Abstract
Estrogen receptors α and β (ERα and ERβ) mediate the actions of estrogens in a variety of normal and cancer target cells. Estrogens differ in their preference for these ERs, and many phytoestrogens bind preferentially to ERβ. To investigate how phytoestrogens such as genistein impact ER-regulated gene expression, we used adenoviral gene delivery of ERβ coupled with ERα depletion with small interfering RNA to generate human breast cancer (MCF-7) cells expressing four complements of ERα and ERβ. We examined the dose-dependent effects of genistein on genome-wide gene expression by DNA microarrays and monitored the recruitment of ERs and coregulators to responsive regions of estrogen-regulated genes. At a low (6 nm) concentration, genistein regulated gene expression much more effectively in cells coexpressing ERα and ERβ than in cells expressing ERα alone, whereas at high concentration (300 nm), genistein induced transcriptome changes very similar to that of 17β-estradiol. We demonstrate that ERβ is preferentially activated by genistein and is recruited to estrogen-responsive genomic sites and that differential occupancy of ERα and ERβ by genistein and 17β-estradiol in turn influences the recruitment patterns of coregulators such as steroid receptor coactivator 3 (SRC3) and receptor-interacting protein 140 (RIP140). Our observations indicate that genistein is a potency-selective ligand for gene expression regulation by ERα and ERβ and that the ability of ERα and ERβ to serve as determinants of gene expression is greatly influenced by the nature of the ligand, by ligand dose, and by the differential abilities of ligand-ER complexes to recruit different coregulators at ER binding sites of hormone-regulated genes.
ESTROGEN RECEPTORS α and β (ERα and ERβ) mediate the actions of diverse estrogens in estrogen target tissues and cells, both normal and cancerous, where the levels and ratios of the two receptors are known to vary substantially (1,2). For example, ERα is the predominant ER found in uterus and liver, whereas ERβ is highly expressed and is almost the exclusive ER in ovarian granulosa cells (3). ERs also mediate the actions of estrogens and selective ER modulators in breast cancers (4,5). Although ERα is usually the predominant ER in breast tumors, 70% of primary breast tumors express ERβ, and most tumors coexpress both ERs, with the levels of ERβ covering a broad range (5,6,7,8,9). The presence of ERα is associated with the proliferative effects of estrogens, whereas the bulk of current evidence implies that ERβ has growth-suppressive activities (10,11,12,13).
Soy-based products have gained much attention lately, in part because increased consumption is thought to contribute to the lower breast cancer incidence in Asian countries relative to the United States (14); however, it is unclear which biologically active components in soy offer these protective effects. In animal studies where soy-supplemented diets were shown to reduce the number of tumors induced by chemical carcinogens, the beneficial effects of soy were attributed to the isoflavones genistein and daidzein (15). However, evidence from clinical trials for efficacy of soy isoflavones for preventing or treating breast cancer is as yet very limited (16). Paradoxically, genistein is reported to have both antitumor benefits as well as growth-stimulatory effects, depending on the concentrations used. Genistein at physiological concentrations stimulates proliferation of ERα-positive breast cancer cells both in vitro and in xenograft animal models (17,18,19) but is growth inhibitory at higher concentrations (IC50 at 6–8 μg/ml) (19,20). It is widely presumed that anti-growth actions by genistein at levels greater than 10 μm are mediated through the inhibition of tyrosine kinases (19,20,21,22).
Growth inhibition by genistein might, however, also stem from its estrogenic properties unrelated to its anti-tyrosine kinase activity observed at extremely high pharmacological doses. Genistein activates ERβ at concentrations lower than required for activation of ERα (3,23), and because ERβ has anti-growth properties, selective activation of ERβ in cells may serve to suppress growth of estrogen-dependent tumor cells.
In this work, we examine the roles of ERα and ERβ as determinants of gene expression in breast cancer cells and the influence of ligand, dose, and chromatin binding in the patterns of cellular gene expression. To this end, we have profiled global gene expression in breast cancer cells containing various complements of ERα and ERβ treated with either 17β-estradiol (E2) or the phytoestrogen ligand genistein. We also examined ligand-induced chromatin binding by the two ERs, when present together or separately, in breast cancer cells. Our studies highlight that the ERβ-preferential gene regulatory effects of genistein are dose dependent and that chromatin binding of ligand-occupied ERs reflects their gene transcriptional output and is associated with differential recruitment of coregulators by ERα and ERβ complexes.
RESULTS
Expression of ERβ in Breast Cancer Cells Influences the Pattern of Estrogen Regulation of Genes Involved in Many Functional Categories
Previous studies by our group and others have demonstrated that coexpression of ERβ with ERα significantly impacted the E2-induced transcriptional response by ERα (24,25,26,27). To further examine the activities of ERβ or ERα, we have used the phytoestrogen genistein because of its ERβ preferential binding and transcriptional potency in reporter gene assays. To assess the impact of ERβ on response profiles of endogenous gene expression to genistein and E2 in MCF-7 cells, which express only ERα, we used adenoviral gene delivery to transduce ERβ expression. We then determined the time-dependent and ligand-specific changes in ER-mediated transcriptional response by profiling MCF-7 cells treated for 4 and 24 h with E2 and two concentrations of genistein using Affymetrix HG-U133A GeneChips. To assess the importance of ER subtype in response to genistein, we examined the dose dependence of genistein transcriptional activity, using low genistein (LG; 6 nm) and high genistein (HG; 300 nm) concentrations, which should activate ERβ preferentially or both ERα and ERβ, respectively.
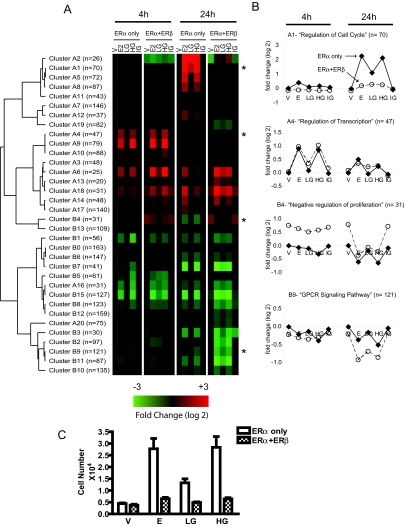
To find genes differentially expressed under our different ligand treatments, we used a multivariate analysis (LIMMA) to assign the statistical significance to the contrasts while controlling for multiple testing. We found 2682 probe sets significantly regulated by one or more treatment conditions after 4- and 24-h treatments. Using the TightCluster algorithm (28) to group genes of like expression profiles, 17 clusters of up-regulated genes and 17 clusters of down-regulated genes emerged (Fig. 1A).
Figure 1.
Gene Clustering Analysis with TightCluster Program Reveals Time-Dependent Regulation and ERβ-Mediated Modulation of Estrogen-Responsive Genes
A, Heat map of average profiles (log base 2) of 32 gene clusters is shown, with n denoting the total gene membership within each cluster. V, 0.1% ethanol vehicle; E2, 6 nm E2; LG, 6 nm genistein; HG, 300 nm genistein; IG, HG plus 1 μm antiestrogen ICI 182,780. B, Average gene expression profiles (in log base 2) of select clusters are shown, with n denoting the total gene membership within each cluster. Samples are grouped by treatment time (4 and 24 h) and ordered by ligand treatment: ERα (♦) and ERα+ERβ (○). Clusters enriched for genes involved in cell cycle progression (cluster A1), regulation of transcription (cluster A4), negative regulation of cell proliferation (cluster B4), and GPCR signaling pathway (cluster B9) are shown. C, Proliferation of MCF-7 cells expressing either ERα alone or coexpressing ERα and ERβ in response to treatment with E2, LG, or HG for 6 d. Cells were seeded at a density of 1000 cells per well and monitored for cell number in quadruplicate samples at d 6.
At the two time points we surveyed (4 and 24 h), the response profiles to HG were almost identical to those elicited by E2, and the gene regulations by HG were all suppressed by the pure antiestrogen ICI 182,780 (treatment IG, ICI plus HG). In contrast, LG preferentially induced transcriptional responses in cells coexpressing ERα and ERβ and elicited little effect on gene expression in cells containing ERα alone. This pattern of response is evident from the cluster heat map (Fig. 1A).
To explore the underlying biological functions of these expression clusters, we used Expression Analysis Systematic Explorer (EASE) software (29) to test for overrepresented gene ontology biological function classes in each expression cluster (Fig. 2). The four clusters shown in Fig. 1B illustrate different patterns of response in terms of direction of regulation (up or down), time (4 and 24 h), influence of ERβ, and response to ligands. Clusters enriched for genes involved in cell cycle regulation (cluster A1) showed up-regulation via ERα that was attenuated by the copresence of ERβ, whereas the gene cluster enriched for G protein-coupled receptor (GPCR) signaling (cluster B9) showed down-regulation by the ligands that was enhanced by ERβ (Fig. 1B). In addition, ERβ increased the expression of genes involved in negative regulation of cell proliferation (cluster B4). Other clusters were associated with RNA processing (clusters A7, A8, A17, and A19) and various transport processes (clusters B10 and B11) (Fig. 2). These findings support and extend previously published results that ERβ expression can positively and negatively modulate distinct ERα-mediated gene expression patterns (24).
Figure 2.
Statistical Analysis of Biological Processes Enriched within Gene Clusters
Gene ontology (GO) biological functional categories significantly enriched (EASE score < 0.05) for 32 gene clusters are mapped and arranged by EASE score. Darker shading denotes more highly significant EASE scores (ranging from 10−12 to 0.05), whereas blank entries denote that the GO term did not reach significance for the gene cluster. Major biological themes present in MCF-7 transcriptomes include transcriptional regulation, mitosis and cell cycle genes, RNA and protein processing, ion homeostasis, and cell adhesion/cytoskeletal rearrangement.
The effects of E2 and genistein on the proliferation of MCF-7 cells containing ERα only or both ERs (Fig. 1C) are consistent with the patterns of gene expression regulation we have observed for these ligands with the gene clusters associated with the cell cycle and proliferation (Fig. 1B, clusters A1 and B4). In parental cells expressing only ERα, we find that HG (300 nm) is as stimulatory as E2 (6 nm), with LG (6 nm) eliciting lesser stimulation of proliferation (Fig. 1C). In cells coexpressing both ERα and ERβ, E2 as well as genistein showed a markedly reduced ability to stimulate cell proliferation. The suppressive effect of ERβ on estrogen-stimulated cell proliferation is as we and others reported previously for E2 (24,30), and it reflects, in ERβ-containing cells, the reduced ability of E2 and genistein to up-regulate genes as-sociated with regulation of cell cycle (Fig. 1B, cluster A1) and the enhanced expression of genes associated with negative regulation of proliferation (Fig. 1B, cluster B4).
High-Concentration (300 nm) Genistein Treatment Elicits Global Gene Expression Changes Nearly Identical to Those of E2 in Cells Expressing ERα Alone or ERα plus ERβ, whereas ERβ Coexpression Greatly Enhances Gene Regulation by Low-Concentration (6 nm) Genistein
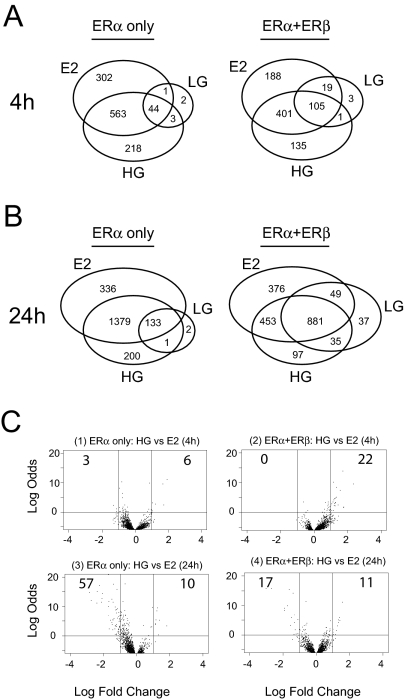
To examine whether genistein elicits a transcriptional response different from E2, we compared probe sets regulated by maximally effective concentrations of E2 (6 nm) and genistein (300 nm, HG). Probe sets specifically regulated in each treatment group were identified using a B-statistic cutoff (B > 0) from pairwise contrasts defined between untreated and treated samples (Fig. 3, A and B).
Figure 3.
At High Concentration, Genistein Elicits Gene Regulation Very Similar to that by E2
Venn diagrams comparing probe sets regulated by E2 (6 nm) or HG (300 nm) in MCF-7 cells expressing ERα alone or ERα+ERβ. A, Genes regulated at 4 h; B, genes regulated at 24 h; C, volcano plots (log odds vs. fold change) of HG-treated vs. E2-treated samples at 4 h in ERα-only cells (panel 1) or ERα+ERβ cells (panel 2) or at 24 h in ERα-only cells (panel 3) or ERα+ERβ cells (panel 4).
We find that the genes regulated by HG are very similar to those regulated by E2 (Fig. 3A). Of the 910 probe sets regulated by E2 at 4 h, 607 were also regulated by HG using B-statistic cutoff greater than 0 (73.6% of all 4-h HG-regulated probe sets). And of the 218 genes induced by HG but not by E2, cotreatment with ICI 182,780 antagonized HG agonism for all except three genes. To further identify probe sets where HG regulation was significantly different from E2, we used the volcano plot to visualize both the significance statistic and fold change of the HG vs. E2 contrast (Fig. 3C). Only nine probe sets from ERα-only cells and 22 probe sets from the ERα+ERβ cells passed our cutoff after 4 h treatment (B-statistic > 0; fold change > 2; Fig. 3C, panels 1 and 2). By the same criteria, few genes were differentially regulated at 24 h by HG vs. E2 (Fig. 3C, panels 3 and 4).
Previous studies using binding and reporter gene-based transactivation assays have demonstrated that genistein preferentially binds to and transactivates ERβ over ERα (3,23). We therefore also explored the impact of ERβ coexpression on genome-wide gene expression using a low concentration of genistein (6 nm, LG) expected to be ERβ preferential. To find genes preferentially regulated by genistein-bound ERβ, we performed statistical analysis using LIMMA and extracted genes with B-statistic score greater than 0.
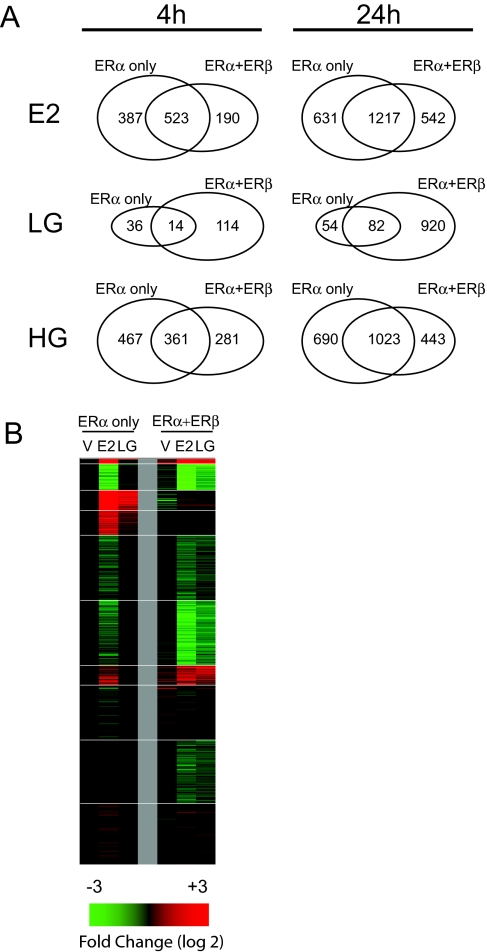
In MCF-7 cells, which express only ERα, LG regulated few genes at 4 and 24 h, compared with E2 (Figs. 3, A and B, and 4). By contrast, in cells expressing ERα+ERβ, LG was considerably more effective in eliciting gene regulation (Fig. 4A). This is illustrated further by the heat map (Fig. 4B) in which the LG panel in the ERα+ERβ cells looks quite similar to that of E2, whereas LG is much less effective in regulating gene expression in the ERα-only cells.
Figure 4.
ERβ Coexpression Enhances Gene Regulation by LG (6 nm)
A, Venn diagrams comparing probe sets regulated by E2 (6 nm), LG (6 nm) or HG (300 nm) in MCF-7 cells expressing ERα alone or ERα+ERβ; B, Heat map depicting 24-h gene expression of 1062 probe sets regulated by control vehicle (V), E2, or LG in MCF-7 cells expressing ERα only or ERα+ERβ.
Examination of Gene Expression by Genistein and Estradiol in Cells Differentially Expressing ERα and ERβ
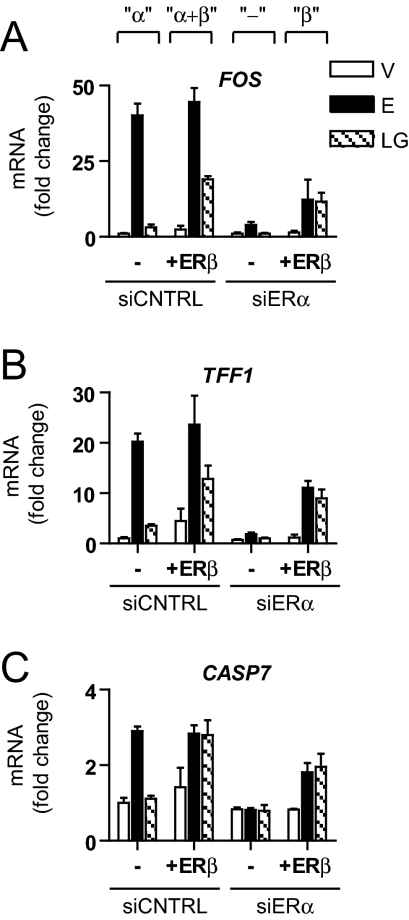
To further examine the ER subtype dependence of genistein and E2 gene regulation, we coupled small interfering RNA (siRNA)-mediated knockdown of endogenous ERα expression with ERβ adenoviral gene delivery. Our siRNA conditions resulted in the depletion of more than 95% of ERα protein, as determined by Western blot (not shown). This enabled us to generate four conditions with varying ER complements in MCF-7 cells: ERα-only (siCNTRL), ERα+ERβ (siCNTRL+ERβ), low ERα (siERα), and ERβ-only (siERα+ERβ). We then monitored the expression of three well-known E2-regulated genes, FOS, TFF1, and CASP7, by quantitative PCR, in response to E2 or genistein (Fig. 5).
Figure 5.
Regulation of Gene Expression by E2 and Genistein in Cells Differentially Expressing ERα and ERβ
A, Transcript levels of estrogen-regulated genes (FOS, TFF1, and CASP7) were assessed by quantitative PCR after 4-h treatment of MCF-7 cells differentially expressing ERα and ERβ. To generate cells with ERα+ERβ or ERβ only vs. ERα only (parental MCF-7 cells), MCF-7 cells were infected with AdGal (control) or AdERβ for 24 h and subsequently transfected with either siCNTRL or siERα for an additional 48 h. Cells were then exposed to control vehicle (V), 6 nm E2, or 6 nm genistein (LG) for 4 h. Data represent average fold change ± sd of triplicate samples and is representative of three separate experiments.
In ERα-only cells, all three genes showed robust stimulation by E2 (black bars), whereas LG (hatched bars) had little effect. By contrast, in ERα+ERβ cells, E2 activity was maintained, and LG evoked substantial stimulation of expression of these three genes. With the knockdown of ERα, gene stimulation was completely lost, but the expression of ERβ at a level equal to that of ERα in the original MCF-7 cells substantially, but not fully, restored gene regulation by all ligands.
Of note, genistein stimulation was comparable between cells expressing ERβ only and ERα+ERβ cells (i.e. α+β ≈ β ≫ α). By contrast, the magnitude of E2 stimulation was reduced in ERβ-only cells relative to ERα+ERβ cells (i.e. α+β ≈ α > β). These findings suggest that genistein regulation of gene expression, under the conditions tested, is largely mediated by ERβ, whereas E2 stimulation can be mediated by either ERα or ERβ.
Recruitment of ERα and ERβ to ER Binding Sites of Estrogen-Responsive Genes by E2 and Genistein
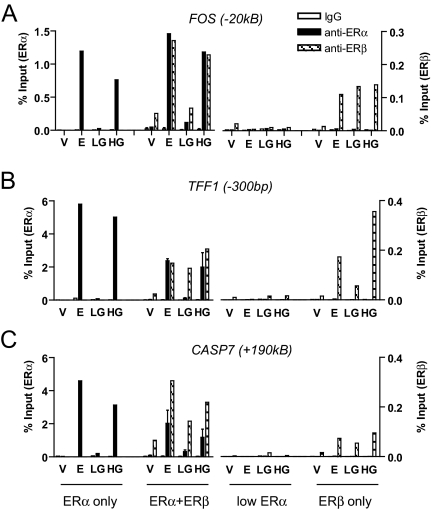
To understand the effects of genistein and E2 on ER-mediated gene regulation, we examined ER recruitment to estrogen-responsive sites using chromatin immunoprecipitation (ChIP) with antibodies against ERα and ERβ. We queried ER binding sites as recently mapped by the ChIP-on-chip method (31,32) and also by ChIP-PET methods (33) to be within 100 kb upstream/downstream of the transcription start site of FOS, TFF1, and CASP7 and hence considered to be potential regulatory sites for these genes (Fig. 6). For each site of interest, we quantitatively measured immunoprecipitated yield normalized to input chromatin. Figure 6 shows recruitment of ERα or ERβ to these sites, under the four cell conditions (cells containing ERα only, ERα+ERβ, low ERα, and ERβ only), after exposure to control vehicle, E2, LG, or HG.
Figure 6.
Recruitment of ERα and ERβ to ER Binding Sites Near Estrogen-Responsive Genes in Response to E2 and Genistein
Changes in chromatin binding by estrogen receptor-containing complexes were measured by quantitative PCR after 45 min ligand treatment with vehicle (V), E2, LG, or HG in MCF-7 cells differentially expressing ERα and/or ERβ. ChIP experiments were performed with ERα- and ERβ-specific antibodies. Genomic locations of estrogen receptor binding sites were from work by Carroll et al. (32) (UCSC Genome Browser). Data are expressed as percentage of input and is representative of three separate experiments.
Overall, the patterns of ERα and ERβ recruitment to these three gene regions were quite similar in response to ligands. In MCF-7 cells expressing only ERα, E2 and HG effectively recruited ERα, whereas LG elicited no ERα recruitment. In cells containing ERβ along with ERα, both ERs were recruited in response to E2 or HG, but of note, only ERβ was recruited by LG. We also observed some ligand-independent ERβ recruitment in cells containing ERα+ERβ, although recruitment of ERβ was greatly increased by ligand. In cells depleted of ERα by siRNA treatment (low ERα cells), recruitment of ERs in response to ligands was completely lost.
In cells expressing ERβ only, recruitment of ERβ in response to ligand showed gene-dependent differences. All three ligand treatments (E2, LG, and HG) elicited good recruitment of ERβ at the FOS binding site, whereas LG was less effective in recruiting ERβ compared with E2 and HG at the TFF1 site. Recruitment of ERβ to the CASP7 site was low with all three ligand treatments, suggesting that recruitment of ERβ to this site is facilitated by the copresence of ERα.
Differential Recruitment of Coregulators Steroid Receptor Coactivator 3 (SRC3) and Receptor-Interacting Protein 140 (RIP140) to ER Binding Sites by ERα and ERβ Ligand Complexes
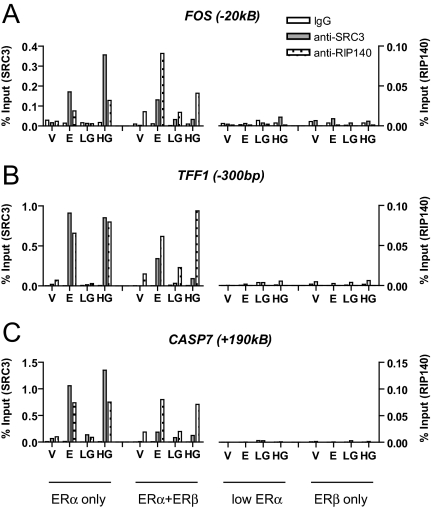
Because the transcriptional activity of chromatin-recruited ERs depends on the recruitment of coregulators, we assessed the recruitment of the p160 coactivator SRC3 and of the putative corepressor RIP140 to the estrogen-responsive sites (Fig. 7). SRC3 was selected because we found it to be the most abundant of the three SRC family members in MCF-7 cells, and RIP140 was evaluated because it has been demonstrated to interact with ERα and modulate estrogen-induced gene activity (34,35).
Figure 7.
Assessment of the Recruitment of SRC3 and RIP140 by ERα- and ERβ-Containing Complexes in Response to Treatment with E2 or Genistein
Chromatin from cells differentially expressing ERα and/or ERβ was immunoprecipitated using SRC3 antibody or RIP140 antibody and quantified using real-time PCR at ER binding regions: A, The −20-kb enhancer of FOS; B, proximal promoter (−300 bp) of TFF1; C, enhancer region (+190 kb) of CASP7. Data are expressed as percentage of input and is representative of three separate experiments.
In parental MCF-7 cells containing ERα only, we observed recruitment of SRC3 and RIP140 in response to E2 and HG, whereas LG elicited no coregulator recruitment, consistent with its low affinity for ERα. As expected, in cells depleted of ERα, there was little to no recruitment of either coregulator. In cells expressing ERα+ERβ, however, SRC3 recruitment by E2 was reduced, but E2 recruitment of RIP140 remained constant relative to ERα-only cells for TFF1 and CASP7 and was increased at the FOS binding site in cells expressing ERα+ERβ. Interestingly, in cells coexpressing ERα and ERβ or expressing ERβ alone, we observed essentially no SRC3 recruitment with genistein at low or high concentration. Although RIP140 recruitment was similar in ERα-only and ERα+ERβ cells with E2 and HG, we observed no recruitment of RIP140 when ERα was depleted by siRNA transfection, and likewise, there was no recruitment of RIP140 in response to E2 or genistein in cells containing ERβ only. It is notable that although RIP140 is not recruited by LG in ERα-only cells, there is substantial RIP140 recruitment by LG in ERα+ERβ cells, suggesting that ERβ may be responsible for this recruitment of RIP140 in cells containing both receptors. Hence, at the sites queried, both SRC3 and RIP140 recruitment mirrored the general pattern of ERα recruitment observed in Fig. 6.
Effects of Knockdown of the Coregulators SRC3 and RIP140 on ER-Mediated Gene Expression
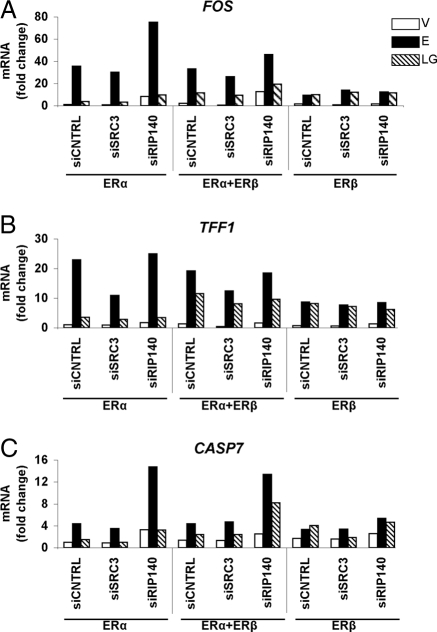
To examine the effects of coregulators on gene expression, we performed siRNA-mediated knockdown of SRC3 and RIP140 in MCF-7 cells expressing different complements of ERα and/or ERβ. We then measured the transcript level of the three genes, FOS, TFF1, and CASP7, by quantitative RT-PCR, in response to E2 or genistein treatment (Fig. 8). Use of the siRNA SMART pools gave 85–90% knockdown of SRC3 and 60–70% knockdown of RIP140 in the different cell types (data not shown).
Figure 8.
Regulation of Gene Expression by E2 and Genistein in Cells Differentially Expressing ERα and ERβ without and with SRC3 and RIP140 Targeted Knockdown
siRNA to SRC3 or RIP140 was transfected into hormone-depleted MCF-7 cells differentially expressing ERα and ERβ, and the transcript level of estrogen-regulated genes (FOS, TFF1, and CASP7) was assessed after 4 h of vehicle or ligand treatment. MCF-7 cells were infected with AdGal (control) or AdERβ for 24 h and subsequently transfected with either siSRC3 or siRIP140 for an additional 48 h. Cells were then exposed to control vehicle (V), 6 nm E2 (E), or 6 nm genistein (LG) for 4 h. Data are the average of two determinations.
In ERα-only cells, E2-stimulated expression of the three genes was reduced by SRC3 knockdown, with TFF1 being reduced to the greatest extent. In contrast, RIP140 knockdown in ERα cells resulted in increased E2-stimulated expression of all three genes (CASP7 and FOS > TFF1). The response to LG in ERα-only cells was very low compared with that of E2, as shown here (and also in Fig. 5), and knockdown of SRC3 had no effect on gene expression under LG treatment in ERα-only cells, relative to siCNTRL, whereas knockdown of RIP140 maintained or increased slightly both the basal and LG response of the three genes. Hence, in ERα-only cells, SRC3 appears to be functioning as a coactivator for E2 and RIP140 as a corepressor on these genes.
In ERα+ERβ cells, LG treatment evoked a greater stimulation of gene expression (cf. Fig. 5), whereas gene response to E2 was similar to that in ERα-only cells (compare siCNTRL values). As in the ERα-only cells, knockdown of SRC3 reduced or maintained expression levels of the three genes in response to E2 or LG. Knockdown of RIP140 enhanced both E2 and LG stimulation of CASP7 and minimally enhanced the response of FOS to these ligands, while having no effect on TFF1. Of particular interest, in cells expressing ERβ only, the targeted knockdown of SRC3 or RIP140 had essentially no effect on E2 or LG stimulation of these three genes. Thus, our findings imply that estrogen action, when only through ERβ, does not rely upon SRC3 or RIP140 and may involve other coregulators. However, when ERα is present, SRC3 functions largely as a coactivator and RIP140 as a corepressor. Our findings also highlight gene and ER subtype-specific differences in regulation of gene expression.
DISCUSSION
Our findings reveal that there are several critical determinants of gene expression regulation by ERα and ERβ: the nature of the ligand, the ligand dose, the presence of ERα and ERβ separately or together in cells, and the recruitment of different coregulators at ER binding sites of hormone-regulated genes by ERα and ERβ ligand complexes. Thus, we observed that a low concentration of the phytoestrogen genistein regulated gene expression preferentially through ERβ and that this preferential regulation of endogenous estrogen-responsive gene expression by ERβ was no longer observed in cells treated with a higher concentration of genistein where ERα and ERβ were equally well recruited to gene regulatory sites and both functioned as effective ligand-regulated transcription factors.
There is much current interest in understanding the effects of genistein in the normal breast and in breast cancers, and reports linking soy consumption and reduced breast cancer incidence have contributed to the marked increase in the consumption of soy-based products in the United States. Many have speculated on the possible mechanisms for genistein action, one possibility being the activation of ERβ to either attenuate ERα activity or induce a set of ERβ-specific antiproliferative genes (36).
In this work, we examined the impact of ERβ coexpression on ERα-regulated gene expression and compared responses with E2 and with two doses of genistein in MCF-7 breast cancer cells expressing different complements of ERα and ERβ. At the lower genistein dose, we observed greater response in cells expressing both ERα and ERβ than in cells expressing only ERα. Preferential activation of ERβ by genistein was lost when genistein dose was increased to 300 nm, whereby genistein activated both ERs and elicited the same transcriptional profile as E2. Regardless of ERβ expression, global analysis of the genistein response at either low or high dose revealed very few genes exclusively induced by genistein and not by E2. Our results suggest that the transcriptional activity of the phytoestrogen ligand genistein at the concentrations tested in our studies is primarily mediated by ERs and is not due to inhibition of other cellular enzymes (e.g. tyrosine kinases), as previously suggested in studies using very high (>10 μm) genistein concentrations (20). In addition, that genistein regulation of gene expression in our studies was completely reversed by the antiestrogen ICI 182,780 further supports our conclusion that this phytoestrogen is acting through the ERs at the 6 and 300 nm concentrations used.
By clustering genes of like expression and testing for enriched biological functions, we found that the presence of ERβ along with ERα attenuated expression of genes involved in growth regulation, with ERβ up-regulating genes important in negative regulation of proliferation (such as cluster B4) and down-regulating genes that enhance cell cycle progression (such as cluster A1). These modulatory effects were observed in E2-treated and HG-treated cells alike, but less so in the LG-treated cells, where ERβ appears to be the chief mediator of genistein action. ERβ also impacted expression of genes encoding proteins associated with RNA processing and metabolism, GPCR signaling, integrin-mediated signaling, and various transport processes.
To examine ERβ activity in the copresence or absence of ERα, we used RNA interference strategies coupled with adenoviral gene delivery to generate cells expressing: ERα alone, ERα+ERβ, no ER, and ERβ alone. We then assessed ligand response in terms of changes in estrogen target gene transcripts and the recruitment of ERs to target gene regulatory regions. We confirmed that although both ERα and ERβ were recruited to estrogen-responsive sites after E2 and HG inductions, only ERβ was recruited to the target sites after LG treatment. The presence of ERβ alone was sufficient to mediate genistein action, and ERβ was still recruited to estrogen binding sites in cells depleted of ERα.
Interestingly, we find that recruitment of the coregulator SRC3 after E2 or HG treatment was diminished in ERα+ERβ cells vs. ERα-only cells. Additionally, in cells expressing ERβ (either ERα+ERβ or ERβ only), LG treatment was unable to induce recruitment of SRC3 to estrogen-responsive sites. Therefore, we conclude that the SRC3 recruitment pattern is influenced by the ER subtype complements at gene regulatory sites and that ERβ recruits SRC3 very poorly.
In contrast, RIP140 recruitment at ER binding sites persisted when ERβ was coexpressed with ERα. RIP140 is a coregulator recruited by agonist-bound ERα, for which repressor effects are well documented (34). Notably, RIP140 can recruit both negative effectors such as histone deacetylases as well as compete for receptor interactions with coactivators such as glucocorticoid receptor-interacting protein 1 (GRIP1) (35). That ERβ disfavors recruitment of a coactivator (SRC3) in cells containing both ER subtypes while maintaining the recruitment of a protein with corepressor activity (RIP140) may contribute to the overall suppressive effect of ERβ on ERα activities including cell proliferation (12,13,24). Of note, both SRC3 and RIP140 were minimally recruited by ERβ to ER binding sites of the regulated genes we examined in response to either genistein or E2, suggesting that ERβ regulation of the expression of these genes involves coregulators other than SRC3 and RIP140. Consistent with this, we observed that knockdown of SRC3 or RIP140 had little impact on the stimulation of gene expression by E2 or genistein in cells expressing ERβ only.
These findings exemplify that the relationships between transcription factor and coregulator recruitments and gene expression are complex and can be ER subtype and gene dependent. We should note that the genomic sites queried here were originally identified through their ability to bind ERα (32,33). Nevertheless, our data show that ERβ can be recruited to these sites. Future studies that define a genome-wide map of ERβ binding sites will be informative in providing additional insights into the relationships between factor recruitment and gene regulation.
In conclusion, our study provides new insights into the cross-modulatory actions of ERα and ERβ in breast cancer cells. We probed ER action through pharmacological means and by gain- and loss-of-expression methods and demonstrated for endogenously regulated genes that the effects of genistein at low nanomolar concentrations were preferentially mediated by ERβ. Although low nanomolar genistein has preferential activities through ERβ, higher concentration of genistein (e.g. 300 nm) activated both ERα and ERβ. Our findings imply that the ratios of the ER subtypes and the concentrations of genistein will be important factors in the estrogen-regulated activities observed in different breast cancers.
MATERIALS AND METHODS
Cell Culture and Adenovirus (Ad) Infection
MCF-7 cells were maintained in MEM (Sigma Chemical Co., St. Louis, MO), supplemented with 5% calf serum (HyClone, Logan, UT), 100 μg/ml penicillin/streptomycin (Invitrogen, Carlsbad, CA), and 25 μg/ml gentamicin (Invitrogen). For estrogen-free experiments, MCF-7 cells were grown in phenol red-free MEM plus 5% charcoal-dextran-treated calf serum for at least 3 d before seeding and then seeded at a density of 3 × 105 cells per 10-cm tissue culture dish (Corning, Corning, NY) for 2 d before adenovirus infection. Cells were infected with either control β-galactosidase Ad (AdGal) or ERβ-containing Ad (AdERβ) for 72 h before estrogen treatment. Conditions used were those described previously (24) to generate MCF-7 cells expressing levels of ERβ equal to that of the endogenously expressed ERα.
siRNA Transfection
To knock down the endogenous ERα in MCF-7 cells, siRNA duplex against the F-domain of ERα was transfected at a final concentration of 20 nm using the DharmaFECT transfection reagent (Dharmacon, Lafayette, CO) as per the manufacturer's recommendations at 48 h before ligand treatment. The siERα forward sequence is UCAUCGCAUUCCUUGCAAAdTdT, and reverse sequence is UUUGCAAGGAAUGCGAUGAdTdT. To knock down endogenous SRC3 or RIP140, we used the SMART pools of siRNA from Dharmacon.
GeneChip Microarrays and Statistical Analysis
Total RNA was harvested for cRNA labeling and hybridization to Affymetrix HG-U133A GeneChips as described previously (10). After washing, the arrays were scanned and analyzed using the GeneChip Operating Software (Affymetrix, Santa Clara, CA). CEL files were processed using the affy and gcrma package protocols in R/BioConductor (37,38). Statistical multivariate analysis was done by the limma package (39). For simplicity's sake, interactions with the time effect were ignored and the CEL files for 4- and 24-h treatments were analyzed separately. The remaining two major effects were defined as ERβ effect (minus and plus ERβ) and ligand effect (vehicle, E2, LG, HG, and ICI+HG). For pairwise comparisons, probe sets were considered significant if the B-statistic was greater than 0 (B-statistic is also known as log odds score).
Several prefiltering steps were taken to reduce the number of false positives in the final analysis. Probe sets with consistently low expression values were discarded (i.e. all calls were absent). Probe sets were also filtered for best overall significance by the F-test statistic. The F-test statistic is calculated from fitting a multivariate model to the data. The entire data set will be available through the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (33) and is accessible through GEO Series accession number GSE9936.
Functional Categorization of Target Genes
Gene functions were curated based on available published data in PubMed (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=PubMed) and the NetAffx databases (http://www.affymetrix.com), and loaded into the GeneSpring software version 7 (Silicon Genetics, Redwood City, CA). Global analysis for overrepresented GenMAPPs (40), KEGG pathways (41), and gene ontology biological function categories was performed using the EASE annotation tool developed by the NIH (29).
Recombinant Ad Preparation
Recombinant Ads were constructed and prepared as described (42). Briefly, replication-deficient Ad serotype 5 was propagated in HEK293 cells (Microbix, Toronto, Ontario, Canada) and then harvested. Approximately 60 μl of the transfected stock was used to reinfect HEK293 cells approaching 70% confluence in 100-mm dishes, and within 2–3 d, more than 90% of the HEK293 cells detached from the dish surface. The cells were harvested as described earlier and used to reinfect up to 20 150-mm dishes of cells using 500 μl of 107 effective plaque-forming units/ml per 150-mm dish. Again, in 2–3 d, more than 90% of the HEK293 cells became detached from the plates. Virus was concentrated up to 1012 effective plaque-forming units/ml, using the CsCl gradient protocol of He et al. (http://www.coloncancer.org/adeasy).
RT-PCR and Quantitative PCR
Total RNA was isolated from MCF-7 cells using TRIzol (Invitrogen) following the manufacturer's recommendations. RNA samples were reverse transcribed by SuperScript II reverse transcriptase (Invitrogen) in 20 μl and subsequently diluted to 500 μl with sterile water. Real-time PCR was performed on the ABI Prism 7900HT using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) according to the manufacturer's recommendations. Briefly, each PCR contained: 1× Master Mix, 5 μl of the diluted cDNA reaction, and 125 nm forward and reverse primers designed to yield 80- to 125-bp amplicons. Relative expression levels were calculated using the ΔΔCT method (43).
Protein Extraction and Western Blot Analysis
Whole-cell extracts were prepared using lysis buffer [20 mm Tris (pH 8.1), 150 mm NaCl, 1% Nonidet P-40, 1% SDS, 5% glycerol], and the Complete-Mini protease inhibitor cocktail tablet (Roche, Indianapolis, IN). Samples were boiled in Laemmli buffer and run on mini-blot SDS-PAGE gels (Bio-Rad, Hercules, CA). Proteins were transferred onto nitrocellulose membrane overnight (20 V constant) and blocked with 5% nonfat milk in Tris-buffered saline solution [10 mm Tris (pH 7.4), 150 mm NaCl, and 0.5% Tween 20] before incubation with primary antibody. Anti-ERα antibodies (polyclonal rabbit HC-20) were from Santa Cruz Biotechnologies (Santa Cruz, CA). Anti-ERβ antibodies (monoclonal mouse, CWK-F12) were produced by our lab (44). Secondary antibodies to the respective primary antibodies were purchased from Zymed Laboratories (South San Francisco, CA).
Cell Proliferation Assays
MCF-7 cells were infected with AdGal or AdERβ in 100-mm dishes (Corning) for 8 h before seeding in phenol red-free MEM supplemented with 5% charcoal-dextran-treated calf serum at 1000 cells per well in 96-well plates (BD Falcon, Franklin Lakes, NJ). The next day, cells received fresh medium and vehicle or ligands every 72 h. Cell growth was monitored using the CellTiter 96 AQueous One Solution Cell Proliferation Assay, according to the manufacturer's recommendations (Promega, Madison, WI).
ChIP Assays
ChIP assays were carried out as described by Metivier and colleagues (45). Confluent 150-mm dishes of MCF-7 cells (∼17 × 106 cells per dish) were treated with control 0.1% ethanol vehicle or ligands (E2, LG, or HG) for 45 min and were then fixed with 1.5% formaldehyde. Fixation was subsequently stopped by 125 mm glycine quench and 2× cold PBS washes. Cross-linked chromatin complexes were collected in nuclei lysis buffer and sonicated three times on ice at setting 8 for 10-sec pulses (output 20 W; Dismembrator model 100; Fisher Scientific, Pittsburgh, PA). To immunoprecipitate chromatin complexes, the following antibodies were used: ERα antibody HC-20 (Santa Cruz Biotechnology); ERβ antibodies combination with equal parts of GeneTex ERβ mouse monoclonal 7B10.7 (GTX70182; GeneTex, San Antonio, TX), Calbiochem mouse monoclonal 10.76 (GR40; Calbiochem, La Jolla, CA), and ABR rabbit polyclonal (PA1–311; ABR Antibodies, Golden, CO); SRC3/AIB1 antibody H-270 (Santa Cruz Biotechnology); and RIP140 antibody (Santa Cruz Biotechnology). We incubated with the specific antibodies and protein A/G agarose beads (Pierce, Rockford, IL) overnight at 4 C followed by washes and elution with 1% SDS. The DNA was recovered by reversing the cross-links with 20 mm NaCl at 65 C overnight and dissolving in a final volume of 100 μl per immunoprecipitation. To quantitatively detect immunoprecipitated chromatin, 1 μl eluent was mixed with SYBR Green Taq mix and 125 nm forward and reverse specific primers directed against known ERα binding sites (32).
Acknowledgments
We thank Ken Chang for his assistance in these studies and appreciate the advice from Lei Liu and Mark Band of the University of Illinois Carver Biotechnology Center.
Footnotes
This work was supported by grants from the National Institutes of Health (R01CA18119 and P01AG024387) and The Breast Cancer Research Foundation. E.C.C. received support from NIH Training Grant T32 ES07326 and the Mary Landfield Fellowship in Cancer Biology. T.H.C. received support from the Singapore Agency for Science Technology and Research (A*STAR).
Disclosure Statement: E.C.C., T.H.C., S.-H.P., W.G.H., J.A.K., and B.S.K. have nothing to declare. B.K. is employed by Wyeth Research.
First Published Online February 7, 2008
Abbreviations: Ad, Adenovirus; ChIP, chromatin immunoprecipitation; EASE, Expression Analysis Systematic Explorer; E2, 17β-estradiol; ER, estrogen receptor; GPCR, G protein-coupled receptor; HG, high genistein; LG, low genistein; RIP140, receptor-interacting protein 140; SRC3, steroid receptor coactivator 3; siRNA, small interfering RNA.
References
- Harris HA 2007 Estrogen receptor-β: recent lessons from in vivo studies. Mol Endocrinol 21:1–13 [DOI] [PubMed] [Google Scholar]
- Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, Enmark E, Pettersson K, Warner M, Gustafsson JA 2001 Mechanisms of estrogen action. Physiol Rev 81:1535–1565 [DOI] [PubMed] [Google Scholar]
- Kuiper GG, Carlsson B, Grandien K, Enmark E, Haggblad J, Nilsson S, Gustafsson JA 1997 Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors α and β. Endocrinology 138:863–870 [DOI] [PubMed] [Google Scholar]
- Hall JM, McDonnell DP 1999 The estrogen receptor β-isoform (ERβ) of the human estrogen receptor modulates ERα transcriptional activity and is a key regulator of the cellular response to estrogens and antiestrogens. Endocrinology 140:5566–5578 [DOI] [PubMed] [Google Scholar]
- Katzenellenbogen BS, Frasor J 2004 Therapeutic targeting in the estrogen receptor hormonal pathway. Semin Oncol 31:28–38 [DOI] [PubMed] [Google Scholar]
- Jarvinen TA, Pelto-Huikko M, Holli K, Isola J 2000 Estrogen receptor beta is coexpressed with ERα and PR and associated with nodal status, grade, and proliferation rate in breast cancer. Am J Pathol 156:29–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurebayashi J, Otsuki T, Kunisue H, Tanaka K, Yamamoto S, Sonoo H 2000 Expression levels of estrogen receptor-α, estrogen receptor-β, coactivators, and corepressors in breast cancer. Clin Cancer Res 6:512–518 [PubMed] [Google Scholar]
- Palmieri C, Cheng GJ, Saji S, Zelada-Hedman M, Warri A, Weihua Z, Van Noorden S, Wahlstrom T, Coombes RC, Warner M, Gustafsson JA 2002 Estrogen receptor β in breast cancer. Endocr Relat Cancer 9:1–13 [DOI] [PubMed] [Google Scholar]
- Saji S, Hirose M, Toi M 2005 Clinical significance of estrogen receptor β in breast cancer. Cancer Chemother Pharmacol 56(Suppl 1):21–26 [DOI] [PubMed] [Google Scholar]
- Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS 2003 Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology 144:4562–4574 [DOI] [PubMed] [Google Scholar]
- Liu MM, Albanese C, Anderson CM, Hilty K, Webb P, Uht RM, Price Jr RH, Pestell RG, Kushner PJ 2002 Opposing action of estrogen receptors α and β on cyclin D1 gene expression. J Biol Chem 277:24353–24360 [DOI] [PubMed] [Google Scholar]
- Paruthiyil S, Parmar H, Kerekatte V, Cunha GR, Firestone GL, Leitman DC 2004 Estrogen receptor β inhibits human breast cancer cell proliferation and tumor formation by causing a G2 cell cycle arrest. Cancer Res 64:423–428 [DOI] [PubMed] [Google Scholar]
- Strom A, Hartman J, Foster JS, Kietz S, Wimalasena J, Gustafsson JA 2004 Estrogen receptor β inhibits 17β-estradiol-stimulated proliferation of the breast cancer cell line T47D. Proc Natl Acad Sci USA 101:1566–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AH, Wan P, Hankin J, Tseng CC, Yu MC, Pike MC 2002 Adolescent and adult soy intake and risk of breast cancer in Asian-Americans. Carcinogenesis 23:1491–1496 [DOI] [PubMed] [Google Scholar]
- Barnes S 1995 Effect of genistein on in vitro and in vivo models of cancer. J Nutr 125:777S–783S [DOI] [PubMed] [Google Scholar]
- Sacks FM, Lichtenstein A, Van Horn L, Harris W, Kris-Etherton P, Winston M 2006 Soy protein, isoflavones, and cardiovascular health: a summary of a statement for professionals from the American Heart Association Nutrition Committee. Circulation 26:1689–1692 [DOI] [PubMed] [Google Scholar]
- Allred CD, Allred KF, Ju YH, Virant SM, Helferich WG 2001 Soy diets containing varying amounts of genistein stimulate growth of estrogen-dependent (MCF-7) tumors in a dose-dependent manner. Cancer Res 61:5045–5050 [PubMed] [Google Scholar]
- Ju YH, Allred CD, Allred KF, Karko KL, Doerge DR, Helferich WG 2001 Physiological concentrations of dietary genistein dose-dependently stimulate growth of estrogen-dependent human breast cancer (MCF-7) tumors implanted in athymic nude mice. J Nutr 131:2957–2962 [DOI] [PubMed] [Google Scholar]
- Wang TT, Sathyamoorthy N, Phang JM 1996 Molecular effects of genistein on estrogen receptor mediated pathways. Carcinogenesis 17:271–275 [DOI] [PubMed] [Google Scholar]
- Akiyama T, Ishida J, Nakagawa S, Ogawara H, Watanabe S, Itoh N, Shibuya M, Fukami Y 1987 Genistein, a specific inhibitor of tyrosine-specific protein kinases. J Biol Chem 262:5592–5595 [PubMed] [Google Scholar]
- Cappelletti V, Fioravanti L, Miodini P, Di Fronzo G 2000 Genistein blocks breast cancer cells in the G2M phase of the cell cycle. J Cell Biochem 79:594–600 [PubMed] [Google Scholar]
- Pagliacci MC, Smacchia M, Migliorati G, Grignani F, Riccardi C, Nicoletti I 1994 Growth-inhibitory effects of the natural phyto-oestrogen genistein in MCF-7 human breast cancer cells. Eur J Cancer 30A:1675–1682 [DOI] [PubMed] [Google Scholar]
- Muthyala RS, Ju YH, Sheng S, Williams LD, Doerge DR, Katzenellenbogen BS, Helferich WG, Katzenellenbogen JA 2004 Equol, a natural estrogenic metabolite from soy isoflavones: convenient preparation and resolution of R- and S-equols and their differing binding and biological activity through estrogen receptors α and β. Bioorg Med Chem 12:1559–1567 [DOI] [PubMed] [Google Scholar]
- Chang EC, Frasor J, Komm B, Katzenellenbogen BS 2006 Impact of estrogen receptor β on gene networks regulated by estrogen receptor α in breast cancer cells. Endocrinology 147:4831–4842 [DOI] [PubMed] [Google Scholar]
- Kian Tee M, Rogatsky I, Tzagarakis-Foster C, Cvoro A, An J, Christy RJ, Yamamoto KR, Leitman DC 2004 Estradiol and selective estrogen receptor modulators differentially regulate target genes with estrogen receptors α and β. Mol Biol Cell 15:1262–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe DG, Secreto FJ, Subramaniam M, Getz BJ, Khosla S, Spelsberg TC 2005 Estrogen receptor α and β heterodimers exert unique effects on estrogen- and tamoxifen-dependent gene expression in human U2OS osteosarcoma cells. Mol Endocrinol 19:1555–1568 [DOI] [PubMed] [Google Scholar]
- Stossi F, Barnett DH, Frasor J, Komm B, Lyttle CR, Katzenellenbogen BS 2004 Transcriptional profiling of estrogen-regulated gene expression via estrogen receptor (ER)α or ERβ in human osteosarcoma cells: distinct and common target genes for these receptors. Endocrinology 145:3473–3486 [DOI] [PubMed] [Google Scholar]
- Tseng GC, Wong WH 2005 Tight clustering: a resampling-based approach for identifying stable and tight patterns in data. Biometrics 61:10–16 [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis Jr G, Sherman BT, Lane HC, Lempicki RA 2003 Identifying biological themes within lists of genes with EASE. Genome Biol 4:R70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C, Edvardsson K, Lewandowski SA, Strom A, Gustafsson JA 2008 A genome-wide study of the repressive effects of estrogen receptor β on estrogen receptor α signaling in breast cancer cells. Oncogene 27:1019–1032 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M 2005 Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33–43 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M 2006 Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38:1289–1297 [DOI] [PubMed] [Google Scholar]
- Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, George J, Kuznetsov VA, Lee YK, Charn TH, Palanisamy N, Miller LD, Katzenellenbogen BS, Ruan Y, Bourque G, Wei C, Liu ET 2007 Whole-genome cartography of estrogen receptor-α binding sites. PLoS Genetics 3:e87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augereau P, Badia E, Fuentes M, Rabenoelina F, Corniou M, Derocq D, Balaguer P, Cavailles V 2006 Transcriptional regulation of the human NRIP1/RIP140 gene by estrogen is modulated by dioxin signalling. Mol Pharmacol 69:1338–1346 [DOI] [PubMed] [Google Scholar]
- Teyssier C, Belguise K, Galtier F, Cavailles V, Chalbos D 2003 Receptor-interacting protein 140 binds c-Jun and inhibits estradiol-induced activator protein-1 activity by reversing glucocorticoid receptor-interacting protein 1 effect. Mol Endocrinol 17:287–299 [DOI] [PubMed] [Google Scholar]
- Bouker KB, Hilakivi-Clarke L 2000 Genistein: does it prevent or promote breast cancer? Environ Health Perspect 108:701–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP 2003 Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31:e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Irizarry R, Gentleman R, Murillo F, Spencer F 2004 A model based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 99:909–917 [Google Scholar]
- Smyth GK 2004 Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:Article3 [DOI] [PubMed] [Google Scholar]
- Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR 2002 GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet 31:19 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S 2000 KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazennec G, Alcorn JL, Katzenellenbogen BS 1999 Adenovirus-mediated delivery of a dominant negative estrogen receptor gene abrogates estrogen-stimulated gene expression and breast cancer cell proliferation. Mol Endocrinol 13:969–980 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001 Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Choi I, Ko C, Park-Sarge OK, Nie R, Hess RA, Graves C, Katzenellenbogen BS 2001 Human estrogen receptor β-specific monoclonal antibodies: characterization and use in studies of estrogen receptor β protein expression in reproductive tissues. Mol Cell Endocrinol 181:139–150 [DOI] [PubMed] [Google Scholar]
- Metivier R, Stark A, Flouriot G, Hubner MR, Brand H, Penot G, Manu D, Denger S, Reid G, Kos M, Russell RB, Kah O, Pakdel F, Gannon F 2002 A dynamic structural model for estrogen receptor-α activation by ligands, emphasizing the role of interactions between distant A and E domains. Mol Cell 10:1019–1032 [DOI] [PubMed] [Google Scholar]