Abstract
Chromosomal translocations are important genetic perturbations frequently associated with hematologic malignancies; characterization of these events has been a rich source of insights into the mechanisms that lead to malignant transformation. The t(10;11)(p13;q14–21) results in a recently identified rare but recurring chromosomal translocation seen in patients with ALL as well as AML, and results in the production of a CALM-AF10 fusion gene. Although the details by which the CALM-AF10 fusion protein exerts its leukemogenic effect remain unclear, emerging data suggests that the CALM-AF10 fusion impairs differentiation of hematopoietic cells, at least in part via an upregulation of HOXA cluster genes. This review discusses the normal structure and function of CALM and AF10; describes the spectrum of clinical findings seen in patients with CALM-AF10 fusions; summarizes recently published CALM-AF10 mouse models; and highlights the role of HOXA cluster gene activation in CALM-AF10 leukemia.
Keywords: CALM-AF10, acute leukemia, chromosomal translocations, t(10;11)
Introduction
Acute leukemia is associated with a wide spectrum of gross chromosomal rearrangements. These acquired mutations include balanced and unbalanced chromosomal translocations, as well as chromosomal inversions, deletions, and amplifications.1–4 The analysis of chromosomal translocations has proven to be especially useful in understanding the biology of hematologic malignancies, leading to improved diagnosis and classification, as well as identification of novel targets for therapy.5 Balanced chromosomal translocations typically result in either dysregulated gene expression or production of a chimeric gene formed from two unrelated genes2, and analysis of numerous recurrent translocations has revealed that many of the genes locates at the translocation breakpoint are transcription factors involved in normal blood cell differentiation.1 In terms of clinical relevance, cloning of the BCR-ABL fusion gene highlighted the role of tyrosine kinases in leukemic transformation and led to the subsequent development of imatinib, a tyrosine kinase inhibitor.6 Furthermore, chromosomal abnormalities are now routinely used to help classify leukemia patients for risk-directed therapy.7
The t(10; 11) translocation was first observed in a patient with diffuse histiocytic lymphoma8; the U937 cell line was established from this patient and has been used extensively as an in vitro model of monocyte differentiation.9 The presence of a reciprocal translocation between chromosomes 10 and 11, a t(10; 11)(p13–14; q14–21) in the U937 cell line was confirmed and subsequently refined by cytogenetic analysis.10, 11 Using positional cloning techniques to narrow the chromosome 10 breakpoint to a 3 cM region, AF10 was identified as a candidate gene on chromosome 10 potentially involved in this translocation. Subsequent studies showed that the AF10 gene was indeed disrupted by the t(10;11) translocation, and identified CALM as the AF10 fusion partner in this recurrent translocation.12
Structure and function of CALM
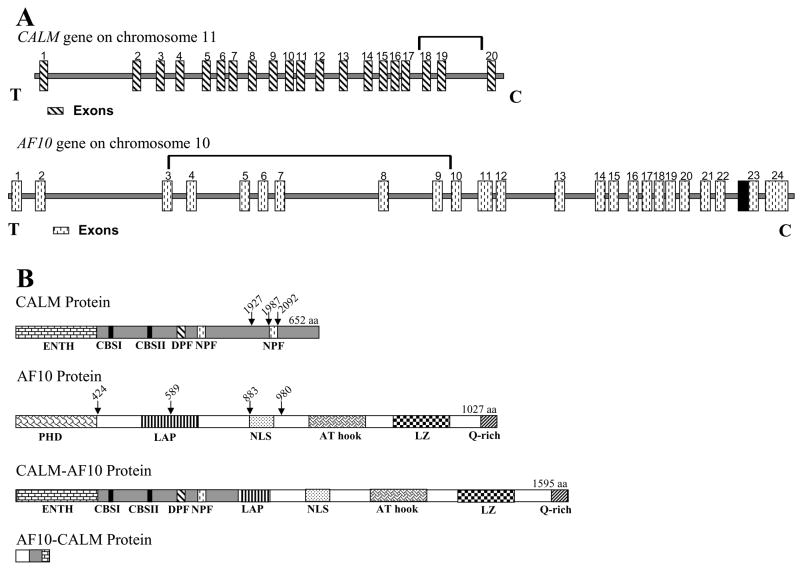
CALM (for Clathrin Assembly Lymphoid Myeloid; also known as PICALM) is located on chromosome 11q23, is ubiquitously expressed, and encodes a 652 aa protein with multiple domains involved in endocytosis (Fig. 1).12 These domains include the epsin N-terminal homology (ENTH) domain (Asp-Pro-Phe), a DPF (ASP-Pro-Phe) motif, an NPF (Asn-Pro-Phe) motif, and type I and type II clathrin-binding sequences (CBS I and II).13–15 The predicted CALM protein is similar to the neuronal specific monomeric clathrin assembly protein AP180, which was first identified in coated vesicles of bovine brain.16, 17 CALM homologues have been identified in rat, mouse and cow.
Figure 1. Structure of CALM, AF10 and CALM-AF10 fusions.
(A) Schematic representation of the human CALM and AF10 genes based on the reference sequences NM_007166 and U13948 respectively. CALM exons 1–20 are indicated by striped rectangles. The bracket spanning introns 18–19 indicate the genomic region where translocations are known to occur within CALM. AF10 exons 1–24 are indicated by stippled rectangles. The bracket spanning introns 3–9 indicate the genomic region where translocations are known to occur within AF10. The black box immediately adjacent to exon 23 represents 124 bp of exon sequence this is present in most ESTs but missing from the reference sequence (U13948). Centromere (C) and telomere (T) orientation is as indicated. (B) Schematic representation of the predicted protein structure for CALM, AF10 and the two chimeric proteins CALM-AF10 and AF10-CALM. Protein domains are indicated in each structure: ENTH (espin N-terminal homology; Asp-Pro-Phe), DPF (ASP-Pro-Phe) motif, NPF (Asn-Pro-Phe) motif, CBS I & II (clathrin binding sequence I & II), PHD (plant homology domain), LAP (leukemia-associated protein), NLS (nuclear localization signal), LZ (leucine zipper), and Q (glutamine)-rich region. Vertical arrows indicate known nucleotide breakpoint regions.
Endocytosis is the active cellular process of removing proteins from the plasma membrane.18 The active formation of transport vesicles that shuttle cytosolic cargo, such as transmembrane and luminal proteins from the plasma membrane and the trans-Golgi network (TGN) to endosomes requires participation of clathrin coated pits.19 Clathrin is composed of three major proteins; a 192 kDa heavy chain bound to two ~30 kDa light chains that forms a structure known as a triskelion.19 In order for coat proteins to function properly in vesicle transport, they link to adaptor proteins (APs).20 The role of APs is complex: First, studies suggest that AP-3 likely functions as a clathrin adaptor protein and aids in signal mediated protein sorting events.21 Secondly, AP1 and AP2 function to attach clathrin to the plasma membrane and help form complexes in the TGN.20 AP180 homologues have been observed in yeast22 and Drosophila23 where they were identified as participants in clathrin binding and endocytosis respectively. Likewise, AP180/CALM homologues identified in C. elegans presumably regulate endocyte vesicle size.23 Finally, point mutations in the CALM gene led to ineffective hematopoiesis, functional iron deficiency, and altered growth in mice, suggesting that CALM may play a role in endocytosis-mediated iron transport.15
In addition to an established role in endocytosis, a recent study suggested that CALM may interact with nuclear proteins.24 In this study, the novel protein CATS (CALM interacting protein, expressed in thymus and spleen) was shown to interact with a region of the CALM protein that is retained in the CALM-AF10 fusion protein and increased the nuclear localization of both CALM and a CALM-AF10 fusion protein. This study further suggested that CATS may be important for malignant transformation mediated by CALM-AF10, through either mis-localization of the CALM-AF10 protein, or via transcriptional regulatory properties of the CATS protein.24
A single case report described an MLL-CALM fusion in a 12-week-old Caucasian female who presented with rapidly progressive AML.25 In this study, the authors used panhandle PCR to determine that MLL was fused to CALM at 11q14–q21. By assaying RNA from the patient’s leukemic cells, fusion transcripts were identified. The breakpoints for this particular translocation were within MLL intron 7 (in the known MLL breakpoint cluster region) and CALM intron 7. This finding is intriguing since, in this case, CALM sequences are present at the 3′ portion of the fusion gene, in contrast to CALM-AF10 fusions, where CALM sequences form the 5′ portion of the fusion gene. In addition the breakpoint lay within CALM intron 7, whereas the breakpoints for CALM-AF10 fusions typically occur within CALM introns 17–19.
Structure and function of AF10
The AF10 (also known as MLLT10) gene is located on chromosome 10p12 and encodes a 109-kDa protein containing 1,027 aa residues, and was initially cloned as an MLL partner gene in the recurrent t(10;11)(p13;q23) translocation.26 It is important to note that the reference sequence for human AF10 (NM_004641 or U13948) is missing 124 nucleotides between exons 22 and 23; these 124 nucleotides are present in most human expressed sequence tags (ESTs) and a homologous 124 nucleotide sequence is present in the mouse reference sequence (NM_010804). Insertion of these 124 nucleotides leads to a frameshift of the COOH terminal portion of the AF10 protein. Studies in both humans and mice have demonstrated that AF10 expression is highest in the testis, and is also expressed in the thymus, ovary, colon, peripheral blood, brain, and kidney.27, 28 AF10 is a member of a small highly conserved protein family that includes the AF17, BR140, and CEZF proteins.27, 29 Although the precise function of AF10 is not known, both structural and functional data suggest that it functions as a transcription factor (Figure 1). The AF10 protein contains PHD (Plant Homeo Domains) fingers, which are structurally conserved domains present in a number of known transcription factors that are involved in chromatin-mediated gene regulation, including the CBP, MLL, TRX, and Drosophila Polycomb group (PCL) proteins.30 In addition to the N-terminal PHD domain, AF10 contains an extended PHD domain also known as a leukemia-associated protein domain (LAP) that functions in homo-oligomerization and is conserved in several other proteins including MLL, AF17, and BR140.29 AF10 also contains an AT-hook motif, a motif that was initially described in the high-mobility group of non-histone chromosomal proteins and other DNA-binding proteins, and is thought to mediate protein binding to cruciform DNA.31 In addition to these domains, AF10 contains a bipartite nuclear localization signal (NLS) and a C-terminus glutamine-rich region. However, due to alternative splicing the latter is not present in all isoforms.29, 32
A role for AF10 in leukemic transformation was proposed as the gene was initially cloned as an MLL chromosomal translocation partner in 3 patients with AML-M5 and an MLL-AF10 fusion.27 An additional report presented two cases of pediatric AML-M5 in which there was paracentric inversion of the 11q region with chromosome 10p12.33 Although pediatric acute megakaryoblastic leukemia (AML-M7) is considered rare, two independent case reports demonstrated fusions between MLL and AF10 in pediatric patients.33, 34 Deletion analysis of the MLL-AF10 fusion protein has demonstrated that the leucine zipper (LZ) motif in the carboxy terminus of AF10 is essential for leukemogenesis 35, and the H3K79 methyltransferase hDOT1L has been shown to be involved in MLL-AF10 mediated leukemogenesis through interaction of hDOT1L and AF10.36
Clinical Features of leukemia with CALM-AF10 fusions
Although the t(10;11)(p13:q14–21) translocation was initially observed in a patient with histiocytic lymphoma, Groupe Français de Cytogénétique Hématologique (GFCH) subsequently identified the t(10;11) translocation as a recurrent reciprocal translocation in patients with T-cell acute lymphoblastic leukemia.37 CALM-AF10 translocations appear to be most frequently associated with T-cell ALL, specifically T-cell ALL of either γ/δ or immature phenotypes. In the largest series of its kind, 20 cases of CALM-AF10 fusion leukemias were identified in a study of 144 patients with T-ALL.38 Patients with a CALM-AF10 fusion ranged from 3–43 years of age (mean 20.7), and were more frequently male (M:F=2.8). WBC at presentation ranged from 3.3–556 × 109/L, and 8 of 20 patients had a mediastinal mass. Although the numbers were small, patients with a CALM-AF10 fusion and an immature phenotype had a poor prognosis, as 10/12 patients either did not respond to therapy or relapsed. In contrast, 8/8 patients with a CALM-AF10 fusion and a TCR γ/δ phenotype were in complete remission at the time of the report. In this series, CALM-AF10 fusions were present in 12/29 cases with an immature TCR γ/δ phenotype and 8/32 cases with a TCR γ/δ phenotype, but in 0/78 α/β or IM β T-ALLs, demonstrating a marked bias toward T-cell ALL of the γ/δ lineage. This study also suggested that the AF10 content of the CALM-AF10 fusion might correlate with the stage of maturation arrest. All but the final four amino acid residues of the protein encoded by the CALM gene are retained in most CALM-AF10 fusion transcripts (Figure 1)12, 38, although due to alternate splicing, 20, 35, or 55 amino acid residues are excluded in some CALM-AF10 fusions. At least four common AF10 fusion sites have been described.38 The resultant CALM-AF10 fusion transcripts can be classified as either 5′ or 3′, depending on how much AF10 sequence is included in the fusion transcript. 5′ fusion transcripts, which retain almost all of the AF10 coding sequence, are associated with TCRγδ T-cells, whereas 3′ fusion transcripts, which contain less AF10 sequence, are associated with more immature T-cells that do not express a TCR on the cell surface.38
CALM-AF10 fusions have also been observed in a variety of myeloid leukemias including AML-M0, M1, M2, M4, M5, and M7 (non-Down’s M7 patients), as well as eosinophilic leukemia and granulocytic sarcoma (Table 1).39–45 Interestingly, most of the AML M0, M1, and M2 cases that have been analyzed show clonal IGH, TCRG, and/or TCRD gene rearrangements, suggesting that the target for malignant transformation may have been a multipotential hematopoietic precursor cell.38, 46 CALM-AF10 translocations have been identified in both children and adults with AML; specific lesions include splenomegaly, hepatomegaly, mediastinal masses, and CNS leukemia.47 Cytochemical and immunophenotypic data varies with the diagnosis but includes markers for lymphoid as well as myeloid antigens.40, 44, 45, 47
Table 1.
Characteristics of patients with the t(10:11) CALM-AF10 translocation
| Patient | Diagnosis | CALM-AF10 Breakpoint(s) | References |
|---|---|---|---|
| U937 | Histiocytic Lymphoma | CALM 2091; AF10 423 | Dreyling, 1996 |
| M/19 | AML-M1 | CALM 1926; AF10 883, CALM 2091; AF10 883 | Bohlander, 2000 |
| M/47 | AML-M0 | CALM 1926; AF10 883 | Bohlander, 2000 |
| F/21 | AML-M1 | CALM 1926; AF10 589, CALM 2091; AF10 589 | Bohlander, 2000 |
| M/22 | T-ALL | CALM 2091; AF10 589 | Bohlander, 2000 |
| F/16 | pre T-ALL | CALM 1926; AF10 883 | Bohlander, 2000 |
| M/5 | pre T-ALL | CALM 2091; AF10 979 | Bohlander, 2000 |
| 1 | ALL | CALM 1926; AF10 980 | Kumon, 1999 |
| 2 | ALL | CALM 2091; AF10 883 | Kumon, 1999 |
| 3 | ALL | CALM 1926; AF10 589 | Kumon, 1999 |
| 4 | AML-M0 | CALM 2091; AF10 589 | Kumon, 1999 |
| M/12 | AmoL | NR | Kobayashi, 1997 |
| F/10 | ALL | NR | Kobayashi, 1997 |
| M/25 | LBL | NR | Kobayashi, 1997 |
| F/30 | Granulocytic sarcoma | NR | Kobayashi, 1997 |
| M/6 | AML-M7 | CALM 1926; AF10 424 | Jones, 2001 |
| F/10 | Acute eosinophilic leukemia | NR | Salmon-Nguyen, 2000 |
| F/17 | T-ALL | NR | Salmon-Nguyen, 2000 |
| M/16 | AML-M4 | CALM 1926; AF10 424 | Nakamura, 2003 |
| M/20 | AML-M4 | NR | Abdou, 2002 |
| F/44 | AML-M0 | CALM 2091; AF10 424 | Carlson, 2000 |
| F/28 | AML-M1 | CALM 2091; AF10 883 | Carlson, 2000 |
| M/41 | AML-M5 | CALM 2091; AF10 979 | Carlson, 2000 |
| M/26 | T-ALL | CALM 2091; AF10 979 | Carlson, 2000 |
| M/12 | T-ALL | CALM 1926; AF10 589 | Carlson, 2000 |
| F/23 | NHL Tcell LBL AML | NR | Carlson, 2000 |
| F/4 | AML-M7 | CALM 2091; AF10 423 | Abdelhaleem, 2007 |
| M/2 | AML-M7 | CALM 2091; AF10 796 | Abdelhaleem, 2007 |
| M/33 | T-ALL TCRγδ | AF10 424 | Macintyre, 2003 |
| M/29 | T-ALL TCRγδ | AF10 589 | Macintyre, 2003 |
| M/20 | T-ALL TCRγδ | AF10 883/979 | Macintyre, 2003 |
| M/15 | T-ALL TCRγδ | AF10 589 | Macintyre, 2003 |
| F/11 | T-ALL TCRγδ | AF10 589 | Macintyre, 2003 |
| M/7 | T-ALL TCRγδ | NR | Macintyre, 2003 |
| M/6 | T-ALL TCRγδ | AF10 589 | Macintyre, 2003 |
| M/3 | T-ALL TCRγδ | AF10 424 | Macintyre, 2003 |
| M/43 | T-ALL IMγ | AF10 589 | Macintyre, 2003 |
| M/37 | T-ALL IMγ | AF10 883/979 | Macintyre, 2003 |
| M/28 | T-ALL IMγ | AF10 883/979 | Macintyre, 2003 |
| M/26 | T-ALL IMγ | AF10 424 | Macintyre, 2003 |
| M/25 | T-ALL IMγ | AF10 883/979 | Macintyre, 2003 |
| F/25 | T-ALL IMδ | AF10 883/979 | Macintyre, 2003 |
| M/24 | T-ALL IMγ | AF10 883/979 | Macintyre, 2003 |
| M/23 | T-ALL IMγ | AF10 424 | Macintyre, 2003 |
| F/20 | T-ALL IMγ | AF10 883/979 | Macintyre, 2003 |
| M/14 | T-ALL IMδ | AF10 883/979 | Macintyre, 2003 |
| F/12 | T-ALL IMδ | AF10 883/979 | Macintyre, 2003 |
| F/12 | T-ALL IMγ | AF10 883/979 | Macintyre, 2003 |
| F/78 | B-ALL | AF10 883/979 | Macintyre, 2003 |
| M/18 | AUL | AF10 883/979 | Macintyre, 2003 |
| M/18 | AUL | AF10 883/979 | Macintyre, 2003 |
| F/29 | AUL | AF10 589 | Macintyre, 2003 |
| M/25 | AML-M1 | AF10 883/979 | Macintyre, 2003 |
| F/16 | AML-M1 | AF10 424 | Macintyre, 2003 |
| M/22 | AML-M0 | CALM 2091; AF10 883 | Deshpande, 2007 |
| F/33 | AML-M2 | CALM 1926; AF10 883 | Deshpande, 2007 |
| F/36 | AML-M1 | CALM 2091; AF10 978 | Deshpande, 2007 |
| M/39 | AUL | CALM 2091; AF10 424 | Deshpande, 2007 |
| M/19 | AML-M1 | CALM 2091; AF10 424 | Deshpande, 2007 |
| M/47 | AML-M1 | CALM 2091; AF10 424 | Deshpande, 2007 |
| F/12 | AML-M1 | CALM 2091; AF10 424 | Deshpande, 2007 |
| F/19 | AML | CALM 2091; AF10 1048 | Deshpande, 2007 |
| F/4 | AML-M5a | NR | Deshpande, 2007 |
| F/12 | AML-M1 | CALM 2091; AF10 424 | Starza, 2006 |
| M/13 | T-ALL | CALM 2091; AF10 424 | Starza, 2006 |
| M/36 | AML-M2 | NR | Starza, 2006 |
| M/27 | AML-M0 | CALM 2091; AF10 424 | Starza, 2006 |
| F/38 | T-ALL | CALM 2091; AF10 589 | Starza, 2006 |
| M/47 | AML-M1 | CALM 2091; AF10 424 | Starza, 2006 |
| M/19 | AML-M1 | NR | Starza, 2006 |
NR; not reported
Mouse Models of CALM-AF10 Leukemia
Animal models have provided tractable systems with which to study a wide range of human diseases, including the recurrent chromosomal translocations involved in hematologic malignancies.48 Given the shared physiology between mice and humans, the mouse has become a standard animal model used to study pathophysiology and explore novel therapeutic approaches. Chromosomal translocations have been studied in mouse models using a variety of techniques including retroviral transduction and bone marrow transplantation as well as gene-targeting and transgenic approaches.48–50 Recently, three groups have published reports that used mice to study CALM-AF10 leukemia; findings from each of these reports are summarized in Table 2.
Table 2.
Summary of CALM-AF10 Mouse Models
| Model | Mouse Strain | Disease | Survival Median (range) | Immunophenotype Markers | Tissue/Organ Involvement | Antigen Receptor Gene Rearrangement | Serial Transplantation | Reference |
|---|---|---|---|---|---|---|---|---|
| Xenograft of siRNA clones | NOD/SCID | AML | 19.5 d (16–23d) control
27d (22–33d) siRNA clones |
CD45 | Spleen, Kidney, Pancreas | not applicable | nd | Okada, 2006 |
| Retroviral infection and bone marrow transplantation | C57Bl/6xC3H | AML | 110 d (46–366d) | Mac1++, Gr1++, B220+/−, MPO++ | Bone Marrow, Spleen, Liver, Lung, Kidney, Brain, Intestine | Igh | yes | Deshpande, 2006 |
| Transgenic | FVB | AML | 12 mo (6–18 mo) | Mac1++, Gr1++, B220 +/−, MPO++, CD24+/−, CD117+/−, CD3 rare +, F4/80 rare + | Bone Marrow, Spleen, Liver, Lung, Kidney, Brain, Lymph node | Igh; Tcrb; Tcrd | nd | Caudell, 2007 |
nd is not done
The first study used a vector based RNA interference system to “knock down” CALM-AF10 expression in the U937 cell line, which expresses a CALM-AF10 fusion protein. The “knockdown” clones proliferated less rapidly in vitro, and showed a modest survival benefit following xeno-transplantation into NOD/SCID mice (median survival 27 vs. 19.5 days post transplant).51
A second study used retroviral transduction and bone marrow transplantation to generate acute leukemia in mice.46 Recipient mice transplanted with bone marrow cells that expressed a CALM-AF10 fusion gene developed disease a median of 110 days following transplantation. Mice with leukemia were anemic and had circulating blasts. Parenchymal infiltration by leukemic myeloid cells was noted in a variety of organs and these cells were positive for the myeloid markers myeloperoxidase and chloracetate esterase. Further analysis of leukemic cells from these mice demonstrated that myeloid cells positive for Mac1 and Gr1 were also positive for the lymphoid marker B220 and had clonal immunoglobulin DH-JH rearrangements, consistent with the observation of clonal IGH gene rearrangements in patients with CALM-AF10 fusions, and suggesting that the target cell for malignant transformation was multi-potential. Interestingly, these authors went on to show that AML could be propagated from a leukemic cell with lymphoid features, as Mac1+ myeloid leukemia could be established by serial transplantation of Mac1+/B220+ or Mac1−/B220+ leukemia cells.46
A third recent study reported a CALM-AF10 transgenic mouse model that developed acute leukemia after a long latency period with incomplete penetrance.52 In this study a CALM-AF10 cDNA was expressed under Vav regulatory elements in the hematopoietic compartment, including thymus, spleen and bone marrow. Depending on the transgenic line, at least 40 or 50% of the F1 generation mice developed acute leukemia, at a median age of 12 months. Prior to the onset of acute leukemia, clinically healthy CALM-AF10 mice displayed a variable, at times severe, inhibition of myeloid and T-cell maturation. CALM-AF10 mice had reduced numbers of CD4+/CD8+ (“double positive”) and increased numbers of immature CD4−/CD8− (“double negative”) cells; interestingly, the proportion of immature CD4−/CD8− thymocytes seemed to increase with age. Leukemic mice typically had enlarged spleens; invasion of parenchymal organs including liver, lung, kidney, and brain with cells that stained positive for myeloid markers such as myeloperoxidase, Mac1, and Gr1. Similar to the findings with the retroviral transduction model described above, many, but not all, of these leukemias expressed both Mac1 and B220, and about half of the leukemic samples had clonal Igh gene rearrangements. Two distinct populations, a B220 “bright” and B220 “dim” population could be detected in the leukemic spleens. The B220 “bright” population was positive for CD19 and IgM, suggesting that these represent contaminating normal B lymphocytes. However, the B220 “dim” population was negative for CD19 and IgM, but positive for CD117 and CD24, suggesting that they represent a progenitor cell not irreversibly committed to the B-cell lineage. This finding is consistent with reports which suggest that B220 is expressed on non-B cell populations progenitor cell populations.45, 53 Additionally, some leukemias arising in CALM-AF10 transgenic mice showed features consistent with T-cell differentiation such as CD3 staining or clonal Tcrb or Tcrd gene rearrangements. Taken together, these data suggest that the B220 expression observed in leukemic cells from CALM-AF10 transgenic mice represent a cell type other than committed B-lineage cells.
The long latency period prior to the onset of leukemia, and the incomplete penetrance of the leukemic phenotype suggest that additional genetic events are needed to complement the CALM-AF10 transgene and complete the process of leukemic transformation. It is tempting to speculate that these proposed additional events may explain the phenotypic differences seen between individual CALM-AF10 mice, in terms of degree of B220 expression, CD3 expression, and clonal rearrangements of antigen receptor genes. In this context, it is interesting to note that some patients with leukemia and CALM-AF10 fusion have clonal IGH and TCRB, TCRG, and TCRD gene rearrangements.38, 46
HOXA cluster genes as CALM-AF10 targets
Mammalian HOX genes are members of the evolutionarily conserved homeodomain family of genes that are structurally organized into four discreet clusters (A, B, C, and D), located on four different chromosomes in mammals, that encode for transcription factors.54, 55 In addition to their well-studied role as executive regulators of anterior-posterior body pattern organization during early embryonic development, HOX genes play a fundamental regulatory role in hematopoietic system organization and development.56 Of the distinct clusters, A, B, and C, but not D are thought to be important for normal hematopoiesis.57 Gene ablation studies have shown specifically that Hoxa9, Hoxc8, and Hoxb6 are necessary for hematopoietic cells to progress though development.57 In addition to their role in normal development, HOX genes also play roles in cell cycle control, cell adhesion, and cell death. Given the central role of HOX genes in normal development, it is not surprising that HOX genes have been implicated in a number of neoplastic and non-neoplastic disease processes.58
Several lines of evidence, based on clinical observations as well as data from experimental animal models, suggest that HOX genes are involved in leukemic transformation. Chromosomal translocations lead to fusions of NUP98 with HOXA9, HOXA11, HOXA13, HOXC11, HOXC13, HOXD11, and HOXD13 in patients with myelodysplastic syndrome (MDS) or AML.59 Chromosomal translocations also lead to ectopic expression of HOXA cluster genes by juxtaposition of TCRA regulatory elements with HOXA coding sequences in some patients with T-cell ALL.60, 61 In addition to these translocations which directly lead to overexpression of HOX genes, a number of recent studies have demonstrated that chromosome translocations which result in MLL fusions lead to unscheduled expression of HOX genes in both AML and T-cell ALL patients.62 In addition to these studies showing HOX gene dysregulation resulting from chromosomal translocations, global gene expression profiling has identified HOX genes as being consistently overexpressed in AML.63, 64 Finally, a number of Hox genes and Hox co-factors, including Hoxa7, Hoxa9, Meis1 and Pbx1 are among the most common upregulated genes in murine leukemias that result from retroviral insertional mutagenesis.65
Direct experimental evidence supporting a role for Hox gene dysregulation in myeloid leukemia comes from several mouse models. Transplantation of bone marrow cells that overexpressed Hoxa5, Hoxa9, or Hoxa10 consistently led to myeloid expansion, and, in collaboration with the Hox cofactor Meis1, AML.57, 58 Transplantation of bone marrow cells that overexpressed Nup98-Hox fusions led to abnormal myeloid differentiation and AML, again in collaboration with Meis1.66–68 Lastly, transgenic mice that expressed a NUP98-HOXD13 fusion showed upregulation of Hoxa7, Hoxa9, and Hoxa10, and developed a myelodysplastic syndrome (MDS) that progressed to acute leukemia in about half of the mice.69
It seems likely that the CALM-AF10 fusion gene, similar to MLL fusions, exerts its leukemic effect, at least in part, through upregulation of HOX genes. HOXA cluster genes, including HOXA5, HOXA9, and HOXA10 were recently shown to be upregulated in the leukemic cells of patients with CALM-AF10 fusions.70 In addition, U937 cells (which express a CALM-AF10 fusion) transduced with a siRNA directed to the CALM-AF10 fusion showed a modest decrease in HOXA5, HOXA7, HOXA9, and HOXA10 expression.51 Consistent with those findings, transgenic mice that expressed a CALM-AF10 transgene showed 8-fold or greater upregulation of Hoxa5, Hoxa7, Hoxa10, Hoxa11, and Meis1 in bone marrow from clinically healthy CALM-AF10 mice, and further upregulation (as much as 500-fold) of these genes in myeloid leukemia cells from CALM-AF10 mice.52
Summary
CALM-AF10 fusions result from a rare but recurring t(10;11) chromosomal translocation seen in both adult and pediatric patients with AML or ALL, and are often associated with a poor prognosis. Recently, it has been shown that CALM-AF10 fusions induce an acute leukemia in mice. Although the exact mechanism by which the CALM-AF10 fusion exerts its leukemogenic effect on hematopoietic cells remains unknown, evolving evidence suggests that the CALM-AF10 fusion leads to upregulation of specific HOXA cluster genes, and interferes with normal hematopoietic differentiation. In the context of a current working model for AML, in which complementary mutations that lead to either impaired differentiation or increased proliferation collaborate to produce a fully leukemic clone,71 it seems likely that the CALM-AF10 fusion functions to impair differentiation. The corollary of this hypothesis is that CALM-AF10 leukemias should acquire spontaneous mutations leading to increased proliferation. The challenge before us is to use this knowledge to develop more effective and less toxic therapies for patients with CALM-AF10 fusions.
Acknowledgments
We would like to thank Drs. Chris Slape, Helge Hartung, Yang Jo Chung, Yue Cheng, R. Mark Simpson, and Siba Samal for their many fruitful and insightful discussions regarding this review. This research was supported by the Intramural Research Program of the NIH, NCI.
References
- 1.Rabbitts TH. Translocations, master genes, and differences between the origins of acute and chronic leukemias. Cell. 1991 Nov 15;67(4):641–644. doi: 10.1016/0092-8674(91)90057-6. [DOI] [PubMed] [Google Scholar]
- 2.Rabbitts TH. Chromosomal translocations in human cancer. Nature. 1994;372(6502):143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 3.Cline MJ. The molecular basis of leukemia. N Engl J Med. 1994 Feb 3;330(5):328–336. doi: 10.1056/NEJM199402033300507. [DOI] [PubMed] [Google Scholar]
- 4.Thandla S, Aplan PD. Molecular biology of acute lymphocytic leukemia. Seminars in Oncology. 1997;24(1):45–56. [PubMed] [Google Scholar]
- 5.Rowley JD. The role of chromosome translocations in leukemogenesis. Seminars in hematology. 1999 Oct;36(4 Suppl 7):59–72. [PubMed] [Google Scholar]
- 6.Rowley JD. Chromosome translocations: dangerous liaisons revisited. Nat Rev Cancer. 2001 Dec;1(3):245–250. doi: 10.1038/35106108. [DOI] [PubMed] [Google Scholar]
- 7.Rubnitz JE, Behm FG, Downing JR. 11q23 rearrangements in acute leukemia. Leukemia. 1996;10(1):74–82. [PubMed] [Google Scholar]
- 8.Sundstrom C, Nilsson K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937) Int J Cancer. 1976 May 15;17(5):565–577. doi: 10.1002/ijc.2910170504. [DOI] [PubMed] [Google Scholar]
- 9.Harris P, Ralph P. Human leukemic models of myelomonocytic development: a review of the HL-60 and U937 cell lines. J Leukoc Biol. 1985 Apr;37(4):407–422. doi: 10.1002/jlb.37.4.407. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi H, Thirman MJ, Rowley JD. U937 cell line has a t(10;11)(p13–14;q14–21) rather than a deletion of 11q. Genes Chromosomes Cancer. 1995 Jul;13(3):217–218. doi: 10.1002/gcc.2870130312. [DOI] [PubMed] [Google Scholar]
- 11.Shipley J, Williams S, O’Byrne A, Kearney L, Jones T, Young B, Dyer M, Catovsky D, Sheer D, Gusterson B. Characterization of a t(10;11)(p13–14;q14–21) in the monoblastic cell line U937. Genes Chromosomes Cancer. 1995 Jun;13(2):138–142. doi: 10.1002/gcc.2870130211. [DOI] [PubMed] [Google Scholar]
- 12.Dreyling MH, Martinez-Climent JA, Zheng M, Mao J, Rowley JD, Bohlander SK. The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Proc Natl Acad Sci U S A. 1996 May 14;93(10):4804–4809. doi: 10.1073/pnas.93.10.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyerholz A, Hinrichsen L, Groos S, Esk PC, Brandes G, Ungewickell EJ. Effect of clathrin assembly lymphoid myeloid leukemia protein depletion on clathrin coat formation. Traffic (Copenhagen, Denmark) 2005 Dec;6(12):1225–1234. doi: 10.1111/j.1600-0854.2005.00355.x. [DOI] [PubMed] [Google Scholar]
- 14.Tebar F, Bohlander SK, Sorkin A. Clathrin assembly lymphoid myeloid leukemia (CALM) protein: localization in endocytic-coated pits, interactions with clathrin, and the impact of overexpression on clathrin-mediated traffic. Mol Biol Cell. 1999 Aug;10(8):2687–2702. doi: 10.1091/mbc.10.8.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klebig ML, Wall MD, Potter MD, Rowe EL, Carpenter DA, Rinchik EM. Mutations in the clathrin-assembly gene Picalm are responsible for the hematopoietic and iron metabolism abnormalities in fit1 mice. Proc Natl Acad Sci U S A. 2003 Jul 8;100(14):8360–8365. doi: 10.1073/pnas.1432634100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahle S, Ungewickell E. Purification and properties of a new clathrin assembly protein. Embo J. 1986 Dec 1;5(12):3143–3149. doi: 10.1002/j.1460-2075.1986.tb04621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takei K, Mundigl O, Daniell L, De Camilli P. The synaptic vesicle cycle: a single vesicle budding step involving clathrin and dynamin. J Cell Biol. 1996 Jun;133(6):1237–1250. doi: 10.1083/jcb.133.6.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherjee S, Ghosh RN, Maxfield FR. Endocytosis. Physiol Rev. 1997 Jul;77(3):759–803. doi: 10.1152/physrev.1997.77.3.759. [DOI] [PubMed] [Google Scholar]
- 19.Schmid SL. Clathrin-coated vesicle formation and protein sorting: an integrated process. Annu Rev Biochem. 1997;66:511–548. doi: 10.1146/annurev.biochem.66.1.511. [DOI] [PubMed] [Google Scholar]
- 20.Robinson MS, Bonifacino JS. Adaptor-related proteins. Curr Opin Cell Biol. 2001 Aug;13(4):444–453. doi: 10.1016/s0955-0674(00)00235-0. [DOI] [PubMed] [Google Scholar]
- 21.Dell’Angelica EC, Klumperman J, Stoorvogel W, Bonifacino JS. Association of the AP-3 adaptor complex with clathrin. Science. 1998 Apr 17;280(5362):431–434. doi: 10.1126/science.280.5362.431. [DOI] [PubMed] [Google Scholar]
- 22.Wendland B, Emr SD. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J Cell Biol. 1998 Apr 6;141(1):71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang B, Koh YH, Beckstead RB, Budnik V, Ganetzky B, Bellen HJ. Synaptic vesicle size and number are regulated by a clathrin adaptor protein required for endocytosis. Neuron. 1998 Dec;21(6):1465–1475. doi: 10.1016/s0896-6273(00)80664-9. [DOI] [PubMed] [Google Scholar]
- 24.Archangelo LF, Glasner J, Krause A, Bohlander SK. The novel CALM interactor CATS influences the subcellular localization of the leukemogenic fusion protein CALM/AF10. Oncogene. 2006 Jul 6;25(29):4099–4109. doi: 10.1038/sj.onc.1209438. [DOI] [PubMed] [Google Scholar]
- 25.Wechsler DS, Engstrom LD, Alexander BM, Motto DG, Roulston D. A novel chromosomal inversion at 11q23 in infant acute myeloid leukemia fuses MLL to CALM, a gene that encodes a clathrin assembly protein. Genes Chromosomes Cancer. 2003 Jan;36(1):26–36. doi: 10.1002/gcc.10136. [DOI] [PubMed] [Google Scholar]
- 26.Chaplin T, Bernard O, Beverloo HB, Saha V, Hagemeijer A, Berger R, Young BD. The t(10;11) translocation in acute myeloid leukemia (M5) consistently fuses the leucine zipper motif of AF10 onto the HRX gene. Blood. 1995 Sep 15;86(6):2073–2076. [PubMed] [Google Scholar]
- 27.Chaplin T, Ayton P, Bernard OA, Saha V, Della Valle V, Hillion J, Gregorini A, Lillington D, Berger R, Young BD. A novel class of zinc finger/leucine zipper genes identified from the molecular cloning of the t(10;11) translocation in acute leukemia. Blood. 1995 Mar 15;85(6):1435–1441. [PubMed] [Google Scholar]
- 28.Linder B, Jones LK, Chaplin T, Mohd-Sarip A, Heinlein UA, Young BD, Saha V. Expression pattern and cellular distribution of the murine homologue of AF10. Biochim Biophys Acta. 1998 Dec 22;1443(3):285–296. doi: 10.1016/s0167-4781(98)00226-7. [DOI] [PubMed] [Google Scholar]
- 29.Linder B, Newman R, Jones LK, Debernardi S, Young BD, Freemont P, Verrijzer CP, Saha V. Biochemical analyses of the AF10 protein: the extended LAP/PHD-finger mediates oligomerisation. J Mol Biol. 2000 Jun 2;299(2):369–378. doi: 10.1006/jmbi.2000.3766. [DOI] [PubMed] [Google Scholar]
- 30.Aasland R, Gibson TJ, Stewart AF. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995 Feb;20(2):56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 31.Aravind L, Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998 Oct 1;26(19):4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Debernardi S, Bassini A, Jones LK, Chaplin T, Linder B, de Bruijn DR, Meese E, Young BD. The MLL fusion partner AF10 binds GAS41, a protein that interacts with the human SWI/SNF complex. Blood. 2002 Jan 1;99(1):275–281. doi: 10.1182/blood.v99.1.275. [DOI] [PubMed] [Google Scholar]
- 33.Morerio C, Rapella A, Tassano E, Rosanda C, Panarello C. MLL-MLLT10 fusion gene in pediatric acute megakaryoblastic leukemia. Leuk Res. 2005 Oct;29(10):1223–1226. doi: 10.1016/j.leukres.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Borkhardt A, Haas OA, Strobl W, Repp R, Mann G, Gadner H, Lampert F. A novel type of MLL/AF10 fusion transcript in a child with acute megakaryocytic leukemia (AML-M7) Leukemia. 1995 Oct;9(10):1796–1797. [PubMed] [Google Scholar]
- 35.DiMartino JF, Ayton PM, Chen EH, Naftzger CC, Young BD, Cleary ML. The AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL-AF10. Blood. 2002 May 15;99(10):3780–3785. doi: 10.1182/blood.v99.10.3780. [DOI] [PubMed] [Google Scholar]
- 36.Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005 Apr 22;121(2):167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 37.t(10;11)(p13–14;q14–21): a new recurrent translocation in T-cell acute lymphoblastic leukemias. Groupe Francais de Cytogenetique Hematologique (GFCH) Genes Chromosomes Cancer. 1991 Nov;3(6):411–415. [PubMed] [Google Scholar]
- 38.Asnafi V, Radford-Weiss I, Dastugue N, Bayle C, Leboeuf D, Charrin C, Garand R, Lafage-Pochitaloff M, Delabesse E, Buzyn A, Troussard X, Macintyre E. CALM-AF10 is a common fusion transcript in T-ALL and is specific to the TCRgammadelta lineage. Blood. 2003 Aug 1;102(3):1000–1006. doi: 10.1182/blood-2002-09-2913. [DOI] [PubMed] [Google Scholar]
- 39.Abdelhaleem M, Beimnet K, Kirby-Allen M, Naqvi A, Hitzler J, Shago M. High incidence of CALM-AF10 fusion and the identification of a novel fusion transcript in acute megakaryoblastic leukemia in children without Down’s syndrome. Leukemia. 2007 Feb;21(2):352–353. doi: 10.1038/sj.leu.2404503. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi H, Hosoda F, Maseki N, Sakurai M, Imashuku S, Ohki M, Kaneko Y. Hematologic malignancies with the t(10;11) (p13;q21) have the same molecular event and a variety of morphologic or immunologic phenotypes. Genes Chromosomes Cancer. 1997 Nov;20(3):253–259. doi: 10.1002/(sici)1098-2264(199711)20:3<253::aid-gcc5>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 41.Salmon-Nguyen F, Busson M, Daniel M, Leblanc T, Bernard OA, Berger R. CALM-AF10 fusion gene in leukemias: simple and inversion-associated translocation (10;11) Cancer Genet Cytogenet. 2000 Oct 15;122(2):137–140. doi: 10.1016/s0165-4608(00)00277-6. [DOI] [PubMed] [Google Scholar]
- 42.Abdou SM, Jadayel DM, Min T, Swansbury GJ, Dainton MG, Jafer O, Powles RL, Catovsky D. Incidence of MLL rearrangement in acute myeloid leukemia, and a CALM-AF10 fusion in M4 type acute myeloblastic leukemia. Leuk Lymphoma. 2002 Jan;43(1):89–95. doi: 10.1080/10428190290000437. [DOI] [PubMed] [Google Scholar]
- 43.Jones LK, Chaplin T, Shankar A, Neat M, Patel N, Samuel DP, Hill AS, Debernardi S, Bassini A, Young BD, Saha V. Identification and molecular characterisation of a CALM-AF10 fusion in acute megakaryoblastic leukaemia. Leukemia. 2001 Jun;15(6):910–914. doi: 10.1038/sj.leu.2402140. [DOI] [PubMed] [Google Scholar]
- 44.Narita M, Shimizu K, Hayashi Y, Taki T, Taniwaki M, Hosoda F, Kobayashi H, Nakamura H, Sadamori N, Ohnishi H, Bessho F, Yanagisawa M, Ohki M. Consistent detection of CALM-AF10 chimaeric transcripts in haematological malignancies with t(10;11)(p13;q14) and identification of novel transcripts. Br J Haematol. 1999 Jun;105(4):928–937. doi: 10.1046/j.1365-2141.1999.01433.x. [DOI] [PubMed] [Google Scholar]
- 45.Bohlander SK, Muschinsky V, Schrader K, Siebert R, Schlegelberger B, Harder L, Schemmel V, Fonatsch C, Ludwig WD, Hiddemann W, Dreyling MH. Molecular analysis of the CALM/AF10 fusion: identical rearrangements in acute myeloid leukemia, acute lymphoblastic leukemia and malignant lymphoma patients. Leukemia. 2000 Jan;14(1):93–99. doi: 10.1038/sj.leu.2401614. [DOI] [PubMed] [Google Scholar]
- 46.Deshpande AJ, Cusan M, Rawat VP, Reuter H, Krause A, Pott C, Quintanilla-Martinez L, Kakadia P, Kuchenbauer F, Ahmed F, Delabesse E, Hahn M, Lichter P, Kneba M, Hiddemann W, Macintyre E, Mecucci C, Ludwig WD, Humphries RK, Bohlander SK, Feuring-Buske M, Buske C. Acute myeloid leukemia is propagated by a leukemic stem cell with lymphoid characteristics in a mouse model of CALM/AF10-positive leukemia. Cancer Cell. 2006 Nov;10(5):363–374. doi: 10.1016/j.ccr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 47.Kumon K, Kobayashi H, Maseki N, Sakashita A, Sakurai M, Tanizawa A, Imashuku S, Kaneko Y. Mixed-lineage leukemia with t(10;11)(p13;q21): an analysis of AF10-CALM and CALM-AF10 fusion mRNAs and clinical features. Genes Chromosomes Cancer. 1999 May;25(1):33–39. doi: 10.1002/(sici)1098-2264(199905)25:1<33::aid-gcc5>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 48.Rabbitts TH. Chromosomal translocation master genes, mouse models and experimental therapeutics. Oncogene. 2001 Sep 10;20(40):5763–5777. doi: 10.1038/sj.onc.1204597. [DOI] [PubMed] [Google Scholar]
- 49.Bernardi R, Grisendi S, Pandolfi PP. Modelling haematopoietic malignancies in the mouse and therapeutical implications. Oncogene. 2002 May 13;21(21):3445–3458. doi: 10.1038/sj.onc.1205313. [DOI] [PubMed] [Google Scholar]
- 50.Forster A, Pannell R, Drynan L, Cano F, Chan N, Codrington R, Daser A, Lobato N, Metzler M, Nam CH, Rodriguez S, Tanaka T, Rabbitts T. Chromosomal translocation engineering to recapitulate primary events of human cancer. Cold Spring Harb Symp Quant Biol. 2005;70:275–282. doi: 10.1101/sqb.2005.70.008. [DOI] [PubMed] [Google Scholar]
- 51.Okada Y, Jiang Q, Lemieux M, Jeannotte L, Su L, Zhang Y. Leukaemic transformation by CALM-AF10 involves upregulation of Hoxa5 by hDOT1L. Nat Cell Biol. 2006 Sep;8(9):1017–1024. doi: 10.1038/ncb1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caudell D, Zhang Z, Chung YJ, Aplan PD. Expression of a CALM-AF10 Fusion Gene Leads to Hoxa Cluster Overexpression and Acute Leukemia in Transgenic Mice. Cancer Res. 2007 Sep 1;67(17):8022–8031. doi: 10.1158/0008-5472.CAN-06-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balciunaite G, Ceredig R, Massa S, Rolink AG. A B220+ CD117+ CD19− hematopoietic progenitor with potent lymphoid and myeloid developmental potential. Eur J Immunol. 2005 Jul;35(7):2019–2030. doi: 10.1002/eji.200526318. [DOI] [PubMed] [Google Scholar]
- 54.Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005 Dec;6(12):893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 55.Garcia-Fernandez J. The genesis and evolution of homeobox gene clusters. Nat Rev Genet. 2005 Dec;6(12):881–892. doi: 10.1038/nrg1723. [DOI] [PubMed] [Google Scholar]
- 56.van Oostveen J, Bijl J, Raaphorst F, Walboomers J, Meijer C. The role of homeobox genes in normal hematopoiesis and hematological malignancies. Leukemia. 1999 Nov;13(11):1675–1690. doi: 10.1038/sj.leu.2401562. [DOI] [PubMed] [Google Scholar]
- 57.Abramovich C, Humphries RK. Hox regulation of normal and leukemic hematopoietic stem cells. Curr Opin Hematol. 2005 May;12(3):210–216. doi: 10.1097/01.moh.0000160737.52349.aa. [DOI] [PubMed] [Google Scholar]
- 58.Grier DG, Thompson A, Kwasniewska A, McGonigle GJ, Halliday HL, Lappin TR. The pathophysiology of HOX genes and their role in cancer. J Pathol. 2005 Jan;205(2):154–171. doi: 10.1002/path.1710. [DOI] [PubMed] [Google Scholar]
- 59.Lam DH, Aplan PD. NUP98 gene fusions in hematologic malignancies. Leukemia. 2001 Nov;15(11):1689–1695. doi: 10.1038/sj.leu.2402269. [DOI] [PubMed] [Google Scholar]
- 60.Soulier J, Clappier E, Cayuela JM, Regnault A, Garcia-Peydro M, Dombret H, Baruchel A, Toribio ML, Sigaux F. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL) Blood. 2005 Jul 1;106(1):274–286. doi: 10.1182/blood-2004-10-3900. [DOI] [PubMed] [Google Scholar]
- 61.Speleman F, Cauwelier B, Dastugue N, Cools J, Verhasselt B, Poppe B, Van Roy N, Vandesompele J, Graux C, Uyttebroeck A, Boogaerts M, De Moerloose B, Benoit Y, Selleslag D, Billiet J, Robert A, Huguet F, Vandenberghe P, De Paepe A, Marynen P, Hagemeijer A. A new recurrent inversion, inv(7)(p15q34), leads to transcriptional activation of HOXA10 and HOXA11 in a subset of T-cell acute lymphoblastic leukemias. Leukemia. 2005 Mar;19(3):358–366. doi: 10.1038/sj.leu.2403657. [DOI] [PubMed] [Google Scholar]
- 62.Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004 Oct;10(10):500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 63.Armstrong SA, Golub TR, Korsmeyer SJ. MLL-rearranged leukemias: insights from gene expression profiling. Seminars in hematology. 2003 Oct;40(4):268–273. doi: 10.1016/s0037-1963(03)00196-3. [DOI] [PubMed] [Google Scholar]
- 64.Mullighan CG, Kennedy A, Zhou X, Radtke I, Phillips LA, Shurtleff SA, Downing JR. Pediatric acute myeloid leukemia with NPM1 mutations is characterized by a gene expression profile with dysregulated HOX gene expression distinct from MLL-rearranged leukemias. Leukemia. 2007 Sep;21(9):2000–2009. doi: 10.1038/sj.leu.2404808. [DOI] [PubMed] [Google Scholar]
- 65.Nakamura T, Largaespada DA, Lee MP, Johnson LA, Ohyashiki K, Toyama K, Chen SJ, Willman CL, Chen IM, Feinberg AP, Jenkins NA, Copeland NG, Shaughnessy JD. Fusion of the nucleoporin gene Nup98 to Hoxa9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nature Genetics. 1996;12(2):154–158. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- 66.Pineault N, Abramovich C, Ohta H, Humphries RK. Differential and common leukemogenic potentials of multiple NUP98-Hox fusion proteins alone or with Meis1. Mol Cell Biol. 2004 Mar;24(5):1907–1917. doi: 10.1128/MCB.24.5.1907-1917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pineault N, Buske C, Feuring-Buske M, Abramovich C, Rosten P, Hogge DE, Aplan PD, Humphries RK. Induction of acute myeloid leukemia in mice by the human leukemia-specific fusion gene NUP98-HOXD13 in concert with Meis1. Blood. 2003 Jun 1;101(11):4529–4538. doi: 10.1182/blood-2002-08-2484. [DOI] [PubMed] [Google Scholar]
- 68.Kroon E, Thorsteinsdottir U, Mayotte N, Nakamura T, Sauvageau G. NUP98-HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. Embo J. 2001 Feb 1;20(3):350–361. doi: 10.1093/emboj/20.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin YW, Slape C, Zhang Z, Aplan PD. NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood. 2005 Jul 1;106(1):287–295. doi: 10.1182/blood-2004-12-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dik WA, Brahim W, Braun C, Asnafi V, Dastugue N, Bernard OA, van Dongen JJ, Langerak AW, Macintyre EA, Delabesse E. CALM-AF10+ T-ALL expression profiles are characterized by overexpression of HOXA and BMI1 oncogenes. Leukemia. 2005 Nov;19(11):1948–1957. doi: 10.1038/sj.leu.2403891. [DOI] [PubMed] [Google Scholar]
- 71.Gilliland DG, Tallman MS. Focus on acute leukemias. Cancer Cell. 2002 Jun;1(5):417–420. doi: 10.1016/s1535-6108(02)00081-8. [DOI] [PubMed] [Google Scholar]