncan MK. Functional conservation of the βB1-crystallin promoter during evolution. Biochim Biophys Acta. 2001;1519:30–38. doi: 10.1016/s0167-4781(01)00201-9. [DOI] [PubMed] [Google Scholar]
- 13.Duncan MK, Roth HJ, Thompson M, Kantorow M, Piatigorsky J. Chicken beta B1 crystallin: gene sequence and evidence for functional conservation of promoter activity between chicken and mouse. Biochim Biophys Acta. 1995;1261:68–76. doi: 10.1016/0167-4781(94)00223-p. [DOI] [PubMed] [Google Scholar]
- 14.Roth HJ, Das GC, Piatigorsky J. Chicken beta B1-crystallin gene expression: presence of conserved functional polyomavirus enhancer-like and octamer binding-like promoter elements found in non-lens genes. Mol Cell Biol. 1991;11:1488–1499. doi: 10.1128/mcb.11.3.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomarev SI, Sundin O, Banerjee-Basu S, Duncan MK, Yang JM, Piatigorsky J. Chicken homeobox gene Prox 1 related to Drosophila prospero is expressed in the developing lens and retina. Dev Dyn. 1996;206:354–367. doi: 10.1002/(SICI)1097-0177(199608)206:4<354::AID-AJA2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 16.Oliver G, Sosa-Pineda B, Geisendorf S, Spana EP, Doe CQ, Gruss P. Prox 1, a prospero-related homeobox gene expressed during mouse development. Mech Dev. 1993;44:3–16. doi: 10.1016/0925-4773(93)90012-m. [DOI] [PubMed] [Google Scholar]
- 17.Yousef MS, Matthews BW. Structural basis of Prospero-DNA interaction: implications for transcription regulation in developing cells. Structure. 2005;13:601–607. doi: 10.1016/j.str.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 18.Ryter JM, Doe CQ, Matthews BW. Structure of the DNA binding region of prospero reveals a novel homeo-prospero domain. Structure. 2002;10:1541–1549. doi: 10.1016/s0969-2126(02)00883-3. [DOI] [PubMed] [Google Scholar]
- 19.Cui W, Tomarev SI, Piatigorsky J, Chepelinsky AB, Duncan MK. Mafs, Prox1, and Pax6 can regulate chicken betaB1-crystallin gene expression. J Biol Chem. 2004;279:11088–11095. doi: 10.1074/jbc.M312414200. [DOI] [PubMed] [Google Scholar]
- 20.Duncan MK, Cui W, Oh DJ, Tomarev SI. Prox1 is differentially localized during lens development. Mech Dev. 2002;112:195–198. doi: 10.1016/s0925-4773(01)00645-1. [DOI] [PubMed] [Google Scholar]
- 21.Wigle JT, Chowdhury K, Gruss P, Oliver G. Prox1 function is crucial for mouse lens-fibre elongation. Nat Genet. 1999;21:318–322. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- 22.Wigle JT, Oliver G. Prox1 function is required for the development of the murine lymphatic system. Cell. 1999;98:769–778. doi: 10.1016/s0092-8674(00)81511-1. [DOI] [PubMed] [Google Scholar]
- 23.Sosa-Pineda B, Wigle JT, Oliver G. Hepatocyte migration during liver development requires Prox1. Nat Genet. 2000;25:254–255. doi: 10.1038/76996. [DOI] [PubMed] [Google Scholar]
- 24.Dyer MA, Livesey FJ, Cepko CL, Oliver G. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet. 2003;34:53–58. doi: 10.1038/ng1144. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi M, Yoshimoto T, Shimoda M, et al. Loss of function of the candidate tumor suppressor prox1 by RNA mutation in human cancer cells. Neoplasia. 2006;8:1003–1010. doi: 10.1593/neo.06595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laerm A, Helmbold P, Goldberg M, Dammann R, Holzhausen HJ, Ballhausen WG. Prospero-related homeobox 1 (PROX1) is frequently inactivated by genomic deletions and epigenetic silencing in carcinomas of the biliary system. J Hepatol. 2007;46:89–97. doi: 10.1016/j.jhep.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 27.Versmold B, Felsberg J, Mikeska T, et al. Epigenetic silencing of the candidate tumor suppressor gene PROX1 in sporadic breast cancer. Int J Cancer. 2007;121:547–554. doi: 10.1002/ijc.22705. [DOI] [PubMed] [Google Scholar]
- 28.Lengler J, Krausz E, Tomarev S, Prescott A, Quinlan RA, Graw J. Antagonistic action of six3 and prox1 at the gamma-crystallin promoter. Nucleic Acids Res. 2001;29:515–526. doi: 10.1093/nar/29.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin JW, Min M, Larrieu-Lahargue F, et al. Prox1 promotes lineage-specific expression of fibroblast growth factor (FGF) receptor-3 in lymphatic endothelium: a role for FGF signaling in lymphangiogenesis. Mol Biol Cell. 2006;17:576–584. doi: 10.1091/mbc.E05-04-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin J, Gao DM, Jiang QF, et al. Prospero-related homeobox (Prox1) is a corepressor of human liver receptor homolog-1 and suppresses the transcription of the cholesterol 7-alpha-hydroxylase gene. Mol Endocrinol. 2004;18:2424–2439. doi: 10.1210/me.2004-0009. [DOI] [PubMed] [Google Scholar]
- 31.Song KH, Li T, Chiang JY. A Prospero-related homeodomain protein is a novel co-regulator of hepatocyte nuclear factor 4alpha that regulates the cholesterol 7alpha-hydroxylase gene. J Biol Chem. 2006;281:10081–10088. doi: 10.1074/jbc.M513420200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuo I, Kitamura M, Okazaki K, Yasuda K. Binding of a factor to an enhancer element responsible for the tissue-specific expression of the chicken alpha A-crystallin gene. Development. 1991;113:539–550. doi: 10.1242/dev.113.2.539. [DOI] [PubMed] [Google Scholar]
- 33.Chen X, Patel TP, Cain WJ, Duncan MK. Production of monoclonal antibodies against prox1. Hybridoma (Larchmt) 2006;25:27–33. doi: 10.1089/hyb.2006.25.27. [DOI] [PubMed] [Google Scholar]
- 34.Taketo M, Schroeder AC, Mobraaten LE, et al. FVB/N: an inbred mouse strain preferable for transgenic analyses. Proc Natl Acad Sci U S A. 1991;88:2065–2069. doi: 10.1073/pnas.88.6.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddan JR, Chepelinsky AB, Dziedzic DC, Piatigorsky J, Golden-berg EM. Retention of lens specificity in long-term cultures of diploid rabbit lens epithelial cells. Differentiation. 1986;33:168–174. doi: 10.1111/j.1432-0436.1986.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 36.Ireland ME, Tran K, Mrock L. Beta-adrenergic mechanisms affect cell division and differentiation in cultured chick lens epithelial cells. Exp Eye Res. 1993;57:325–333. doi: 10.1006/exer.1993.1131. [DOI] [PubMed] [Google Scholar]
- 37.Sandaltzopoulos R, Becker PB. Solid phase DNaseI footprinting: quick and versatile. Nucleic Acids Res. 1994;22:1511–1512. doi: 10.1093/nar/22.8.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duncan MK, Haynes JI, II, Cvekl A, Piatigorsky J. Dual roles for Pax-6: a transcriptional repressor of lens fiber cell-specific beta-crystallin genes. Mol Cell Biol. 1998;18:5579–5586. doi: 10.1128/mcb.18.9.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y, Stopka T, Golestaneh N, et al. Regulation of alphaA-crystallin via Pax6, c-Maf, CREB and a broad domain of lens-specific chromatin. EMBO J. 2006;25:2107–2118. doi: 10.1038/sj.emboj.7601114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 41.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kopp J, Schwede T. The SWISS-MODEL Repository of annotated three-dimensional protein structure homology models. Nucleic Acids Res. 2004;32:D230–D234. doi: 10.1093/nar/gkh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiner SJ, Kollman PA, Nguyren DT, Case DA. An all atom force field for simulations of proteins and nucleic acids. J Comput Chem. 1986;7:220–252. doi: 10.1002/jcc.540070216. [DOI] [PubMed] [Google Scholar]
- 44.Chen R, Weng Z. Docking unbound proteins using shape complementarity, desolvation, and electrostatics. Proteins. 2002;47:281–294. doi: 10.1002/prot.10092. [DOI] [PubMed] [Google Scholar]
- 45.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 46.Chen R, Weng Z. A novel shape complementarity scoring function for protein-protein docking. Proteins. 2003;51:397–408. doi: 10.1002/prot.10334. [DOI] [PubMed] [Google Scholar]
- 47.Li L, Chen R, Weng Z. RDOCK: refinement of rigid-body protein docking predictions. Proteins. 2003;53:693–707. doi: 10.1002/prot.10460. [DOI] [PubMed] [Google Scholar]
- 48.Wilmarth PA, Taube JR, Riviere MA, Duncan MK, David LL. Proteomic and sequence analysis of chicken lens crystallins reveals alternate splicing and translational forms of beta B2 and beta A2 crystallins. Invest Ophthalmol Vis Sci. 2004;45:2705–2715. doi: 10.1167/iovs.04-0131. [DOI] [PubMed] [Google Scholar]
- 49.Duncan MK, Haynes JI, II, Piatigorsky J. The chicken βA4- and βB1-crystallin-encoding genes are tightly linked. Gene. 1995;162:189–196. doi: 10.1016/0378-1119(95)00363-b. [DOI] [PubMed] [Google Scholar]
- 50.Cook T, Pichaud F, Sonneville R, Papatsenko D, Desplan C. Distinction between color photoreceptor cell fates is controlled by prospero in Drosophila. Dev Cell. 2003;4:853–864. doi: 10.1016/s1534-5807(03)00156-4. [DOI] [PubMed] [Google Scholar]
- 51.Zinovieva RD, Duncan MK, Johnson TR, Torres R, Polymeropoulos MH, Tomarev SI. Structure and chromosomal localization of the human homeobox gene Prox 1. Genomics. 1996;35:517–522. doi: 10.1006/geno.1996.0392. [DOI] [PubMed] [Google Scholar]
- 52.Fagerholm PP, Philipson BT, Lindstrom B. Normal human lens—the distribution of proteins. Exp Eye Res. 1981;33:615–620. doi: 10.1016/s0014-4835(81)80101-7. [DOI] [PubMed] [Google Scholar]
- 53.Wistow G, Bernstein SL, Wyatt MK, et al. Expressed sequence tag analysis of adult human lens for the NEIBank Project: over 2000 non-redundant transcripts, novel genes and splice variants. Mol Vis. 2002;8:171–184. [PubMed] [Google Scholar]
- 54.Ireland ME. A novel model for studying lens fiber elongation. In Vitro Cell Dev Biol Anim. 1996;32:382–385. doi: 10.1007/BF02722996. [DOI] [PubMed] [Google Scholar]
- 55.Krausz E, Augusteyn RC, Quinlan RA, et al. Expression of crystallins, Pax6, filensin, CP49, MIP, and MP20 in lens-derived cell lines. Invest Ophthalmol Vis Sci. 1996;37:2120–2128. [PubMed] [Google Scholar]
- 56.Hong YK, Harvey N, Noh YH, et al. Prox1 is a master control gene in the program specifying lymphatic endothelial cell fate. Dev Dyn. 2002;225:351–357. doi: 10.1002/dvdy.10163. [DOI] [PubMed] [Google Scholar]
- 57.Wigle JT, Harvey N, Detmar M, et al. An essential role for Prox1 in the induction of the lymphatic endothelial cell phenotype. EMBO J. 2002;21:1505–1513. doi: 10.1093/emboj/21.7.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nagai H, Li Y, Hatano S, et al. Mutations and aberrant DNA methylation of the PROX1 gene in hematologic malignancies. Genes Chromosomes Cancer. 2003;38:13–21. doi: 10.1002/gcc.10248. [DOI] [PubMed] [Google Scholar]
- 59.Liu YW, Gao W, Teh HL, Tan JH, Chan WK. Prox1 is a novel coregulator of Ff1b and is involved in the embryonic development of the zebra fish interrenal primordium. Mol Cell Biol. 2003;23:7243–7255. doi: 10.1128/MCB.23.20.7243-7255.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petrova TV, Makinen T, Makela TP, et al. Lymphatic endothelial reprogramming of vascular endothelial cells by the Prox-1 homeobox transcription factor. EMBO J. 2002;21:4593–4599. doi: 10.1093/emboj/cdf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hassan B, Li L, Bremer KA, Chang W, Pinsonneault J, Vaessin H. Prospero is a panneural transcription factor that modulates homeodomain protein activity. Proc Natl Acad Sci U S A. 1997;94:10991–10996. doi: 10.1073/pnas.94.20.10991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choksi SP, Southall TD, Bossing T, et al. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 63.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci U S A. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]