Abstract
Prolonged cold exposure induces nonshivering thermogenesis primarily through β-adrenergic- and cAMP-mediated regulation of uncoupling protein-1 (UCP1) in brown adipose tissue. Molecular mechanisms involved in this induction of Ucp1 gene transcription consists of an intricate interplay between many nuclear receptors in coordination with coactivators/corepressors. Recently, it has been shown that members of the nuclear receptor-4A (NR4A) family of orphan nuclear receptors (Nur77, Nurr1, and NOR-1) are highly responsive to cAMP-second messenger pathways. Here we have identified a new regulatory motif in the Ucp1 promoter that binds NR4As to stimulate Ucp1 gene transcription. Upon cold exposure of mice, or β-agonist treatment of mouse and human adipocytes, the expression of NR4A nuclear receptors is rapidly induced, with NOR-1 being the most robust, and this precedes increases in Ucp1 expression. A dominant-negative mutant Nur-77 receptor that prevents the transcriptional activity of NR4A receptors blocked β-adrenergic receptor-stimulated Ucp1 gene transcription. By gel shift and chromatin immunoprecipitation assays, we defined the sequence (−5.64 kb) in the Ucp1 promoter to which NOR-1 binds. In transient reporter assays, this element significantly augments the activity of a 3.7-kb Ucp1 promoter. These results extend our understanding of the combinatorial complexity in the signaling pathways that control this tissue-specific gene.
OBESITY RESULTS FROM an imbalance in energy homeostasis between caloric intake and expenditure. Because it appears that central mechanisms of energy balance tend to favor intake and preservation of caloric load (1), the identification of cellular mechanisms that can cause net energy expenditure could be advantageous as targets for obesity therapy. One of the most well understood cellular processes of sustained negative energy balance is the uncoupling of mitochondrial respiration in brown adipose tissue (BAT) due to a regulated proton leak in the inner mitochondrial membrane through uncoupling protein 1 (UCP1) (2). As a result of this process, fuel oxidation in brown adipocytes becomes uncoupled from ATP synthesis, and energy is dissipated as heat. This adaptive thermogenic response is driven by the sympathetic nervous system in response to cold temperature as well as diet, to activate β-adrenergic receptors (β-ARs) in white and brown adipocytes. These in turn regulate the activity of lipases as well as nuclear transcription factors (2,3). In the case of the Ucp1 gene, many elements of the signal transduction cascade emanating from the β-ARs that are involved in its regulation have been identified, but questions remain (reviewed in Ref. 3).
BAT-specific and adrenergic control of Ucp1 gene transcription converge on a 221-bp cis-acting regulatory sequence, located −2.31 kb from the transcription start site, to which a number of different transcription factors and cofactors bind in response to hormonal cues (3,27). As such, this enhancer is a regulatory nexus that permits a multiplicity of outcomes through additive and synergistic interactions. In addition, a proximal cAMP-responsive element 4 (CRE4) in the Ucp1 promoter has been shown to contribute to the adrenergic response (4).
Recently, a family of orphan nuclear receptors [nuclear receptors 4A (NR4A)] have been shown to regulate glucose metabolism in liver and muscle (5,6). The NR4A family consists of three members: NR4A1 (Nur77), NR4A2 (Nurr1), and NR4A3 (NOR-1) (7). NR4A receptors are immediate-early genes that are regulated by many physiological stimuli including growth factors, hormones, and inflammatory signals. These receptors are highly expressed in a wide variety of metabolically demanding and energy-dependent tissues, such as skeletal muscle, adipose, heart, kidney, T cells, liver, and the brain (7). In the C2C12 skeletal muscle cell line, Nur77 was rapidly induced by β-AR agonists with subsequent regulation of genes involved in energy expenditure and lipid homeostasis (8). It has also been shown that in 3T3-L1 adipocytes, all three NR4As were rapidly and transiently increased upon initiating their differentiation (9), but more recent evidence indicates that they are not required for differentiation per se (10). Together these studies suggest that NR4A may be involved in the balance of metabolic fuel use in ways not previously appreciated.
In this study, we show that in white and brown adipocytes, the expression of NR4A receptors increases in response to β-AR stimulation of cAMP levels, with NOR-1 being the most robustly increased. Using a variety of approaches, we demonstrate that it is one of the necessary elements in the concerted transcriptional program to increase Ucp1 gene transcription, and we identify the promoter element through which NOR-1 mediates this effect.
RESULTS
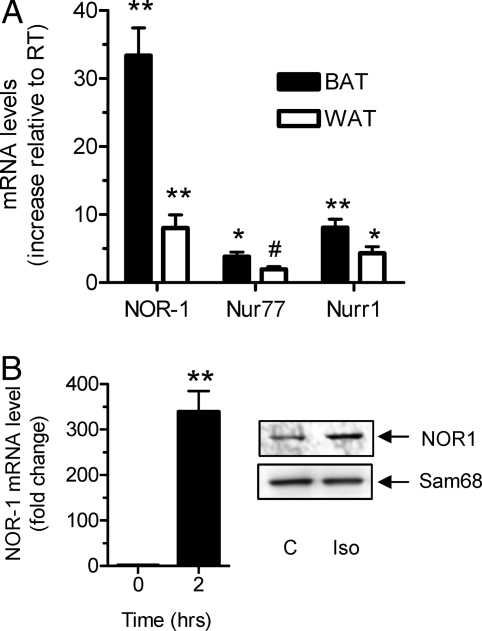
In many cell types, the three NR4A family members, Nur77, Nurr1, and NOR-1, are often regulated in parallel (6,11). Therefore, we first determined their expression in vivo in BAT and white adipose tissue (WAT) under conditions that would activate β-AR signaling. Because cold temperature exposure stimulates the sympathetic nervous system and release of noradrenaline activates β-ARs, mice were acutely placed at 4 C. As shown in Fig. 1A, there was a significant increase in transcripts for all three NR4A family members, with NOR-1 exhibiting the most robust response. To more directly address a role for β-ARs in this process, mouse HIB-1B brown adipocytes were treated with 1 μm of the synthetic β-AR agonist isoproterenol (Iso) for 2 h to mimic sympathetic nervous system activation. There was a similar increase in NOR-1 mRNA, which was also reflected by increased NOR-1 protein as shown in Fig. 1B.
Figure 1.
Expression of NR4A Family Members in BAT and WAT of Cold-Exposed Mice
A, Real-time PCR analysis of NOR-1, Nurr1, and Nurr77 genes in BAT and WAT of mice at 4 C expressed relative to those of mice at thermoneutrality. The results are mean ± sem of three mice in each group; **, P < 0.001; *, P < 0.01; #, P < 0.05. B, Levels of NOR-1 from control or 2-h Iso-treated HIB-1B cells as determined by RT-PCR (left) and Western blotting of nuclear extracts (right).
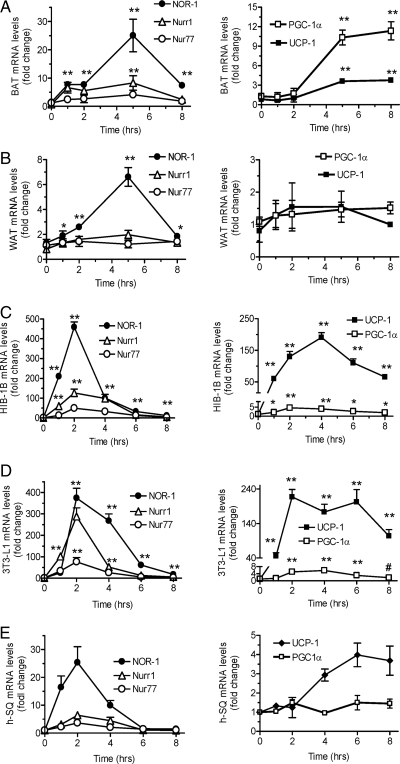
A previous brief report suggested that Nur77 might be involved in the inhibition of Ucp1 gene transcription (12). We explored the regulation of NR4A transcription factors in adipose tissue and how they might control the expression of the Ucp1 gene or other genes involved in thermogenesis. The expression of NOR-1, Nur77, and Nurr1 as well as UCP1 and the transcriptional coactivator peroxisome proliferator-activated receptor-γ (PPARγ) coactivator-1α (PGC-1α) were measured as a function of time in BAT (Fig. 2A) and WAT (Fig. 2B) of cold-exposed mice as well as in cultured mouse HIB-1B (Fig. 2C), 3T3-L1 (Fig. 2D), and human sc adipocytes (Fig. 2E) that had been treated with Iso. As shown in the left panels of Fig. 2, all NR4A family members were induced at 1–2 h of stimulation; particularly NOR-1, which was the most highly induced in all samples. Note that the responses were always more pronounced in BAT and brown adipocytes (Fig. 2, A and C) than in WAT and white adipocytes (Fig. 2, B–E). In addition, the pattern of induction of all NR4A mRNAs was transient, displaying a rapid decline by 5–6 h. When comparing their peak increase against the expression of Ucp1 and PGC-1α, shown in the right panels of Fig. 2, the initial rise in NR4A more often than not preceded that of Ucp1 and PGC-1α.
Figure 2.
Time Course of β-AR-Stimulated NR4A Gene Expression
Real-time PCR analysis of gene expression in BAT (A), WAT (B), HIB-1B (C), 3T3-L1 (D), and human (E) sc adipocytes (h-SQ). BAT and WAT are from mice exposed to 4 C for indicated times (n = 3). HIB-1B, 3T3-L1, and human adipocytes were treated with 1 μm Iso for indicated times. The results are mean ± sem of three experiments; **, P < 0.001; *, P < 0.01. Basal cycle threshold (Ct) values for UCP1 were as follows: BAT, 21; WAT, 28; HIB-1B, 28; 3T3-L1, 31; h-SQ, 32. Basal Ct values for PGC-1α were as follows: BAT, 21; WAT, 25; HIB-1B, 23; 3T3-L1, 24; h-SQ, 25. Basal Ct values for NOR-1 were as follows: BAT, 26; WAT, 27; HIB-1B, 27; 3T3-L1, 30; h-SQ, 30. Basal Ct values for Nur77 were as follows: BAT, 22; WAT, 23; HIB-1B, 24; 3T3-L1, 23: h-SQ, 25. Basal Ct values for Nurr1 were as follows: BAT, 25; WAT, 28; HIB-1B, 28; 3T3-L1, 31; h-SQ, 30.
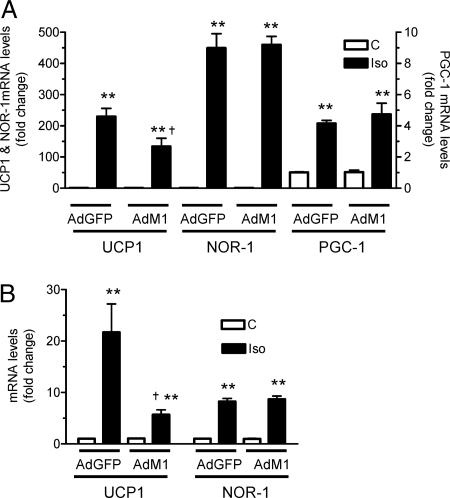
The next series of experiments sought to more directly address the question of whether NR4A regulate Ucp1 gene expression. Because NOR-1 was consistently the most robustly induced in both adipose tissues and cultured adipocytes, most of the following studies focused on NOR-1, realizing that these three orphan nuclear receptors have been shown to have redundant function in many instances (6,11). The first experiment used a dominant-negative approach. 6EB2 CARΔ1 brown adipocytes and 3T3-L1 CARΔ1 white adipocytes, which were engineered to express the Coxsackie virus receptor (13), were infected with an adenoviral vector expressing a dominant-negative mutant of Nur77 (Nur77-M1) that lacks the AF-1 activation domain (6). This mutant receptor functions as a pan antagonist of the transcriptional activity of all three NR4A family members. Adenoviral green fluorescent protein (GFP) was expressed as a negative control. In cells infected with Nur77-M1, Iso-stimulated Ucp1 gene expression was significantly reduced in both brown (40%, Fig. 3A) and white (60%, Fig. 3B) adipocytes. Because the efficiency of infection of the differentiated 6EB2 CARΔ1 and 3T3-L1 CARΔ1 adipocytes was routinely 50–60%, this suggests that the suppression of UCP1 in the cells expressing the mutant Nur77 construct was probably much greater. There was also no effect of Nur77-M1 on the increase of NOR-1 or PGC-1α transcripts stimulated by Iso (Fig. 3, A and B; in the case of 3T3-L1 CARΔ1 adipocytes, there was no effect of Iso on PGC-1α; not shown). Therefore, our results collectively are consistent with a stimulatory role of at least NOR-1 on Ucp1 gene transcription.
Figure 3.
Suppression of Iso-Stimulated Ucp1 Gene Expression by Dominant-Negative Nur77-M1
Differentiating 6Eb2 CARΔ1 (A) and 3T3-L1 CARΔ1 (B) adipocytes were infected with adenoviruses (Ad) for either GFP or Nur77-M1 as detailed in Materials and Methods. Cells were treated with 1 μm Iso for 2 h, and RNA was isolated. UCP-1, NOR-1, and PGC-1α transcripts were measured by real-time PCR. The results shown are mean ± sem of three independent experiments; **, P < 0.001; *, P < 0.01; #, P < 0.05.
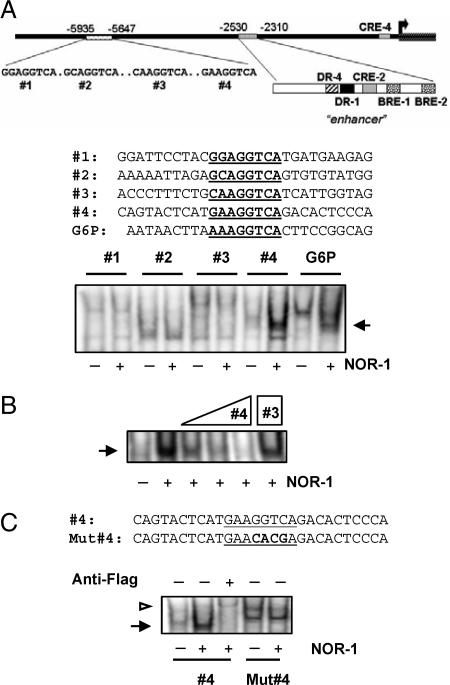
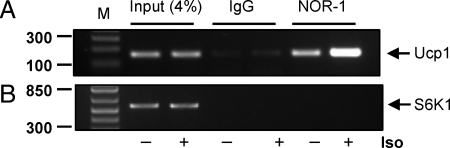
To determine whether NOR-1 directly binds to a regulatory site in the Ucp1 promoter, we searched for potential nerve growth factor-responsive element (NBRE) binding site(s) up to 7 kb 5′ to the start site of transcription. Figure 4A shows a schematic illustration of this region that includes the important enhancer containing a number of essential regulatory elements (3,4,14). This cartoon also shows several candidate NBRE sequences that we identified. EMSA with oligonucleotides containing these potential binding sites were performed using nuclear extracts from COS-7 cells in which we expressed Flag-tagged NOR-1. Among the four potential sites at −5.6 kb of the Ucp1 promoter (1–4 in Fig. 4A) oligo 4 strongly bound NOR-1. Binding of NOR-1 to the NBRE sequence from the G6P promoter (6) served as a positive control. There were two weakly homologous sites in the enhancer; however, none could bind to NOR-1 (data not shown). The specificity of binding to probe sequence 4 is shown by competitive binding of the unlabeled probe in the reaction, where increasing amounts up to 100× of sequence 4 progressively eliminated all binding (Fig. 4B, lane 5). There was no competition for this binding when 100× of sequence 3 (Fig. 4B, lane 6) was used. The identity of NOR-1 as the binding species was confirmed by addition of anti-Flag antiserum, which resulted in the slower-migrating supershifted band (white arrowhead in Fig. 4C). Also shown in Fig. 4C, binding of NOR-1 to sequence 4 was lost when point mutations were introduced. These results together show that NOR-1 can bind to an NBRE in the UCP1 promoter. These in vitro findings were extended to chromatin immunoprecipitation assays to assess the occupancy of this distal region of the Ucp1 promoter by NOR-1 in intact cells. HIB-1B brown adipocytes were treated with Iso (1 μm) for 2 h. As shown in Fig. 5A, NOR-1 was found on this distal portion of the UCP1 promoter under basal conditions, but it was significantly increased (2.4-fold) in response to Iso. PCR amplification with primers to a negative control, the S6K1 gene, demonstrated the specificity of the assay (Fig. 5B). We also observed a weak binding of Nur77 to this distal site of UCP1 promoter (data not shown).
Figure 4.
NOR-1 Binds to NBRE No. 4 in the UCP1 Promoter
A, EMSA with nuclear extracts (5 μg), prepared from COS-7 cells transfected with NOR-1-Flag vector, that had been incubated with radiolabeled sequences numbered 1–4 or the NBRE from G6P as a positive control. B, Cold probes from NBRE 4 (10×, 50×, and 100×) or NBRE 3 (100×) was included in the binding reactions using NBRE 4 as radiolabeled probe. C, A mutant oligonucleotide for NBRE 4 (Mut#4) or anti-Flag antiserum (1 μl) for super-shift was included in the binding reactions of NBRE 4. A representative image from three independent experiments is shown.
Figure 5.
Recruitment of NOR-1 to the UCP1 Gene Promoter
Differentiated HIB-1B cells were treated with Iso (1 μm) for 2 h. NOR-1 was immunoprecipitated as described in Materials and Methods. Binding of NOR-1 to the UCP1 promoter (A) or S6K1 gene (B) was detected by PCR. A representative image from two independent experiments is shown.
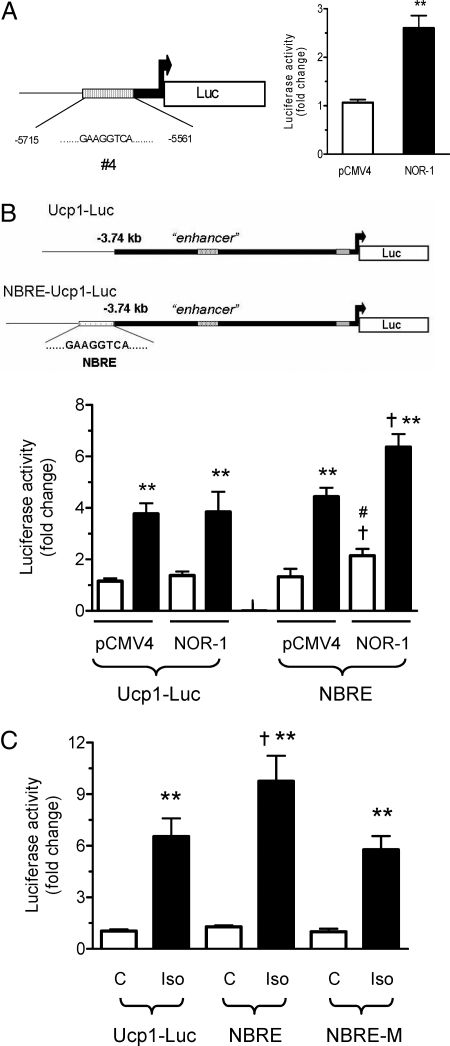
To establish the functional relevance of this binding, a small (150-bp) piece of the UCP1 promoter containing the NBRE (sequence 4) was inserted into two different reporter constructs to test for responsiveness to NOR-1. First, as a simple functional test, the NBRE was placed within the context of a minimal TATA-box promoter construct driving a luciferase reporter (NBRE-Luc) and introduced into COS-7 cells with or without a NOR-1 expression plasmid. Figure 6A shows that NOR-1 significantly increased luciferase activity by 2.5-fold as compared with cotransfection with empty vector pCMV4. Next the NBRE was inserted immediately 5′ to the 3.7-kb mouse UCP1 promoter (4), designated NBRE-Ucp1-Luc, and its activity was measured in either COS-7 cells or HIB-1B cells. Using COS-7 cells, which contain negligible levels of the prerequisite transcription factors for the expression of the Ucp1 gene, we reconstituted cAMP-inducible UCP1 enhancer activity by transfecting expression plasmids encoding PPARγ, retinoid X receptor-α (RXRα), PGC-1α, and providing the PPARγ agonist rosiglitazone, as done previously (15). Under these conditions, we tested the effect of cotransfected NOR-1 on the Ucp-1 reporter vectors UCP1-Luc and NBRE-Ucp1-Luc. As shown in Fig. 6B, luciferase activity was increased in both of the reporter vectors in response to Fsk. However, when cotransfected with NOR-1, there was a significant increase in the Fsk-stimulated activity of NBRE-Ucp1-Luc as compared with Ucp1-Luc. In addition, even in the absence of Fsk, NBRE-Ucp1-Luc showed a significantly higher basal activity than Ucp1-Luc with NOR-1 cotransfection. Finally as shown in Fig. 6C, when transfected into HIB-1B brown adipocytes, Iso increased Ucp1-Luc reporter activity by 6.5-fold, whereas for NBRE-Ucp1-Luc there was a more than 9-fold increase in activity. However, this increase in luciferase activity was eliminated when the Ucp1 NBRE sequence (GAAGGTCA) was mutated (changes in italic) to GAACACGA in the NBRE-UCP1-Luc plasmid (NBRE-M-Ucp1-Luc; Fig. 6C). Together these results illustrate that the increase in UCP1 promoter activity in NBRE-Ucp1-Luc was due to the binding of NR4A family members to the Ucp1 NBRE sequence. From our results collectively, we can conclude that NOR-1 participates in the cAMP-dependent increase in transcription of the Ucp1 gene.
Figure 6.
Functional Activity of the NBRE from the UCP1 Promoter in Transient Reporter Assays
A, COS-7 cells were transfected with NBRE-Luc with NOR-1 expression vector, and luciferase activity was measured after 48 h. B, Reconstitution of UCP1 promoter activity in COS-7 cells and effect of NOR-1. COS-7 cells were cotransfected with either Ucp1-Luc or NBRE-Ucp1-Luc (denoted as NBRE) and expression vector for pCMV-NOR-1 as described in Materials and Methods. Cells were treated with Fsk (10 μm) during the final 6 h of transfection. Luciferase activity was normalized to β-galactosidase activity (lower panel). C, HIB-1B cells were transfected with either Ucp1-Luc or NBRE-Ucp1-Luc (denoted as NBRE) or NBRE-M-Ucp1-Luc (denoted as NBRE-M). Cells were treated or not with Iso (1 μm) during the final 4 h of transfection. Luciferase activity was normalized to β-galactosidase activity. The results shown are means ± sem of three independent experiments; **, P < 0.001; *, P < 0.01; †, P < 0.01 as compared with Iso-treated samples; #, P < 0.05 as compared with control untreated samples.
DISCUSSION
The NR4A orphan nuclear receptors were first cloned as early response genes to growth factors, and previous literature confined their importance to immune function and neuronal development (7). More recently, NR4A have been shown to be regulators of genes involved in glucose metabolism in liver and skeletal muscle (5,6). Here we show that they are also part of the metabolic regulation of adipose tissue by augmenting the β-adrenergic stimulation of Ucp1 gene transcription. In response to cold exposure, expression of NR4A receptors, particularly NOR-1 (NR4A3), is robustly induced in BAT and WAT. This response is mimicked by β-adrenergic stimulation in cultured adipocytes. By a variety of approaches, we clearly establish that NR4A receptors bind to the Ucp1 promoter and contribute to the overall magnitude of the cAMP-dependent transcription of the Ucp1 gene. The increased expression of NR4A receptors seen here in adipocytes in response to β-AR-stimulated increases in cAMP levels are similar to that observed in other tissues by β-ARs or other Gs-coupled receptors (6,16,17). Several CREs have been noted in the NOR-1 promoter that may be responsible for these cAMP-dependent increases in expression (18).
Of the four candidate binding sites for NOR-1 in the Ucp1 promoter, sequence 4 is the most similar to the consensus binding site for NR4A receptors. This NBRE in the Ucp1 promoter is located distal to the small highly conserved enhancer region (−2.31 to −2.53 kb) (3,19), which is now well established to be densely packed with a number of sequences shown to be critically important for the cAMP- and brown fat-specific expression of Ucp1. A number of transcription factors are recruited to this enhancer region that serve to either stimulate (PPARγ, PPARα, RXRα, activating transcription factor-2, PGC-1α) (3) or inhibit (liver X receptor-α, nuclear receptor interacting protein 140) UCP1 gene transcription (20,27). Recently, estrogen-related receptor-α also has been shown to bind to the Ucp1 enhancer (20) but its role in adipose tissue biology is complex and remains to be clarified (21,22). Like the distal NBRE that resides outside of the enhancer region, a cAMP response element-binding protein binding site in the proximal portion of the UCP1 promoter (CRE4) is an important contributor to overall Ucp1 gene transcription in response to cAMP (4). Therefore, although the enhancer is a critical component for transcriptional activation, other regions of the promoter such as CRE4 and the NBRE are also important contributors and amplify the response.
Several features of the NR4A family make it difficult to evaluate their functions in cells as well as in whole-animal physiology. In most studies of Nur77, Nurr1, and NOR-1, their expression is often regulated by similar stimuli (7). They are also shown to have redundant function in several tissues, including liver, macrophages, hypothalamic and pituitary systems, and T cells (6,7,11,17). Despite evidence for significant roles for one or the other in various cell types (7), Nur77−/− and NOR-1−/− mice were reported to have a minimal phenotype (23,24), suggesting that their absence was likely compensated for by other family members (7). Only when both Nur77 and NOR-1 were jointly eliminated was a phenotype of acute myeloid leukemia observed (25). Therefore, in the case of Ucp1 gene transcription, adipose tissue-specific deletion of all three NR4A family members may be required to elucidate their exact role.
Our finding of a new player in the cAMP-dependent control of the Ucp1 gene illustrates the complex regulatory control on the expression of this mitochondrial uncoupler. These studies also extend the realm of tissues in which NR4A receptors regulate energy metabolism to all three major organ systems involved in the management of metabolic balance: liver, muscle, and adipose. Because cAMP-dependent increases in NR4A occur in white adipocytes, where mitochondrial uncoupling by UCP1 is at best a very minor aspect of their metabolism, the identity of other target genes in white fat regulated by NR4A could lend important new insight into β-AR stimulation of adipose tissue fuel mobilization or adipokine production and release.
MATERIALS AND METHODS
Animal Experiments
Ten- to 12-wk-old C57BL/6J mice were acclimated to thermoneutrality (28 C) for 3 d and then transferred to a 4 C environment for up to 8 h. After euthanasia, the interscapular BAT and WAT were rapidly collected and frozen in liquid nitrogen. All animal experiments were approved by Institutional Animal Care and Use Committees of The Hamner Institutes for Health Sciences in accordance with National Institutes of Health Guidelines for the Care and Use of Laboratory Animals.
Chemicals and Plasmids
The plasmid pCMV4-NOR1 was generated by cloning the complete NOR-1 coding sequence into pCMV4-Flag vector (Stratagene, Palo Alto, CA). NBRE-Luc, Ucp1-Luc, and NBRE-Ucp1-Luc were constructed in pLuc-MCS vector (Stratagene). SB202190 and PD098059 were from Calbiochem (La Jolla, CA). Isobutylmethylxanthine, insulin, Iso, dexamethasone, and Fsk were from Sigma Chemical Co. (St. Louis, MO). Tris-glycine gels (4–20%) were from NOVEX (Invitrogen, Carlsbad, CA). A protease inhibitor cocktail (catalog item 11697498001) was purchased from Roche Applied Science (Indianapolis, IN).
Cell Culture and Transfections
HIB-1B brown preadipocytes were cultured in DMEM with 10% fetal bovine serum (FBS). At 80% confluency, they were differentiated into adipocytes by addition of rosiglitazone (1 μm) for 4–5 d. Transfections of HIB-1B cells were performed in six-well plates with 1 μg of either Ucp1-Luc or NBRE-Ucp1-Luc and 3 μl Lipofectamine 2000 (Invitrogen). Where indicated, during the final 6 h of the transfection, the cells were treated with Iso (1 μm).
The 3T3-L1 and 3T3-L1 CARΔ1 cells (13) were differentiated 1 d after confluence in DMEM containing 10% FBS with the addition of 0.5 mm isobutylmethylxanthine, 0.4 μg/ml dexamethasone, 5 μg/ml insulin, and 1 μm rosiglitazone to the medium. Three days later, cells received fresh DMEM plus 10% FBS medium without additives and replenished with same every 3 d. Infection of 3T3-L1 CARΔ1 (on d 3 of differentiation) was done with adenoviruses expressing either GFP or activation domain mutant (Nur77-M1) at a multiplicity of infection of 300, which routinely resulted in 50–60% of cell infection. Forty eight hours after infection, the cells were harvested for RNA analysis.
6Eb2 and 6Eb2 CARΔ1 cells were differentiated as described previously (26). 6Eb2 CARΔ1 cells were infected with adenoviruses as described above on d 5, and cells were harvested after 48 h as above.
COS-7 cells were maintained in DMEM with 10% FBS and transfected with NBRE-Luc (0.5 μg) and pCMV-NOR-1 (0.5 μg) and 3 μl Lipofectamine 2000 in six-well plates. Forty-eight hours after transfection, cells were collected for luciferase assays (Promega, Madison, WI). For the reconstitution of UCP1 promoter activity, Ucp1-Luc or NBRE-Ucp1-Luc (0.8 μg) was cotransfected with pCMV-NOR-1 (0.4 μg), pECMV-PGC-1α (0.4 μg), PPARγ (0.2 μg), RXRα (0.2 μg), and β-galactosidase (0.1 μg). Rosiglitazone (1 μm) was added to COS-7 cells at the initiation of the transfections. Cells were treated with Fsk (10 μm) during the last 6 h of transfection.
Subcutaneous human preadipocytes were from Zen-Bio Inc. (Research Triangle Park, NC). These were pooled samples from healthy, nondiabetic women [n = 6; average body mass index 26.98 (range 24.96–29.13) kg/m2; average age 43.7 (range 29–63) yr] and differentiated into adipocytes according to the supplier's protocol.
RNA Isolation and Analysis
Total RNA was extracted from cultured cells or tissues using Tri reagent (Sigma). RT-PCR was performed on an ABI Prism 7700 Sequence Detector (PerkinElmer, Norwalk, CT) using TaqMan probes or SYBR-Green reagent and specific primers (Integrated DNA Technologies, Coralville, IA) as indicated. Expression levels for all genes were normalized to the mean value of internal control GAPDH.
EMSA
Nuclear extracts were prepared from COS-7 cells that had been transfected with Flag-NOR-1 vector by TransFactor Extraction Kit from BD Biosciences (San Diego, CA). Double-stranded DNA probes were end-labeled with [γ32P]ATP and T4 polynucleotide kinase. The binding reactions (10 μl) contained 5 μg nuclear protein in a buffer composed of 25 mm HEPES (pH 7.9), 0.5 mm EDTA, 0.5 mm dithiothreitol, 1% Nonidet P-40, 5% glycerol, 50 mm NaCl, and 1μg poly (dΙ-dC). Reaction mixtures were incubated at room temperature for 15 min, and DNA-protein complexes were resolved on a 6% DNA retardation gel for 40 min at 180 V. Specificity of binding interactions was determined by addition of 1 μl anti-Flag antibody (Sigma) to the binding reaction mixtures 10 min before the addition of labeled oligo, followed by an additional 15 min of incubation before applying to the gel. Gels were dried and subjected to autoradiography.
Immunoblotting
Nuclear extract were prepared from Iso-treated differentiated cells as described above. Proteins were resolved by SDS-PAGE and transferred onto nitrocellulose membranes (Bio-Rad, Hercules, CA), which were then incubated at room temperature for 1 h in a blocking buffer composed of Tris-buffered saline containing 0.1% Tween 20 (TBS-T) and 4% BSA, followed by incubation for 1 h with the indicated antibodies in the blocking buffer. After being washed 3 times for 5 min each with TBS-T, the membrane was incubated with alkaline phosphatase-conjugated anti-sheep IgG (Sigma) antibodies followed by washing with TBS-T. Immunoreactive bands were visualized with enhanced chemical fluorescence substrate (GE Healthcare, Piscataway, NJ).
Chromatin Immunoprecipitation Assays
Chromatin immunoprecipitation assays were performed using a commercial kit (Upstate, Lake Placid, NY) according to manufacturers' protocol. Briefly, differentiated HIB-1B cells were treated with Iso (1 μm) for 2 h. For the cross-linking step, formaldehyde (37%, vol/vol) was added directly to the culture medium to achieve a final concentration of 1% and incubated for 10 min at 37 C. Cells were rinsed twice with cold PBS, harvested, and then lysed in 0.4 ml SDS buffer [1% SDS, 10 mm EDTA, 50 mm Tris (pH 8.1)]. The lysates were sonicated to shear genomic DNAs and were centrifuged at 16,000 × g for 10 min. The supernatants were diluted 10-fold with a buffer [24 ml 0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl (pH 8.1), 167 mm NaCl] in the presence of protease inhibitors (1 mm phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, and 1 μg/ml pepstatin A). An aliquot (75 μl) of a 50% slurry of salmon sperm DNA/protein A agarose was added, incubated for 30 min at 4 C, and then centrifuged briefly. This precleared supernatant was incubated with antibodies against NOR-1 or IgG overnight at 4 C with gentle mixing. Sixty microliters of the salmon sperm DNA/protein A agarose slurry were added and incubated for 1 h at 4 C to collect the antibody/histone/DNA complex by gentle and brief centrifugation. After reversion of cross-links, the presence of target DNA fragments was detected by PCR. Primers (forward CCTCCCTGCAAACTCAAAAG, and reverse, CAACTCTGCCCGTGTTTTCT) were used to amplify the UCP1 distal site.
Acknowledgments
We thank Zen-Bio, Inc. for human adipocyte cultures, Peter Tontonoz for the 3T3-L1 CARΔ1 cells and the Nur77-M1 adenovirus, J. Simon C. Arthur for the Nur77 and NOR-1 antisera, and Bruce Spiegelman for 6Eb2 and 6Eb2 CARΔ1 cells.
Footnotes
Disclosure Statement: The authors have nothing to disclose.
First Published Online January 31, 2008
Abbreviations: β-AR, β-Adrenergic receptor; BAT, brown adipose tissue; CRE4, cAMP-responsive element 4; FBS, fetal bovine serum; Fsk, forskolin; GFP, green fluorescent protein; Iso, isoproterenol; NBRE, nerve growth factor-responsive element; PGC-1α, PPARγ coactivator-1α; PPARγ, peroxisome proliferator-activated receptor-γ; RXRα, retinoid X receptor-α; TBS-T, Tris-buffered saline containing 0.1% Tween 20; UCP1, uncoupling protein-1; WAT, white adipose tissue.
References
- Morton GJ, Cummings DE, Baskin DG, Barsh GS, Schwartz MW 2006 Central nervous system control of food intake and body weight. Nature 443:289–295 [DOI] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J 2004 Brown adipose tissue: function and physiological significance. Physiol Rev 84:277–359 [DOI] [PubMed] [Google Scholar]
- Collins S, Cao W, Robidoux J 2004 Learning new tricks from old dogs: β-adrenergic receptors teach new lessons on firing up adipose tissue metabolism. Mol Endocrinol 18:2123–2131 [DOI] [PubMed] [Google Scholar]
- Kozak UC, Kopecky J, Teisinger J, Enerback S, Boyer B, Kozak LP 1994 An upstream enhancer regulating brown-fat-specific expression of the mitochondrial uncoupling protein gene. Mol Cell Biol 14:59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao LC, Zhang Z, Pei L, Saito T, Tontonoz P, Pilch PF 2007 Nur77 coordinately regulates expression of genes linked to glucose metabolism in skeletal muscle. Mol Endocrinol 21:2152–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L, Waki H, Vaitheesvaran B, Wilpitz DC, Kurland IJ, Tontonoz P 2006 NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat Med 12:1048–1055 [DOI] [PubMed] [Google Scholar]
- Maxwell MA, Muscat GE 2006 The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal 4:e002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell MA, Cleasby ME, Harding A, Stark A, Cooney GJ, Muscat GE 2005 Nur77 regulates lipolysis in skeletal muscle cells. Evidence for cross-talk between the β-adrenergic and an orphan nuclear hormone receptor pathway. J Biol Chem 280:12573–12584 [DOI] [PubMed] [Google Scholar]
- Fu M, Sun T, Bookout AL, Downes M, Yu RT, Evans RM, Mangelsdorf D 2005 A nuclear receptor atlas: 3T3-L1 adipogenesis. Mol Endocrinol 19:2437–2450 [DOI] [PubMed] [Google Scholar]
- Au WS, Payne VA, O'Rahilly S, Rochford JJ 2008 The NR4A family of orphan nuclear receptors are not required for adipogenesis. Int J Obes (Lond) 32:388–392 [DOI] [PubMed] [Google Scholar]
- Pei L, Castrillo A, Tontonoz P 2006 Regulation of macrophage inflammatory gene expression by the orphan nuclear receptor Nur77. Mol Endocrinol 20:786–794 [DOI] [PubMed] [Google Scholar]
- Kanzleiter T, Schneider T, Walter I, Bolze F, Eickhorst F, Heldmaier G, Klaus S, Klingenspor M 2005 Evidence for Nr4a1 as a cold-induced effector of brown fat thermogenesis. Physiol Genomics 24:37–44 [DOI] [PubMed] [Google Scholar]
- Orlicky DJ, DeGregori J, Schaack J 2001 Construction of stable coxsackievirus and adenovirus receptor-expressing 3T3-L1 cells. J Lipid Res 42:910–915 [PubMed] [Google Scholar]
- Sears IB, MacGinnitie MA, Kovacs LG, Graves RA 1996 Differentiation-dependent expression of the brown adipocyte uncoupling protein gene: regulation by peroxisome proliferator-activated receptor γ. Mol Cell Biol 16:3410–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Robidoux J, Puigserver P, Daniel KW, Medvedev AV, Bai X, Floering LM, Spiegelman BM, Collins S 2004 p38 MAP kinase is the central regulator of cAMP-dependent transcription of the brown fat uncoupling protein-1 gene. Mol Cell Biol 24:3057–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearen MA, Ryall JG, Maxwell MA, Ohkura N, Lynch GS, Muscat GE 2006 The orphan nuclear receptor, NOR-1, is a target of β-adrenergic signaling in skeletal muscle. Endocrinology 147:5217–5227 [DOI] [PubMed] [Google Scholar]
- Humphries A, Weller J, Klein D, Baler R, Carter DA 2004 NGFI-B (Nurr77/Nr4a1) orphan nuclear receptor in rat pinealocytes: circadian expression involves an adrenergic-cyclic AMP mechanism. J Neurochem 91:946–955 [DOI] [PubMed] [Google Scholar]
- Rius J, Martinez-Gonzalez J, Crespo J, Badimon L 2004 Involvement of neuron-derived orphan receptor-1 (NOR-1) in LDL-induced mitogenic stimulus in vascular smooth muscle cells: role of CREB. Arterioscler Thromb Vasc Biol 24:697–702 [DOI] [PubMed] [Google Scholar]
- Cassard-Doulcier A-M, Gelly C, Fox N, Schrementi J, Raimbault S, Klaus S, Forest C, Bouillaud F, Ricquier D 1993 Tissue-specific and β-adrenergic regulation of the mitochondrial uncoupling protein gene: control by cis-acting elements in the 5′-flanking region. Mol Endocrinol 7:497–506 [DOI] [PubMed] [Google Scholar]
- Debevec D, Christian M, Morganstein D, Seth A, Parker M, White R 2007 Receptor interacting protein 140 regulates expression of uncoupling protein 1 in adipocytes through specific peroxisome proliferator activated receptor isoforms and estrogen-related receptor-α. Mol Endocrinol 21:1581–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Sladek R, Carrier J, Bader JA, Richard D, Giguere V 2003 Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor α. Mol Cell Biol 23:7947–7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villena JA, Hock MB, Chang WY, Barcas JE, Giguere V, Kralli A 2007 Orphan nuclear receptor estrogen-related receptor α is essential for adaptive thermogenesis. Proc Natl Acad Sci USA 104:1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SL, Wesselschmidt RL, Linette GP, Kanagawa O, Russell JH, Milbrandt J 1995 Unimpaired thymic and peripheral T cell death in mice lacking the nuclear receptor NGFI-B (Nur77). Science 269:532–535 [DOI] [PubMed] [Google Scholar]
- Ponnio T, Burton Q, Pereira FA, Wu DK, Conneely OM 2002 The nuclear receptor Nor-1 is essential for proliferation of the semicircular canals of the mouse inner ear. Mol Cell Biol 22:935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullican SE, Zhang S, Konopleva M, Ruvolo V, Andreeff M, Milbrandt J, Conneely OM 2007 Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat Med 13:730–735 [DOI] [PubMed] [Google Scholar]
- Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM 2006 Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab 3:333–341 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang Y, Yehuda-Shnaidman E, Medvedev AV, Kumar N, Daniel KW, Robidoux J, Czech MP, Mangelsdorf DJ, Collins S 2008 Liver X receptor α is a transcriptional repressor of the uncoupling protein 1 gene and the brown fat phenotype. Mol Cell Biol 28:2187–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]