SUMMARY
SUMOylation is essential for cell cycle regulation in invertebrates; however, its functions during the mammalian cell cycle are largely uncharacterized. Mammals express three SUMO paralogues, SUMO-1, SUMO-2 and SUMO-3 (SUMO-2 and SUMO-3 are 96% identical and referred to as SUMO-2/3). We found that SUMO-2/3 localize to centromeres and condensed chromosomes, whereas SUMO-1 localizes to the mitotic spindle and spindle midzone, indicating that SUMO paralogues regulate distinct mitotic processes in mammalian cells. Consistent with this, global inhibition of SUMOylation caused a prometaphase arrest due to defects in targeting the microtubule motor protein, CENP-E, to kinetochores. CENP-E was found to be modified specifically by SUMO-2/3 and to possess SUMO-2/3 polymeric chain-binding activity essential for kinetochore localization. Our findings indicate that SUMOylation is a key regulator of the mammalian cell cycle, with SUMO-1 and SUMO-2/3 modification of different proteins regulating distinct processes.
INTRODUCTION
SUMOs (small ubiquitin-related modifiers), are ~100 amino acid proteins that are post-translationally and covalently conjugated to other proteins (Johnson, 2004; Kerscher et al., 2006). Although invertebrates express one SUMO, vertebrates express three paralogues: SUMO-1, SUMO-2 and SUMO-3. Human SUMO-2 and SUMO-3 are ~96% identical to each other (and are referred to collectively as SUMO-2/3), whereas they share only ~45% identity with SUMO-1. All three SUMOs are covalently conjugated to other proteins through a common enzyme cascade involving the same E1 activating and E2 conjugating enzymes (Johnson, 2004). In addition, all three paralogues are generally thought to affect modified proteins through related mechanisms involving effects on protein structure and function and/or changes in protein-protein or nucleic acid interactions. Several lines of evidence, however, indicate that SUMO-2/3 have protein targets, signaling properties and functions that are unique from those of SUMO-1. Proteomic studies, for example, have identified distinct but partially overlapping subsets of SUMO-1 and SUMO-2/3 modified proteins (Rosas-Acosta et al., 2005; Vertegaal et al., 2006). In addition, SUMO-2/3 conjugation is preferentially up-regulated in response to cell stress and SUMO-2/3 are more mobile within the nucleus relative to SUMO-1 (Ayaydin and Dasso, 2004; Saitoh and Hinchey, 2000). Despite these general observations, however, specific functional differences between SUMO-1 and SUMO-2/3 remain to be identified.
Over 200 proteins have been identified as SUMO-1 or SUMO-2/3 substrates through biochemical and proteomic approaches, implicating SUMOylation as a regulator of a wide range of functions largely associated with the nucleus (Seeler and Dejean, 2003). Genetic studies in particular have identified roles for SUMOylation in regulating chromosome segregation and progression through mitosis. In yeast and Drosophila, SUMOylation is essential for mitotic chromosome condensation, sister chromatid cohesion, kinetochore function and mitotic spindle elongation (Watts, 2007). Although it can be presumed that multiple different proteins are SUMOylated at distinct stages to regulate these diverse mitotic events, few relevant SUMO substrates have been identified. Known substrates include topoisomerase II, Pds5, Ndc10 and Bir1 (Azuma et al., 2003; Bachant et al., 2002; Montpetit et al., 2006; Stead et al., 2003). Notable, however, is that few studies have identified mitotic functions or specific protein targets for SUMOylation in mammalian cells.
Here, we demonstrate that SUMO-1 and SUMO-2/3 conjugation to distinct subsets of proteins is essential for progression through mitosis in mammalian cells. Inhibition of SUMOylation caused cells to arrest at prometaphase due to a defect in the association of CENP-E, a SUMO-2/3 substrate and SUMO-2/3 polymeric chain binding protein, to kinetochores. Our findings reveal a paralogue-specific role for SUMO-2/3 in regulating the association of CENP-E with kinetochores and demonstrate that SUMOylation, like ubiquitination and phosphorylation, is a key regulator of mitosis.
RESULTS
Differential regulation of SUMO-1 and SUMO-2/3 modification
To identify and characterize potentially unique functions of mammalian SUMO-2/3, we immunized mice with human SUMO-2 and produced monoclonal antibodies (mAbs). Immunoblot and immunofluorescence analysis demonstrated that mAb 8A2 recognized both SUMO-2 and SUMO-3, but not SUMO-1 (Figure S1). The previously isolated mAb, 21C7 (Matunis et al., 1996), reacted specifically with SUMO-1 (Figure S1).
Using these mAbs, we performed immunofluorescence microscopy on HeLa cells and detected significant differences in SUMO-1 and SUMO-2/3 localization at different stages of the cell cycle (Figure 1A). To enhance detection, cells were permeabilized with digitonin prior to fixation. This treatment had no effect on the overall localization patterns observed when cells were fixed prior to permeabilization (data not shown), but allowed for clearer detection of SUMO-1 and SUMO-2/3 associations with underlying cell structures. Using these conditions, SUMO-1 was evident at the cytoplasmic face of the nuclear envelope of interphase cells, but SUMO-2/3 was not. In prophase, little if any SUMO-1 was detected in the region of condensed mitotic chromosomes, but as cells progressed to metaphase, SUMO-1 co-localized with the mitotic spindle, as previously reported (Joseph et al., 2002; Matunis et al., 1996). In anaphase, SUMO-1 was no longer detected on the mitotic spindle, but was seen at the spindle midzone. During telophase, SUMO-1 continued to localize to the midzone and ultimately to the cleavage furrow as cells divided. SUMO-1 reappeared at the nuclear envelope as it reformed during late telophase.
Figure 1.
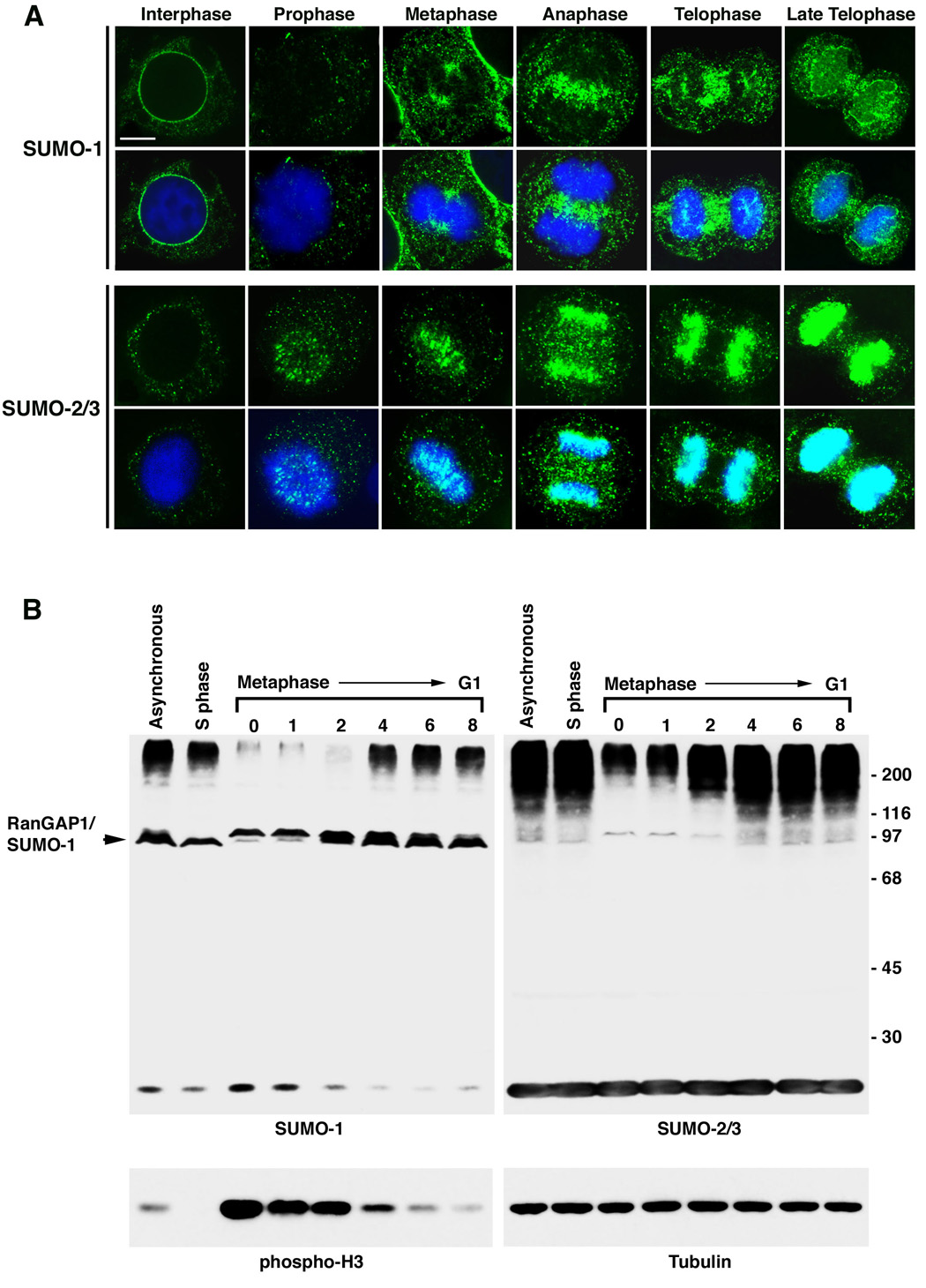
SUMO-1 and SUMO-2/3 conjugates localize to distinct sub-cellular domains during mitosis and are differentially regulated. A. HeLa cells were permeabilized with digitonin, fixed with formaldehyde and analyzed by immunofluorescence confocal microscopy using SUMO-1 or SUMO-2/3 specific mAbs. Indicated cell cycle stages were defined by DNA staining with DAPI. Bar equals 5 µm. B. HeLa cells were arrested in S phase using thymidine, or synchronized in mitosis by a thymidine and nocodazole block followed by release from nocodazole for the indicated times (hr). Cell lysates were separated by SDS-PAGE and analyzed by immunoblotting with SUMO-1 or SUMO-2/3 specific mAbs. As markers for mitosis and protein loading, lysates were probed with antibodies against phospho-histone H3 and α-tubulin.
Remarkably, SUMO-2/3 localized to distinct structures during mitosis compared to SUMO-1 (Figure 1A). During prophase and metaphase, SUMO-2/3 localized to foci on condensed chromosomes. As cells progressed from metaphase to anaphase, SUMO-2/3 appeared to spread along the length of the chromosomes. By late anaphase, the SUMO-2/3 signal covered the condensed chromosomes and the signal increased in intensity as cells progressed through telophase and cytokinesis.
To investigate whether changes in the repertoire of SUMO-1 and SUMO-2/3 modified proteins could be detected as cells progress through the cell cycle, we performed immunoblot analysis using lysates from synchronized cells (Figure 1B). The profiles of SUMO-1 and SUMO-2/3 modified proteins did not differ noticeably between asynchronous cells and cells in S phase. In contrast, a dramatic decrease in high molecular mass SUMO-1 modified proteins was detected in mitotic cells. This decrease persisted until cells re-entered G1. Notably, levels of SUMO-1 modified RanGAP1 remained stable throughout the cell cycle. High molecular mass SUMO-2/3 modified proteins also decreased in overall abundance following nocodazole treatment. However, in contrast to SUMO-1, the levels of SUMO-2/3 conjugates gradually increased as cells progressed through mitosis following nocodazole release. This increase in SUMO-2/3 conjugates correlated with the observed increase in SUMO-2/3 labeling of chromosomes detected in anaphase and telophase (Figure 1A). Collectively, these results indicate that SUMO-1 and SUMO-2/3 are conjugated to distinct subsets of proteins as cells progress through the cell cycle and that their conjugation to these proteins is differentially regulated.
SUMO-2/3 modified proteins at centromeres and kinetochores
To further investigate SUMO-2/3 localization on mitotic chromosomes, cells were analyzed by confocal immunofluorescence microscopy using antibodies specific for SUMO-2/3 and CENP-B, a marker for centromeric heterochromatin (Cooke et al., 1990) (Figure 2A). CENP-B staining appeared as a bar-like signal connecting the centromeres of paired sister chromatids and overlapped significantly with SUMO-2/3 localization. Cells were also co-labeled with antibodies to CENP-C (Saitoh et al., 1992), which appeared as pairs of foci that partially overlapped with the SUMO-2/3 signal, indicating a partial juxtaposition of SUMO-2/3 with the inner kinetochore plate (Figure 2A).
Figure 2.

SUMO-2/3 modified proteins distinct from topoisomerase IIα localize to the centromere and inner kinetochore plate of mitotic chromosomes. A. HeLa cells were permeabilized with digitonin and fixed with formaldehyde. Cells were labeled with SUMO-2/3 mAbs and CENP-B or CENP-C antibodies and analyzed by immunofluorescence confocal microscopy. Bar equals 10 µm. B. Purified mitotic chromosomes were separated by SDS-PAGE and analyzed by immunoblotting with SUMO-2/3, SUMO-1 and Topoisomerase IIα (Topo IIα) specific mAbs. C. HTETOP cells were cultured in the absence of tetracycline (−Tet) to allow topoisomerase IIα expression, or in the presence of tetracycline (+Tet) to inhibit expression. Cells were permeabilized with digitonin, fixed with formaldehyde and analyzed by immunofluorescence microscopy using topoisomerase IIα or SUMO-2/3 specific mAbs. Bar equals 10 µm.
To characterize the profile of SUMOylated proteins associated with mitotic chromosomes, chromosomes were purified from colcemid-arrested cells and associated proteins were analyzed by immunoblotting (Figure 2B). Antibodies specific for SUMO-2/3, revealed a ladder of high molecular mass conjugates (>120 kDa). Unconjugated SUMO-2/3 was not detected (not shown), indicating that free monomeric SUMO-2/3 are not associated with mitotic chromosomes. In contrast to SUMO-2/3, only a single major band corresponding in molecular mass to modified RanGAP1 was detected with antibodies to SUMO-1.
Topoisomerase IIα is the major SUMO-2/3 modified protein associated with mitotic chromosomes formed in Xenopus egg extracts (Azuma et al., 2003). Prometaphase chromosome preparations from HeLa cells were therefore probed with antibodies against topoisomerase IIα (Figure 2B). Topoisomerase IIα antibodies reacted with a protein of ~160 kDa that correlates with the predicted molecular mass of unmodified topoisomerase IIα but not with higher molecular mass bands indicative of SUMOylation. We also analyzed SUMO-2/3 labeling of mitotic chromosomes in HTETOP cells where the topoisomerase IIα gene expression is controlled by a tetracycline responsive promoter (Carpenter and Porter, 2004). In the absence of topoisomerase IIα expression, we observed strong SUMO-2/3 labeling of centromeres and mitotic chromosomes, similar to labeling in cells expressing topoisomerase IIα (Figure 2C). These findings are consistent with topoisomerase IIα being only transiently modified by SUMO-2/3 at the metaphase to anaphase transition (Azuma et al., 2003) and demonstrate that additional SUMO-2/3 modified proteins distinct from topoisomerase IIα are associated with mitotic chromosomes in mammalian cells.
Inhibition of SUMOylation blocks cell cycle progression
Based on their localizations during mitosis, we sought to investigate how SUMO-1 and SUMO-2/3 modifications might control chromosome segregation in mammalian cells. The modification of most SUMO substrates is dynamic, with the levels of modified proteins determined by the relative rates of conjugation and isopeptidase-mediated deconjugation (Johnson, 2004). We therefore tested whether over-expression of SUMO-specific isopeptidases could inhibit the accumulation of SUMOylated proteins in cultured cells. Cells were transiently transfected with GFP-tagged SENP2, a SUMO-specific isopeptidase normally associated with nuclear pore complexes (Hang and Dasso, 2002; Zhang et al., 2002), and the effect on SUMO-1 and SUMO-2/3 modified proteins was determined by immunoblot analysis (Figure 3A). In untransfected cells, high molecular mass SUMO-1 and SUMO-2/3 conjugates were detected. In contrast, high molecular mass SUMO-1 and SUMO-2/3 conjugates were absent in GFP-SENP2 transfected cells, demonstrating that SENP2 over-expression inhibits the accumulation of SUMOylated proteins. Notably, SUMO-1 modified RanGAP1 was not affected by SENP-2 over-expression, possibly due to protective interactions with Nup358 (Zhang et al., 2002).
Figure 3.
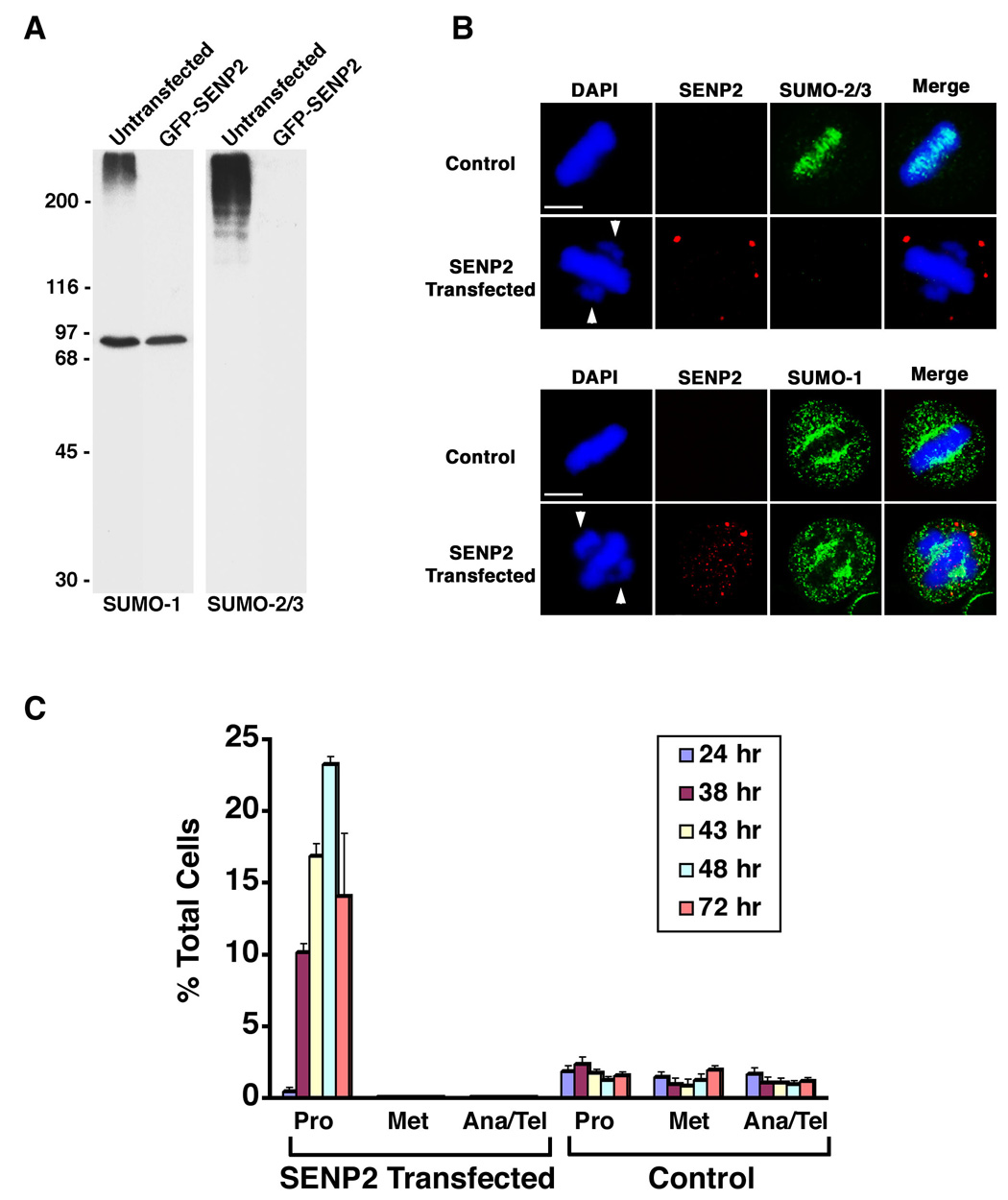
Over-expression of SENP2 inhibits SUMOylation and causes a prometaphase arrest. A. GFP-SENP2 transfected HeLa cells were purified by flow cytometry 48 hr after transfection and lysates were analyzed by immunoblotting with SUMO-1 or SUMO-2/3 specific mAbs. Equal numbers of untransfected cells were analyzed as controls. B. Myc-tagged SENP2 and control transfected HeLa cells were permeabilized with digitonin and fixed with formaldehyde 48 hr after transfection. Cells were double labeled with anti-Myc antibodies and SUMO-2/3 or SUMO-1 specific mAbs. Arrows indicate unaligned chromosome pairs at spindle poles. Bars equal 10 µm. C. The fraction of total cells present at each stage of mitosis following transfection with Myc-tagged SENP2 (n ≥ 135 cells for each time point) or empty vector (Control) (n ≥ 633 cells for each time point) was determined by DAPI staining and fluorescence microscopy (Pro = prophase and prometaphase; Met = metaphase; Ana/Tel = anaphase and telophase). The plotted values are the means plus standard deviation (SD) from a minimum of 3 experiments.
To investigate the effects of SENP2 over-expression on SUMO-1 and SUMO-2/3 localization, immunofluorescence microscopy was performed following transfection (Figure 3B). Analysis with SUMO-2/3 specific antibodies revealed a loss of SUMO-2/3 association with mitotic chromosomes in SENP2 over-expressing cells, consistent with the loss of SUMO-2/3 modified proteins seen by immunoblotting. In contrast, SUMO-1 staining was nearly identical in SENP2 over-expressing and control cells. SUMO-1 and RanGAP1 both localized to the spindle (Figures 3B and S2J), indicating that RanGAP1 is the major SUMO-1 modified protein associated with spindles.
We next investigated whether inhibition of SUMOylation affected chromosome segregation and/or cell cycle progression. Cells were transfected with myc-tagged SENP2 and the fraction of cells at each stage of mitosis was determined at different time points by DAPI staining and fluorescence microscopy (Figure 3C). Significantly, a dramatic increase in mitotic cells was observed 24 to 48 hours post-transfection. At 48 hours, ~25% of SENP2 over-expressing cells were arrested in prometaphase. DAPI staining revealed a defect in chromosome congression, with significant numbers of chromosome pairs failing to align at the metaphase plate and concentrating at the spindle poles (Figure 3B). Transfected cells in anaphase or later stages of mitosis were not detected and the percentage of prometaphase arrested cells decreased at later times. Time-lapse photomicroscopy revealed that arrested cells fail to exit mitosis and undergo apoptosis (not shown).
Inhibition of SUMOylation affects CENP-E localization
The chromosome congression defect observed in SENP2 over-expressing cells could result from a defect in kinetochore assembly and/or function. We therefore investigated the effects of SENP2 over-expression on the localization of individual centromere and kinetochore-associated proteins by immunofluorescence microscopy. The localizations of the centromere-associated proteins Aurora B, CENP-B and Survivin were not affected by SENP-2 over-expression (Figures 4A, S2A and B). Similarly, the kinetochore-associated proteins Hec1, CENP-C, CENP-F, Nup107 and Nup96 were all properly localized in SENP-2 over-expressing cells (Figures 4B, S2C–F). The only protein clearly mislocalized in SENP2 over-expressing cells was the outer kinetochore-associated protein CENP-E (Figure 4C). In control prophase cells, CENP-E was detected at kinetochores of paired sister chromatids. In contrast, CENP-E was undetectable at kinetochores in SENP-2 over-expressing cells. Immunoblot analysis, and attempts to overcome SENP2-induced prometaphase arrest using proteasome inhibitors, indicated that CENP-E loss at kinetochores was not due to degradation, but rather to a defect in kinetochore localization (Figure S3).
Figure 4.

Inhibition of SUMOylation blocks CENP-E association with kinetochores and results in persistent activation of the spindle checkpoint. HeLa cells were transfected with Myc-tagged SENP2 or empty vector. Cells were fixed with paraformaldehyde and permeabilized with Triton X-100 48 hr after transfection and analyzed by immunofluorescence microscopy following labeling with DAPI and the indicated antibodies. Bars equal 10 µm. A. Analysis with anti-Myc and Aurora B antibodies. B. Analysis with anti-Myc and Hec1 antibodies. C. Analysis with anti-Myc and CENP-E antibodies. D. Analysis with anti-Myc and BubR1 antibodies.
A defect in CENP-E kinetochore localization is consistent with the chromosome congression defect seen in SENP2 over-expressing cells, as loss or inhibition of CENP-E also causes cells to arrest at prometaphase with unaligned chromosome pairs (McEwen et al., 2001). To further establish the link between SENP2 over-expression and CENP-E mislocalization, we assayed for the localizations of the spindle checkpoint proteins BubR1, Mad2 and Bub1 (Figure 4D, S2G–I). These checkpoint proteins interact with kinetochores independently of CENP-E. However, they are not released from kinetochores of unaligned chromosome pairs that accumulate in cells lacking functional CENP-E (McEwen et al., 2001). In control prophase cells, BubR1, Mad2 and Bub1 were detected at the kinetochores of all paired sister chromatids. In SENP2 over-expressing cells, BubR1, Mad2 and Bub1 signals were weak or undetectable on the kinetochores of chromosomes aligned at the metaphase plate. However, strong signals for all three proteins were detected on unaligned chromosomes at the spindle poles. These results are consistent with the loss of CENP-E representing the major kinetochore defect affecting SENP2 over-expressing cells.
To confirm that mislocalization of CENP-E is directly related to inhibition of SUMOylation, we used small interfering RNAs to inhibited expression of the essential SUMO E2 conjugating enzyme, Ubc9. Depleting cells of Ubc9 resulted in a significant decrease in high molecular mass SUMO-1 and SUMO-2/3 conjugates and, most significantly, resulted in a cell cycle arrest identical in nature to the arrest observed in SENP2 over-expressing cells (Figure S4). Notably, as in SENP2 over-expressing cells, the observed chromosome congression defect correlated with a mislocalization of CENP-E. These findings demonstrate that inhibition of SUMOylation results in chromosome congression defects and persistent activation of the spindle checkpoint due to mislocalization of CENP-E.
Essential interactions between CENP-E and SUMO-2/3 polymeric chains
Several mechanisms could explain the requirement for SUMOylation in targeting CENP-E to kinetochores. We sought to investigate possible mechanisms by first determining whether CENP-E is a SUMO substrate. To simplify this analysis, we focused on an ~700 amino acid carboxyl-terminal fragment of CENP-E (amino acids 1958–2701 and referred to as the tail domain) that is necessary and sufficient for kinetochore localization (Chan et al., 1998). A plasmid coding for a FLAG-tagged CENP-E tail domain was transiently transfected into cells in the presence and in the absence of a plasmid coding for SENP2 (Figure 5A). Without SENP2 over-expression, the CENP-E tail domain localized to kinetochores as revealed by co-localization with CENP-B. In contrast, and similar to endogenous CENP-E, localization of the tail domain to kinetochores was inhibited in SENP2 over-expressing cells.
Figure 5.
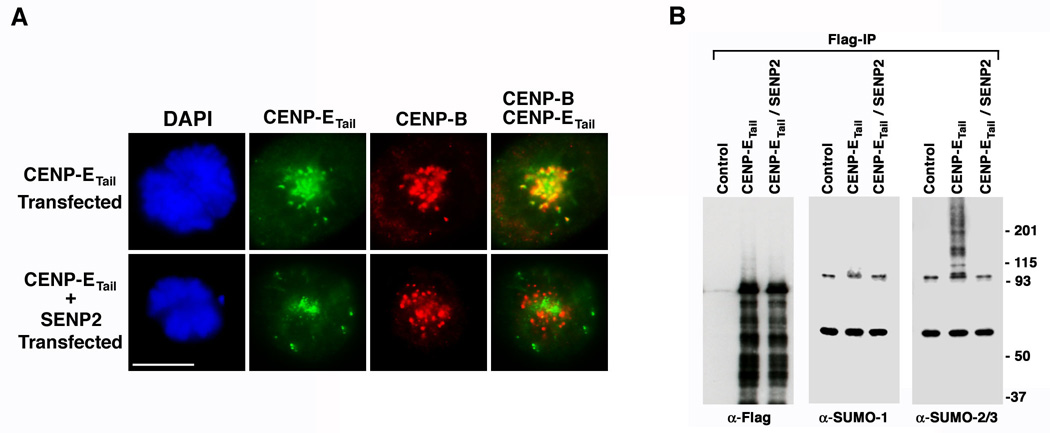
Localization of the CENP-E tail domain to kinetochores correlates with its SUMO-2/3 modification. A. HeLa cells were transfected with a plasmid coding for FLAG-tagged CENP-E tail domain alone or together with a plasmid coding for Myc-tagged SENP2. Cells were fixed with paraformaldehyde and permeabilized with Triton X-100 48 hr after transfection and analyzed by immunofluorescence microscopy after labeling with DAPI and anti-FLAG and CENP-B antibodies. Bar equals 10 µm. B. 293 cells were transfected with empty plasmid (Control), plasmid coding for FLAG-tagged CENP-E tail domain alone, or plasmid coding for FLAG-tagged CENP-E tail domain together with plasmid coding for Myc-tagged SENP2. The FLAG-tagged CENP-E tail domain was immunopurified in the presence of 1% Empigen BB and analyzed by immunoblotting with FLAG, SUMO-1 or SUMO-2/3 specific antibodies as indicated.
To determine whether the CENP-E tail domain is a substrate for SUMOylation, cells were transiently transfected with a plasmid coding for a FLAG-tagged tail domain alone or together with a plasmid coding for SENP2. Immunopurifications were performed under conditions that disrupt protein-protein interactions, and purified proteins were analyzed by immunoblot analysis (Figure 5B and S5A). Equal amounts of FLAG-tagged CENP-E tail domain were immunopurified from cells transfected with or without SENP2 and a majority of the full-length protein migrated according to its unmodified 80kDa form (proteins lower than 80 kDa likely represent degradation products). No high molecular mass forms of the CENP-E tail domain were detected upon immunoblotting with SUMO-1 specific antibodies. In contrast, a ladder of high molecular mass bands migrating between 95 kDa and the top of the gel was observed following immunoblotting with SUMO-2/3 specific antibodies. This ladder of CENP-E conjugates was not detected in cells co-expressing SENP2, demonstrating a correlation between SUMO-2/3 modification of the CENP-E tail domain and its localization to kinetochores. SUMOylation of the CENP-E tail domain was verified in vitro (Figure S7). In an attempt to map the SUMO-2/3 modification sites in CENP-E, we mutated 8 lysine residues in the tail domain that exist within consensus SUMOylation sites. Expression of this mutant tail domain in vivo, however, revealed that it is both modified by SUMO-2/3 and targeted to kinetochores (Figure S6). Because the tail domain contains 87 additional lysine residues and appears to be modified at multiple sites, we sought an alternative approach to investigate the relationship between SUMO-2/3 modification and CENP-E kinetochore targeting.
Recent studies have highlighted the importance of both covalent SUMOylation and non-covalent SUMO-binding in targeting proteins to PML nuclear bodies (Lin et al., 2006; Shen et al., 2006). Analysis of the CENP-E tail domain revealed a sequence motif that closely matches the conserved SUMO-interacting motif (SIM) found in other SUMO-binding proteins (Figure 6A) (Hecker et al., 2006). To investigate whether this motif mediates interactions with SUMO, in vitro binding assays were performed using wild type CENP-E tail domain and a mutant in which two consensus hydrophobic residues in the SIM were mutated (Figure 6A and B). In binding assays with monomeric SUMO-1 or SUMO-2, no interactions were detected. Because SUMO-1 and SUMO-2/3 may also exist as polymers in vivo, we performed binding assays with polymeric chains. Strikingly, the wild type CENP-E tail domain bound specifically to SUMO-2 polymeric chains and this interaction was significantly reduced (> 8 fold) by mutations in the SIM. Based on both in vivo and in vitro analyses, SIM mutations had no effect on covalent SUMOylation of the CENP-E tail domain (Figures S5B and S7).
Figure 6.

The association of CENP-E with kinetochores is dependent on non-covalent binding to SUMO-2/3 polymeric chains. A. Sequence alignment of a putative SIM within the CENP-E tail domain and known SIMs in PML and Daxx. Two isoleucine residues, at positions 2307 and 2308, were mutated to lysines to generate the SIM mutant CENP-E tail domain. B. Wild type (WT) and SIM mutant CENP-E tail domains were expressed in vitro in the presence of [35S]methionine and incubated with glutathione sepharose beads containing GST, SUMO-1, SUMO-2, SUMO-1 polymeric chains or SUMO-2 polymeric chains. Bound proteins were analyzed by SDS-PAGE and autoradiography. Input represents 50% of the total protein used in each binding assay. Relative binding activity was quantified by phosphorimage analysis. The relative binding activity of the SIM mutant was normalized to that of WT. Error bars indicate SD from 3 experiments. C. HeLa cells were transfected with FLAG-tagged wild type (WT) and SIM mutant CENP-E tail domain constructs and analyzed by immunofluorescence microscopy 48 hr after transfection. Cells were double labeled with anti-FLAG and CENP-B antibodies. Shown are prophase cells expressing wild type and SIM mutant CENP-E tail domains. Bar equals 10 µm. D. Late telophase and post-mitotic cells expressing FLAG-tagged SIM mutant CENP-E tail domain were analyzed by immunofluorescence microcopy with anti-FLAG antibodies (left two panels). Midbody localization of endogenous CENP-E was detected in untransfected cells with anti-CENP-E antibodies (right two panels). DNA was detected using DAPI. Bar equals 10 µm. E. HeLa cell lysates were prepared 48 hr following transfection with FLAG-tagged wild type (WT) and SIM mutant CENP-E tail domain constructs and analyzed by immunoblotting with anti-FLAG and tubulin antibodies. F. The fraction of cells present at each stage of mitosis 48 hr after transfection with FLAG-tagged wild type (WT) (n ≥ 105 cells) or SIM mutant (n ≥ 273 cells) CENP-E tail domains was determined by DAPI staining and fluorescence microscopy (Pro = prophase and prometaphase; Met = metaphase; Ana/Tel = anaphase and telophase; Multi-Nuc = multi-nucleated cells). The plotted values are the means plus SD from 3 experiments.
To determine whether non-covalent interactions between CENP-E and polymeric SUMO-2/3 are important for CENP-E localization, cells were transfected with plasmids coding for FLAG-tagged wild type and SIM mutant CENP-E tail domains and analyzed by immunofluorescence microscopy (Figure 6C–F). The wild type CENP-E tail domain localized to kinetochores in a majority of mitotic cells (>90%, n=51) and caused a prometaphase arrest due to displacement of endogenous full-length CENP-E (Chan et al., 1998). In contrast, the majority of cells transfected with the SIM mutant tail domain (>88%, n=51) showed no detectable kinetochore localization despite expression levels similar to wild type protein. Moreover, expression of the SIM mutant caused no mitotic arrest, consistent with inefficient kinetochore localization. The SIM mutant CENP-E tail domain was detected at the spindle midzone and midbody during late stages of mitosis, possibly affecting cytokinesis and accounting for an increase in multi-nucleated cells. Localization of the SIM mutant to the midbody mirrored localization of endogenous CENP-E (Figure 6D), suggesting that the protein was properly folded. We also examined the localization of full-length CENP-E containing SIM mutations and found that, like the mutant tail domain, it failed to localize properly to kinetochores (Figure 8S).
The finding that non-covalent interactions with SUMO-2/3 polymeric chains are essential for CENP-E localization prompted us to identify other kinetochore-associated proteins that are covalently modified by SUMO-2/3. We specifically investigated whether BubR1 and/or Nuf2 could be modified by SUMO-2/3, as both are implicated in CENP-E kinetochore targeting (Chan et al., 1998; Liu et al., 2007). Cells were transiently transfected with plasmids coding for FLAG-tagged BubR1 or Nuf2 and immunopurifications were performed using a FLAG-specific antibody. Immunoblot analysis revealed that both BubR1 and Nuf2 are modified by SUMO-2/3 but not by SUMO-1 (Figures 7A and 7B). Notably, SUMO-2/3 modification of BubR1 and Nuf2 was inhibited by SENP2 over-expression. Thus, non-covalent interactions between CENP-E and SUMO-2/3 polymeric chains (conjugated to kinetochore-associated proteins that may include BubR1 and Nuf2) are essential for CENP-E localization and function at kinetochores.
Figure 7.
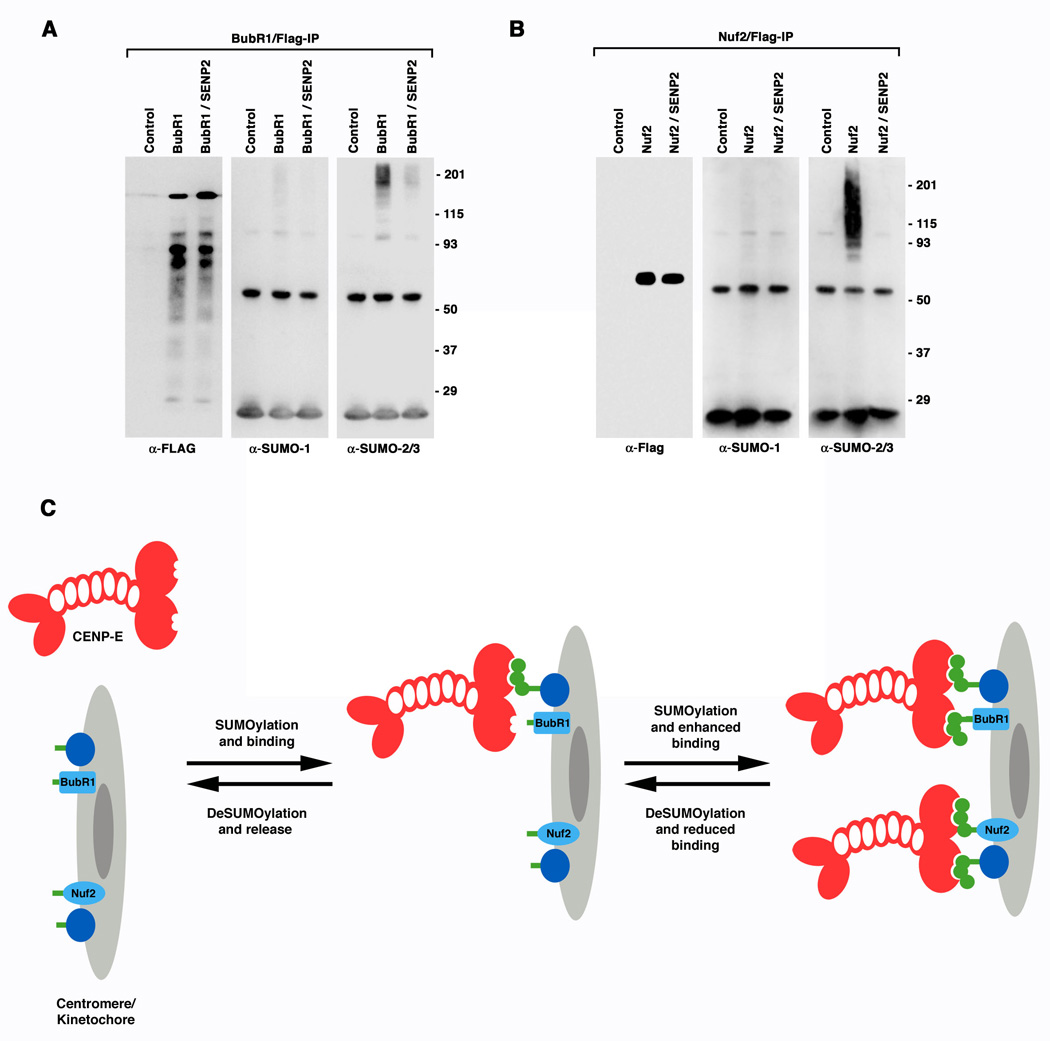
BubR1 and Nuf2 are specifically modified by SUMO-2/3 in vivo, suggesting a model for SUMO-2/3 dependent targeting of CENP-E to kinetochores. A. 293 cells were transfected with empty plasmid (Control), FLAG-tagged BubR1 plasmid alone, or FLAG-tagged BubR1 and Myc-tagged SENP2 plasmids together. FLAG-tagged BubR1 was immunopurified and analyzed by immunoblotting with FLAG, SUMO-1 or SUMO-2/3 specific antibodies as indicated. B. SUMOylation of Nuf2 was analyzed as described for BubR1. C. A proposed model for the roles of SUMO-2/3 modification and polymeric chain binding in regulating the association of CENP-E with kinetochores. CENP-E is depicted as a dimer containing SUMO-2/3 polymeric chain interacting motifs (notches) within the carboxyl-terminal tail domain. The kinetochore/centromere is depicted as having associated proteins, including BubR1 and Nuf2, with sites for covalent SUMO-2/3 modification (green bars). The relative levels of SUMOylation and deSUMOylation at the kinetochore define the stability of CENP-E association. Although not depicted, CENP-E may also be covalently modified by SUMO-2/3 polymeric chains. SUMOylation of CENP-E could further contribute to kinetochore association by mediating interactions with other SIM-containing proteins at the kinetochore.
DISCUSSION
By investigating the properties and functions of SUMO-1 and SUMO-2/3 modification in mammalian cells using antibodies to endogenous proteins, we found that SUMO-1 and SUMO-2/3 are conjugated to unique subsets of proteins, are differentially regulated and control distinct aspects of cell cycle progression. Global inhibition of SUMOylation revealed an essential paralogue-specific role for SUMO-2/3 modification in regulating the interaction of CENP-E with kinetochores, and thus the alignment of chromosomes at the metaphase plate. This and other essential functions of SUMOylation, including SUMO-1-dependent targeting of RanGAP1 to the mitotic spindle (Joseph et al., 2002) and SUMO-2/3 regulation of topoisomerase II activity at the metaphase to anaphase transition (Azuma et al., 2003), indicate that SUMOylation of multiple distinct factors regulates progression through mitosis. Thus, SUMOylation, like ubiquitination and phosphorylation, is a key mitotic regulator.
Unique signaling through SUMO-1 and SUMO-2/3
Our observations that SUMO-1 and SUMO-2/3 localize to different sub-cellular structures in both interphase and mitotic cells indicate that they are conjugated to unique subsets of proteins and regulate unique cellular functions. Further supporting this conclusion, we also found that SUMO-1 and SUMO-2/3 conjugation and deconjugation are differentially regulated with changes in the cell cycle. Understanding how proteins are selectively modified by SUMO-1 or SUMO-2/3, and how their modifications are temporally regulated through the cell cycle, are important future questions.
Their unique protein targets and differential regulation also suggest that SUMO-1 and SUMO-2/3 are distinct signals specifying distinct fates. What these fates are, and how they are defined, are also important questions. By analogy to ubiquitination, SUMO-1 and SUMO-2/3 may specify distinct fates by mediating interactions with different down-stream binding partners (Pickart and Fushman, 2004). Supporting this, SUMO-binding proteins with paralogue-specific binding affinities have been identified (Hecker et al., 2006). In addition, monomeric and polymeric SUMO paralogues may represent distinct signals. Consistent with this, we found that CENP-E contains a SUMO-interacting motif that specifically binds SUMO-2/3 polymeric chains.
SUMO-2/3 modulates CENP-E kinetochore association
CENP-E is a plus-end directed microtubule motor protein associated with the outer kinetochore plate and fibrous corona of mitotic chromosomes (Cooke et al., 1997; Wood et al., 1997; Yao et al., 1997). By connecting kinetochores and spindle microtubules, CENP-E functions in aligning chromosomes at the metaphase plate and in silencing the mitotic checkpoint. Several kinetochore-associated proteins interact directly with CENP-E, including BubR1, CENP-F and Nuf2, and likely mediate the association of CENP-E with kinetochores (Chan et al., 1998; Liu et al., 2007). Notably, however, CENP-E’s association with kinetochores is dynamic. Although concentrated at kinetochores in prophase and metaphase, CENP-E is also associated with spindle microtubules (Cooke et al., 1997; Yao et al., 1997). In addition, the relative concentrations of CENP-E at kinetochores varies with microtubule binding, such that leading kinetochores having more CENP-E relative to lagging kinetochores (Yao et al., 1997). Most dramatically, the bulk of CENP-E redistributes from kinetochores to the spindle midzone and midbody during anaphase and telophase (Cooke et al., 1997; Yao et al., 1997). Thus, one or more mechanisms modulate the stability of CENP-E’s association with kinetochores.
We have identified SUMO-2/3 modification as an essential modulator of CENP-E’s interactions with kinetochores. Upon inhibition of SUMOylation, SUMO-2/3 modified proteins were lost at centromeres and kinetochores. Concomitantly, a defect in CENP-E kinetochore localization was detected. We found that the loss of CENP-E at kinetochores upon inhibition of SUMOylation was due to a defect in CENP-E localization, not degradation. We specifically found that non-covalent interactions between CENP-E and SUMO-2/3 polymeric chains are essential for kinetochore association. Based on this, we propose that covalent SUMOylation of kinetochore-associated proteins, and non-covalent interactions between CENP-E and these modified proteins, regulate CENP-E localization (Figure 7C). This model is similar to one proposed for the assembly and interaction of proteins with PML nuclear bodies (Lin et al., 2006; Shen et al., 2006). Importantly, the model provides a mechanism for producing a gradient of affinities between CENP-E and kinetochores through regulated SUMOylation and deSUMOylation of kinetochore-associated proteins. Notably, we found that BubR1 and Nuf2, proteins implicated in facilitating interactions between CENP-E and kinetochores, are modified by SUMO-2/3 in vivo. Thus, SUMOylation of BubR1 and Nuf2 may be directly linked to CENP-E localization. In addition, SUMO-2/3 modification of the CENP-E tail domain itself was detected in vitro and in vivo. Future work will determine if this modification, and the modifications of BubR1 and Nuf2, directly regulates CENP-E localization. Identifying factors that regulate SUMOylation and deSUMOylation at centromeres and kinetochores is also of interest. Whether SENP2 normally deconjugates proteins at kinetochores is currently not known.
Where previous studies detected defects in chromosome segregation and/or cytokinesis upon inhibition of SUMOylation (Hayashi et al., 2002; Nacerddine et al., 2005), ours are the first to detect defects in CENP-E localization. This may reflect differences in the absolute requirement for CENP-E to sustain spindle checkpoint signaling in different cell types. In primary mouse fibroblasts, CENP-E is essential for spindle checkpoint activation and its depletion results in chromosome segregation defects without mitotic arrest (Putkey et al., 2002). In contrast, depletion or inhibition of CENP-E in cultured HeLa cells results in persistent activation of the spindle checkpoint and prometaphase arrest (Schaar et al., 1997; Yao et al., 2000).
In S. cerevisiae, at least four kinetochore-associated proteins are modified by SUMO, including Ndc10, Bir1, Ndc80 and Cep1 (Montpetit et al., 2006). SUMOylation of Ndc10 targets it to the mitotic spindle in anaphase. The effects of SUMOylation on Bir1, Ndc80 and Cep1 are less well understood. Notably, however, the mammalian homolog of Ndc80 (Hec1) is part of a protein complex containing Nuf2. It is also noteworthy that the dynamic association of the mammalian homolog of Bir1 (Survivin) with centromeres is regulated by modification with K63-linked polyubiquitin chains, possibly through a mechanism similar to that proposed for SUMOylation (Vong et al., 2005). Thus, reversible ubiquitination and SUMOylation both regulate of the dynamic protein-protein interactions required for proper kinetochore and centromere function. Also like ubiquitination, SUMOylation likely regulates multiple mitotic events through modification of multiple different proteins. Understanding the full range of events regulated by SUMOylation will require identification of the full repertoire of proteins modified by SUMO-1 and SUMO-2/3 during mitosis.
METHODS
Antibodies
The SUMO-2/3 specific monoclonal antibody, 8A2, was generated by immunizing mice with full-length recombinant human SUMO-2. Hybridoma production and screening were performed as described for the SUMO-1 specific monoclonal antibody, 21C7 (Matunis et al., 1996).
Other antibodies used in this study were obtained from the following sources: anti-PML, Dr. Paul Freemont, Imperial College London; anti-CENP-B (Ra 764); anti-CENP-C (Ra 554), Dr. Ann Pluta, National Institutes of Health; anti-CENP-E (HpX), Dr. Don Cleveland, University of California at San Diego; anti-Bub1 and anti-BubR1, Dr. Tim Yen, Foxchase; anti-MAD2, Dr. E.D. Salmon, University of North Carolina; CREST human auto-antibodies, Dr. B.R. Brinkley, Baylor College of Medicine; anti-Survivin and anti-Aurora B, Dr. Yixian Zheng, Carnegie Institution of Washington; anti-ubiquitin, Dr. Cecile Pickart, Johns Hopkins University; anti-Myc antibodies, Cell Signaling Technology, Beverly, MA; anti-RanGAP1 (Matunis et al., 1996); anti-Hec1 and anti-CENP-F, BD Biosciences, San Jose, CA; anti-phospho-histone H3 (Ser 10), Upstate, Lake Placid, NY; anti-α tubulin (DM1A), Sigma-Aldrich, St. Louis, MO; and anti-topoisomerase IIα, TopoGEN, Columbus, OH.
Cell culture, synchronization and transfection
HeLa and 293 cells were maintained at 37°C in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum, 10 mM HEPES (pH 8.0) and 1% penicillin-streptomycin. HeLa cells were synchronized in mitosis by culturing in the presence of 2 mM thymidine for 19 hr, release from thymidine for 3 hr, then culturing in the presence of 100 ng/ml nocodazole for 13 hr. Cells were released from the nocodazole block by washing and then culturing in fresh medium for the indicated times. S phase cells were obtained by culturing in the presence of 2 mM thymidine for 35 hr, washing with PBS and lysing in SDS sample buffer. The HTETOP cell line was maintained and cultured in the presence or absence of tetracycline as previously described (Carpenter and Porter, 2004).
For transfection, HeLa or 293 cells were cultured to ~60–70% confluency and transfected with the indicated plasmids using Lipofectamine-Plus reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Controls were performed using identical conditions and empty plasmids.
Immunofluorescence microscopy
HeLa cells cultured on glass coverslips were prepared using three approaches, as indicated. In the first approach, cells were fixed for 30 min at room temperature using 2% formaldehyde in PBS, and then permeabilized in −20 °C acetone for 5 min. In the second approach, cells were permeabilized prior to fixation using 20 µg/ml digitonin (Sigma-Aldrich, St. Louis, MO) in buffer containing 20 mM HEPES-KOH (pH 7.3), 110 mM potassium acetate, 2 mM magnesium acetate, 10 mM N-ethylmaleimide (NEM) and protease inhibitors (1 mM PMSF, 20 µg/ml aprotinin, and 5 µg/ml leupeptin, antipain, and pepstatin A) for 5 min at room temperature. Cells were subsequently fixed using 2% formaldehyde in PBS for 30 min at room temperature. In the third approach, cells were fixed with 3.5% paraformaldehyde in PBS for 7 min then permeabilized with 0.2% Triton X-100 in PBS for 30 min. Immunostaining was done as previously described (Matunis et al., 1996). Confocal images were collected using an UltraVIEW spinning disk confocal microscope and acquisition software (Perkin Elmer, Wellesley, MA).
Chromosome purification
HeLa cells were grown in suspension and arrested at prometaphase by culturing in the presence of 100 ng/ml colcemid for 16 hr. Mitotic chromosomes were purified as described (Lewis and Laemmli, 1982). Chromosome fractions were digested with Benzonase® Nuclease (Novagen, La Jolla, CA) in PBS with 0.02% Tween-20, 5 mM MgCl2 and protease inhibitors at 30 °C for 10 min prior to addition of SDS sample buffer.
Flow cytometry
24 hr post-transfection with GFP-SENP2, HeLa cells were trypsinized and resuspended in DMEM without serum. GFP-positive and negative cells were sorted and counted using a MoFlo™ High Performance Cell Sorter (Dako, Glostrup, Denmark). Collected cells were lysed in SDS sample buffer and analyzed by SDS-PAGE and immunoblotting.
Immunopurification
48 hr after transfection, 293 cells were lysed by sonication in buffer containing 20 mM HEPES pH 8.0, 150 mM NaCl, 2 mM EDTA, 2 mM MgCl2, 1% Empigen (Calbiochem, San Diego, CA), protease inhibitors, and 10 mM NEM. Cell lysates were clarified by centrifugation at 20,000 × g for 10 min at 4 °C and incubated with anti-FLAG beads (Sigma-Aldrich, St. Louis, MO) at 4 °C for 5 hr. Antibody beads were washed six times with lysis buffer and bound proteins were eluted with SDS sample buffer. Immunopurified proteins were separated by SDS-PAGE and analyzed by immunoblot analysis. Immunopurifications carried out in the presence of SDS yielded identical results (Figure S5).
For immunopurification of BubR1 and Nuf2, transfected cells were lysed by sonication in buffer containing 1% SDS, 1× RIPA buffer (1% Triton X-100, 1% sodium deoxycholate, 300 mM NaCl, 20 mM HEPES pH 8.0, 2 mM EDTA and 2 mM MgCl2), protease inhibitors and 20 mM NEM. Lysates were heated at 95°C for 10 min, chilled and diluted with 1× RIPA buffer to adjust the final SDS concentration to 0.1%. Immunopurifications were performed using anti-FLAG beads.
RNA interference
HeLa cells were transfected with Ubc9 siRNAs (Lin et al., 2003) (40 pmol/ 3.5 cm well) using Oligofectamine, according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). An siRNA against firefly luciferase (Dharmacon, Lafayette, CO) was used as a control.
SUMO-binding assays
GST, GST-SUMO-1, GST-SUMO-2, GST-SUMO-1 polymeric chains, and GST-SUMO-2 polymeric chains were immobilized on glutathione-sepharose beads (GE Healthcare, Piscataway, NJ). Equal protein loading and stability was verified by SDS-PAGE before and after binding (not shown). Wild type and SIM mutant CENP-E tail domains were transcribed and translated in rabbit reticulocyte lysate in the presence of [35S]methionine according to the manufacturer’s protocol (Promega, Madison, WI). Translated proteins were incubated in binding buffer (75 mM NaCl, 10 mM Tris-HCl pH 7.4, 0.05% Tween 20) with beads containing immobilized proteins for 1 hr and the beads were subsequently washed 5 × with binding buffer. Bound proteins were eluted using SDS sample buffer and analyzed by SDS-PAGE and autoradiography. Relative SUMO-binding activities of wild type and SIM mutant proteins were quantified using a FUJIFILM FLA-7000 Image Reader (FUJIFILM Life Science, Stamford, CT).
SUMO polymeric chain synthesis was performed at 37 °C for 2 hr in 20 µl reactions containing 1.0 µg Aos1/Uba2, 0.8 µg Ubc9, 1 mM ATP, 40 U/ml creatine phosphokinase, 10 mM phosphocreatine, 1.2 µg/ml inorganic pyrophosphatase, 20 mM HEPES-KOH (pH 7.3), 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM DTT and 1 mM EGTA. Reactions contained either 1.0 µg of GST-SUMO-1 and 2.0 µg of SUMO-1 or 1.0 µg of GST-SUMO-2 and 2.0 µg of SUMO-2.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Don Cleveland, Ann Pluta, Ted Salmon, Yixian Zheng, William Brinkley and Paul Freemont for antibodies. We also thank members of the Matunis laboratory and Drs. Mary Dasso and Robert Cohen for insightful discussions. This work was supported by a grant from the National Institutes of Health (GM060980 to M.J.M.) and the Lang Fellowship Fund (X.-D. Zhang).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ayaydin F, Dasso M. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol Biol Cell. 2004;15:5208–5218. doi: 10.1091/mbc.E04-07-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Arnaoutov A, Dasso M. SUMO-2/3 regulates topoisomerase II in mitosis. J Cell Biol. 2003;163:477–487. doi: 10.1083/jcb.200304088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell. 2002;9:1169–1182. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- Carpenter AJ, Porter AC. Construction, characterization, and complementation of a conditional-lethal DNA topoisomerase IIalpha mutant human cell line. Mol Biol Cell. 2004;15:5700–5711. doi: 10.1091/mbc.E04-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan GK, Schaar BT, Yen TJ. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J Cell Biol. 1998;143:49–63. doi: 10.1083/jcb.143.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke CA, Bernat RL, Earnshaw WC. CENP-B: a major human centromere protein located beneath the kinetochore. J Cell Biol. 1990;110:1475–1488. doi: 10.1083/jcb.110.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke CA, Schaar B, Yen TJ, Earnshaw WC. Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma. 1997;106:446–455. doi: 10.1007/s004120050266. [DOI] [PubMed] [Google Scholar]
- Hang J, Dasso M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J Biol Chem. 2002;277:19961–19966. doi: 10.1074/jbc.M201799200. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Seki M, Maeda D, Wang W, Kawabe Y, Seki T, Saitoh H, Fukagawa T, Yagi H, Enomoto T. Ubc9 is essential for viability of higher eukaryotic cells. Exp Cell Res. 2002;280:212–221. doi: 10.1006/excr.2002.5634. [DOI] [PubMed] [Google Scholar]
- Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1-and SUMO2-interacting motifs. J Biol Chem. 2006;281:16117–16127. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Joseph J, Tan SH, Karpova TS, McNally JG, Dasso M. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J Cell Biol. 2002;156:595–602. doi: 10.1083/jcb.200110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Lewis CD, Laemmli UK. Higher order metaphase chromosome structure: evidence for metalloprotein interactions. Cell. 1982;29:171–181. doi: 10.1016/0092-8674(82)90101-5. [DOI] [PubMed] [Google Scholar]
- Lin DY, Huang YS, Jeng JC, Kuo HY, Chang CC, Chao TT, Ho CC, Chen YC, Lin TP, Fang HI, et al. Role of SUMO-Interacting Motif in Daxx SUMO Modification, Subnuclear Localization, and Repression of Sumoylated Transcription Factors. Mol Cell. 2006;24:341–354. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Lin X, Liang M, Liang YY, Brunicardi FC, Feng XH. SUMO-1/Ubc9 promotes nuclear accumulation and metabolic stability of tumor suppressor Smad4. J Biol Chem. 2003;278:31043–31048. doi: 10.1074/jbc.C300112200. [DOI] [PubMed] [Google Scholar]
- Liu D, Ding X, Du J, Cai X, Huang Y, Ward T, Shaw A, Yang Y, Hu R, Jin C, Yao X. Human NUF2 interacts with centromere-associated protein E and is essential for a stable spindle microtubule-kinetochore attachment. J Biol Chem. 2007;282:21415–21424. doi: 10.1074/jbc.M609026200. [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Chan GK, Zubrowski B, Savoian MS, Sauer MT, Yen TJ. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol Biol Cell. 2001;12:2776–2789. doi: 10.1091/mbc.12.9.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montpetit B, Hazbun TR, Fields S, Hieter P. Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J Cell Biol. 2006;174:653–663. doi: 10.1083/jcb.200605019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Putkey FR, Cramer T, Morphew MK, Silk AD, Johnson RS, McIntosh JR, Cleveland DW. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev Cell. 2002;3:351–365. doi: 10.1016/s1534-5807(02)00255-1. [DOI] [PubMed] [Google Scholar]
- Rosas-Acosta G, Russell WK, Deyrieux A, Russell DH, Wilson VG. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol Cell Proteomics. 2005;4:56–72. doi: 10.1074/mcp.M400149-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- Saitoh H, Tomkiel J, Cooke CA, Ratrie H, 3rd, Maurer M, Rothfield NF, Earnshaw WC. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- Schaar BT, Chan GK, Maddox P, Salmon ED, Yen TJ. CENP-E function at kinetochores is essential for chromosome alignment. J Cell Biol. 1997;139:1373–1382. doi: 10.1083/jcb.139.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead K, Aguilar C, Hartman T, Drexel M, Meluh P, Guacci V. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J Cell Biol. 2003;163:729–741. doi: 10.1083/jcb.200305080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics. 2006 doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- Vong QP, Cao K, Li HY, Iglesias PA, Zheng Y. Chromosome alignment and segregation regulated by ubiquitination of survivin. Science. 2005;310:1499–1504. doi: 10.1126/science.1120160. [DOI] [PubMed] [Google Scholar]
- Watts FZ. The role of SUMO in chromosome segregation. Chromosoma. 2007;116:15–20. doi: 10.1007/s00412-006-0079-z. [DOI] [PubMed] [Google Scholar]
- Wood KW, Sakowicz R, Goldstein LS, Cleveland DW. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 1997;91:357–366. doi: 10.1016/s0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- Yao X, Abrieu A, Zheng Y, Sullivan KF, Cleveland DW. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat Cell Biol. 2000;2:484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- Yao X, Anderson KL, Cleveland DW. The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J Cell Biol. 1997;139:435–447. doi: 10.1083/jcb.139.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Saitoh H, Matunis MJ. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol Cell Biol. 2002;22:6498–6508. doi: 10.1128/MCB.22.18.6498-6508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


