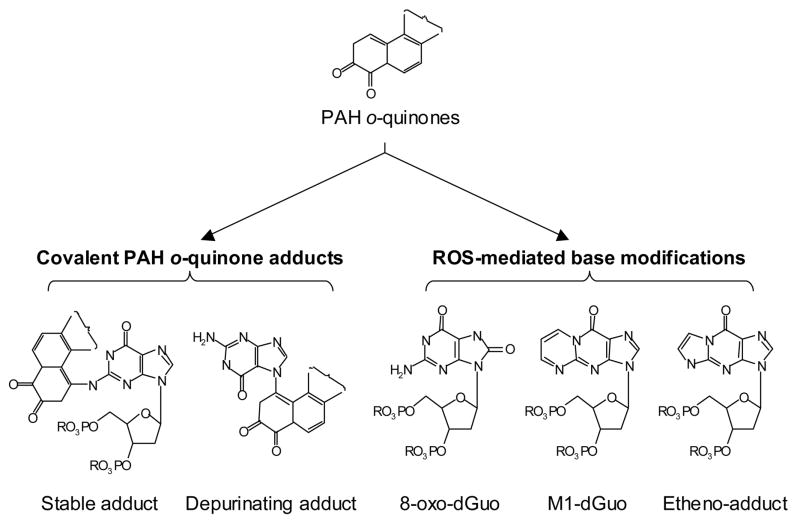
Abstract
Reactive and redox-active polycyclic aromatic hydrocarbon (PAH) o-quinones produced by Aldo-Keto Reductases (AKRs) have the potential to cause depurinating adducts leading to the formation of abasic sites and oxidative base lesions. The aldehyde reactive probe (ARP) was used to detect these lesions in calf thymus DNA treated with three PAH o-quinones (BP-7,8-dione, 7,12-DMBA-3,4-dione and BA-3,4-dione) in the absence and presence of redox-cycling conditions. In the absence of redox-cycling a modest amount of abasic sites were detected indicating the formation of a low level of covalent o-quinone depurinating adducts (> 3.2 × 106 dNs). In the presence of NADPH and CuCl2, the three PAH o-quinones increased the formation of abasic sites due to ROS derived lesions destabilizing the N-glycosidic bond. The predominant source of AP sites, however, was revealed by coupling the assay with human 8-oxoguanine glycosylase (hOGG1) treatment showing that 8-oxo-dGuo was the major lesion caused by PAH o-quinones. The levels of 8-oxo-dGuo formation were independently validated by HPLC-ECD analysis. Apyrimidinic sites were also revealed by coupling the assay with Escherichia coli (Endo III) treatment showing that oxidized pyrimidines were formed but to a lesser extent. Different mechanisms were responsible for the formation of the oxidative lesions depending on whether Cu(II) or Fe(III) was used in the redox-cycling conditions. In the presence of Cu(II)-mediated PAH o-quinone redox-cycling, catalase completely suppressed the formation of the lesions but mannitol and sodium benzoate were without effect. By contrast, sodium azide, which acts as •OH and 1O2 scavenger, inhibited the formation of all oxidative lesions, suggesting that the ROS responsible was 1O2. However, in the presence of Fe(III)-mediated PAH o-quinone redox-cycling the •OH radical scavengers and sodium azide consistently attenuated their formation, indicating that the ROS responsible was •OH.
Keywords: ARP assay, oxidized pyrimidines, singlet oxygen, hydroxyl radical, N-glycosidic bond cleavage
Introduction
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous environmental pollutants present in automobile exhaust, furnace gases and smoked foods (1,2) and they are also known tobacco carcinogens which are suspected causative agents in human lung cancer (3). PAHs are procarcinogens and require metabolic activation to electrophiles that react with DNA leading to mutation (4–6). One prominent group of PAH electrophiles are PAH o-quinones produced by human Aldo-Keto Reductases (AKRs) (6,7).
PAH o-quinones are reactive Michael-acceptors and produce significant amounts of reactive oxygen species (ROS) (8,9) and can give rise to covalent DNA adducts as well as oxidative DNA lesions (Scheme 1). For example, BP-7,8-dione produces N2 or N6 stable adducts with 2′-dGuo and 2′-dAdo by Michael addition which can hydrate and cyclize, respectively (10,11). In addition, BP-7,8-dione reacts with guanine to form N7-depurinating guanine adducts in vitro (12). PAH o-quinones can also generate ROS to form 8-oxo-dGuo in vitro (13–15). Park et al (15) have recently shown that in the presence of Cu(II) and cellular reducing equivalent (e.g. NADPH), nanomolar concentrations of PAH o-quinones produced significant amounts of 8-oxo-dGuo and that the oxidant responsible was singlet oxygen (1O2). The ROS produced by PAH o-quinones may also oxidize deoxyribose leading to the formation of a base propenal which can react to form M1-dGuo adducts (16). Additionally, the ROS produced may attack PUFA leading to the formation of lipid peroxides which decompose to electrophiles e.g. 4-hydroxy-2-nonenal and 4-oxo-2-nonenal to form etheno-adducts and heptano-etheno-adducts, respectively (17,18). Which of these adducts dominate in DNA exposed to PAH o-quinones in vitro and in vivo remains to be clarified.
Scheme 1.
Spectrum of DNA modifications that PAH o-quinones can cause in DNA
Although the formation of covalent PAH-DNA adducts is a known mechanism that contributes to PAH carcinogenesis, the formation of PAH-mediated oxidative DNA damage and its link to carcinogenesis has been less well studied. PAH-exposed sites can produce significant amounts of oxidative DNA damage in vitro and in vivo (19–21). Recently, areca-quid chewing in combination with smoking was observed to increase the incidence of human oral squamous cell carcinoma (OSCC) (22). Exposure of human buccal cells to hydroxychavicol (a major ingredient of areca-quid) induced AKR1C1 and subsequent treatment with benzo[a]pyrene caused a decrease in bulky stable adducts with a concomitant increase in 8-oxo-dGuo. In addition, a significant increase in 1N-ε-ethenoguanine adducts in the urine of smokers has been detected by LC/MS (23). Thus, both direct and indirect evidence suggests that PAH-mediated ROS may contribute to PAH carcinogenesis.
The AKR pathway diverts PAH trans-dihydrodiols to PAH o-quinones which may be responsible for these PAH-induced oxidative lesions. Previously, BP-7,8-dione was found to be a weak acting mutagen of the tumor suppressor gene p53. However, BP-7,8-dione became a potent mutagen in the nanomolar range when redox-cycling conditions were included and the dominant mutatons observed in p53 were G to T transversions which were abolished by ROS scavengers suggesting that they were oxidatively generated (24). The mutational pattern seen in p53 by BP-7,8-dione was reminiscent of that seen with lung cancer patients (24).
In this paper, three PAH o-quinones including BP-7,8-dione, benz[a]anthracene-3,4-dione (BA-3,4-dione) and 7,12-dimethylbenz[a]anthracene-3,4-dione (7,12-DMBA-3,4-dione) were tested for their ability to produce AP sites. We show that in the absence of redox-cycling PAH o-quinones produced a modest number of heat-labile depurinating adducts. In the presence of NADPH and Cu(II) three PAH o-quinones produced significant amounts of the AP sites, 8-oxo-dGuo and oxidized pyrimidines. This represents the first report of PAH o-quinone generated ROS to cause abasic sites and form oxidized pyrimidines. The mechanism responsible for formation of three types of oxidative DNA damage generated by PAH o-quinone was different depending on whether Cu(II) or Fe(III) was added to the redox-cycling conditions. Those lesions caused in the presence of Cu(II)-mediated PAH o-quinone redox-cycling were produced by singlet oxygen whereas those lesions caused in the presence of Fe(III)-mediated redox-cycling were produced by hydroxyl radical.
Materials and methods
Caution
all PAH o-quinones are potentially hazardous and should be handled in accordance with NIH Guidelines for the Laboratory Use of Chemical Carcinogens.
Chemicals and Reagents
BP-7,8-dione, 7,12-DMBA-3,4-dione, and BA-3,4-dione were synthesized according to published methods (25,26). All compounds were analyzed for purity and identity by LC/MS before use. ARP, standard ARP-DNA conjugate and DNA binding solution were purchased from Dojindo Molecular Technologies Inc. (Gaithersburg, MA). Streptavidin-HRP conjugate and 3,3′,5,5′-tetramethylbenzidine (TMB) were obtained from Amersham Biosciences Co. (Piscataway, NJ). Human 8-oxoguanine glycosylase (hOGG1) and Escherichia coli endonuclease III (Endo III) were acquired from New England BioLabs Inc. (Beverly, MA). Standard 8-oxo-dGuo was obtained from Cayman Chemical Co. (Ann Arbor, MI). Calf thymus DNA, DNase I (type II), alkaline phosphatase (type III from Escherichia coil), catalase (17,600 units/mg from bovine liver), deferoxamine mesylate (desferal), sodium azide, cupric chloride, ferric chloride, hydrogen peroxide and methoxyamine hydrochloride were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO). β–Nicotinamide adenine dinucleotide phosphate, reduced form (NADPH), was obtained from Boehringer Manheim Biochemicals (Indianapolis, IN). All other chemicals were of the highest grade available and all solvents for HPLC-ECD analysis were HPLC grade.
Preparation of methoxyamine-treated calf thymus DNA
A dried calf thymus DNA pellet was dissolved in 10 mM potassium phosphate buffer (pH 6.5) containing 0.1 mM desferal overnight at 4 °C. The concentration of the hydrated DNA was determined by measuring the absorbance at 260 nm and was adjusted to 0.64 mg/mL. The DNA solution was mixed with an equal volume of 0.2 M methoxyamine hydrochloride (pH 8.0) and was incubated for 2 h at 37 °C to block the endogenous AP sites present in commercial calf thymus DNA. The DNA solution-exposed to methoxyamine hydrochloride was then divided into 80 μg aliquots and was precipitated by adding 5 M NaCl to yield a final concentration of 1 M and two volumes of ice-cold ethanol. To remove excess unbound methoxyamine, the methoxyamine-treated DNA pellet was washed with 70% ethanol 3 times and the pellets were kept at −20 °C until used.
Depurination of heat-labile DNA adducts
The methoxyamine-treated calf thymus DNA pellet was dissolved in the 10 mM potassium phosphate buffer (pH 6.5) containing 0.5–10 μM PAH o-quinone and 8% DMSO and was incubated for 3 h at 37 °C. Excess PAH o-quinone was completely removed by chloroform-isopropyl alcohol (24:1 v/v) extraction and the DNA was precipitated using NaCl and 100% ethanol and subsequently washed twice with 70% ethanol to remove excess NaCl salt. The DNA pellet was re-suspended in Chelex-treated water (< pH 7.0) and was incubated for 2 h at 70 °C. The heat-treated DNA was rapidly cooled and left on ice for 1 h. The DNA was then precipitated and washed with ethanol. The samples were also kept at −20 °C for ARP tagging.
Formation of AP sites in calf thymus DNA in the presence of PAH o-quinones
The methoxyamine-treated calf thymus DNA was treated with PAH o-quinones as follows. Briefly, the DNA pellet was dissolved in 250 μL of potassium phosphate buffer (pH 6.5) containing 0.5–10 μM PAH o-quinone, and 180 μM NADPH and plus 8% DMSO as cosolvent. Other incubations contained either 10 μM CuCl2 or 10 μM FeCl3. Samples were incubated for 3 h at 37 °C. The effects of various scavenging agents on the formation of AP sites in the PAH o-quinone-treated calf thymus DNA were also measured. After incubation, the samples were extracted twice with an equal volume of chloroform and isopropyl alcohol (24:1 v/v) to remove PAH o-quinones and proteins. The DNA was then precipitated and washed twice with ethanol. The resultant DNA samples were kept at −20 °C for ARP tagging.
Enzymatic treatments with hOGG1 and Endo III
To measure the levels of 8-oxo-dGuo and oxidized pyrimidines generated by PAH o-quinones under Cu(II)- and Fe(III)-mediated redox-cycling conditions, the PAH o-quinone-exposed calf thymus DNA was first allowed to react with methoxyamine to eliminate the additional AP sites generated during the quinone treatment. Then, hOGG1 and Endo III were used for removing the oxidatively modified bases to reveal AP sites. Aliquots of DNA (1.2 μg) were treated with 0.6 unit of hOGG1 in 10 μL of enzyme reaction buffer containing 10 mM Tris-HCl, 50 mM NaCl, 10 mM MgCl2 and 1 mM dithiothreitol (pH 8.0) or aliquots of DNA (1.2 μg) were treated with 1.2 unit of Endo III in 10 μL of enzyme reaction buffer containing 20 mM Tris-HCl, 1 mM dithiothreitol and 1 mM EDTA (pH 7.9) and the samples were then incubated for 1.5 h at 37 °C. At the end of the incubation, an equal volume of ARP was added to the enzyme-DNA mixture to label the AP sites produced by enzymatic base excision. As a negative control, incubations were also performed with the same buffer containing DNA or hOGG1 and endonuclease III alone. The resultant AP sites from both enzymatic treatments were quantified using the ARP microtiter plate assay.
ARP tagging
AP sites were tagged with biotinylated ARP according to the manufacture’s protocol (Dogindo Molecular Technologies, Inc.) with minor modifications. A DNA sample (1.2 μg) was incubated with 20 μL of 5 mM ARP reagent in 10 mM potassium phosphate buffer (pH 6.5) for 1 h at 37 °C. At that time, 400 μL of 10 mM Tris-HCl buffer (pH 8.0) containing 1 mM EDTA was added to the ARP-DNA mixture and the solution was transferred into a filtration tube and centrifuged for 15 min at 3,500 × g to purifiy the ARP-tagged DNA. The ARP-tagged DNA retained in the tube was solubilized with 400 μL of the Tris-EDTA buffer by pipetting and was further washed by centrifugation. The washing was repeated 3 times. The resultant DNA was then dissolved in an equal volume of Tris-EDTA buffer and the DNA concentration in the samples was measured at 260 nm.
Measurement of AP sites using a microtiter plate assay
An aliquot of the ARP-tagged DNA (30 μL ) was diluted with 150 μL of the Tris-EDTA buffer and 60 μL of the diluted ARP-tagged DNA was added to each well of a 96-well microtiter plate. The DNA binding solution (100 μL) was added to each well and the plate was incubated in the dark at room temperature overnight. The DNA binding solution was then discarded and the plate was washed 6 times with phosphate buffered saline (pH 7.4) containing 0.8% saline and 0.1% tween 20 (PBST). An aliquot of streptavidin-HRP solution (1 in 2000 dilution) in PBST (150 μL) was added to each well and the plate was incubated in the dark for 1 h at 37 °C to permit formation of the biotin-streptavidin complex. Excess streptavidin-HRP solution was discarded and the plate was thoroughly washed 6 times with PBST. An aliquot of HRP substrate, 3,3′,5,5′-tetramethylbenzidine (TMB) was added to each well and incubated for 1 h at 37 °C for color development. At that time, an aliquot (60 μL) from each well was transferred to a well of a new plate, and mixed with 60 μL of 1 M sulfuric acid and the absorbance at 450 nm was read using an ELISA reader (EL808, BioTek Instruments, Inc). Each assay was performed in triplicate and all experiments were repeated at least 3 times.
The number of AP sites in the treatment group was quantified by using a standard ARP-tagged DNA conjugate (Dojindo Molecular Technologies, Inc.) which has a known amount of AP sites that range from 0 per 40 per 1 × 105 base pairs of calf thymus DNA. The ARP-DNA standard curve was replicated for each sample analysis.
Detection of 8-oxo-dGuo by HPLC-ECD
To validate the ARP microtiter plate assay for the detection of 8-oxo-dGuo estimated values were compared with those obtained using a fully validated HPLC-ECD protocol. The quantitative digestion of calf thymus DNA was conducted according to our previous protocol (15). Briefly, the sample DNA was dissolved in 200 μL of 10 mM Chelex-treated Tris-HCl buffer (pH 7.4) containing 100 mM MgCl2 and 0.1 M desferal was added to yield a final concentration of 0.1 mM to prevent adventitious formation of 8-oxo-dGuo during DNA hydrolysis. The sample was digested with 1 unit of DNase I for 1.5 h at 37 °C. After the pH was adjusted to 9.0 with 0.2 M glycine-acetate buffer (pH 10.0), the mixture was digested with 0.025 unit of phosphodiesterase I for 1.5 h. The pH was then adjusted to 8.6 with 50 mM Tris-HCl buffer (pH 7.4) containing 50 mM MgCl2 and the mixture was further digested with 0.1 unit of alkaline phosphatase for 1.5 h. Finally, the DNA digest was adjusted again to pH 5.5 with 0.5 M acetic acid and stored at −20 °C for HPLC-ECD analysis.
The amounts of the 8-oxo-dGuo in the DNA digests was measured by HPLC-ECD using a Coularray detector (ESA Inc., Chelmsford, MA). The level of 8-oxo-dGuo detected was expressed as 8-oxo-dGuo per 105 dGuo as previously reported (15). The four channel electrochemical detector had potential settings of 200 – 300 to detect 8-oxo-dGuo and 700 – 800 for the detection of dGuo. A mobile phase of 100 mM sodium acetate (pH 5.2) containing 5% methanol (v/v) was used for developing the YMC basic S column (3 μm; 4.6 mm × 150 mm).
Statistical analysis
The Wilcoxon rank-sum and Kruskal-Wallis non-parametric tests were used to compare the levels of depurinating adducts, AP sites, oxidized pyrimidines and 8-oxo-dGuo formed under various experimental conditions. Linear regression analysis was used to determine whether there was a concentration-dependency on lesion formation for each quinone and whether there was a dependency on lesion formation with ROS scavengers; in most cases the regression models also included interaction terms to test whether effects were constant across various conditions. All analyses were performed using SAS 9.1.2. Effects that exhibited p-values less than 0.05 were considered statistically significant.
Results
Formation of heat-labile depurinating DNA adducts by PAH o-quinones
Three PAH o-quinones; BP-7,8-dione (a bay region quinone), BA-3,4-dione and 7,12-DMBA-3,4-dione (nonmethylated and methylated bay region quinones from the same structural series) were examined for their ability to produce abasic sites in the absence of redox-cycling. Previously, McCoull et al (12) reported that heat-labile N7-guanine depurinating adducts were detected in calf thymus DNA treated with naphthalene-1,2-dione, phenanthrene-1,2-dione and BP-7,8-dione using LC/ESI/MS, confirming that PAH o-quinone can produce the unstable DNA adducts in DNA. However, the levels were never quantified. We now show that the unstable DNA adducts produced by PAH o-quinones are removed by the heat-mediated depurination and the resulting AP sites can be quantitatively measured by the ARP assay (27,28) as heat-labile DNA adducts.
In the absence of redox-cycling, all three PAH o-quinones modestly increased the level of the unstable adducts in a concentration-dependent manner (Figure 1). The basal level of AP sites observed in methoxyamine-treated calf thymus DNA obtained was about 5 AP sites per 106 dNs (Figure 1). The addition of heat treatment (70 °C) raised the basal level to 18 AP sites per 106 dNs. At the concentration of 10 μM PAH o-quinones, the level of unstable DNA adducts increased to 25.1 heat-labile DNA adducts per 106 dNs for BP-7,8-dione, 24.2 adducts per 106 dNs for 7,12-DMBA-3,4-dione and 21.1 adducts per 106 dNs for BA-3,4-dione, respectively.
Figure 1.
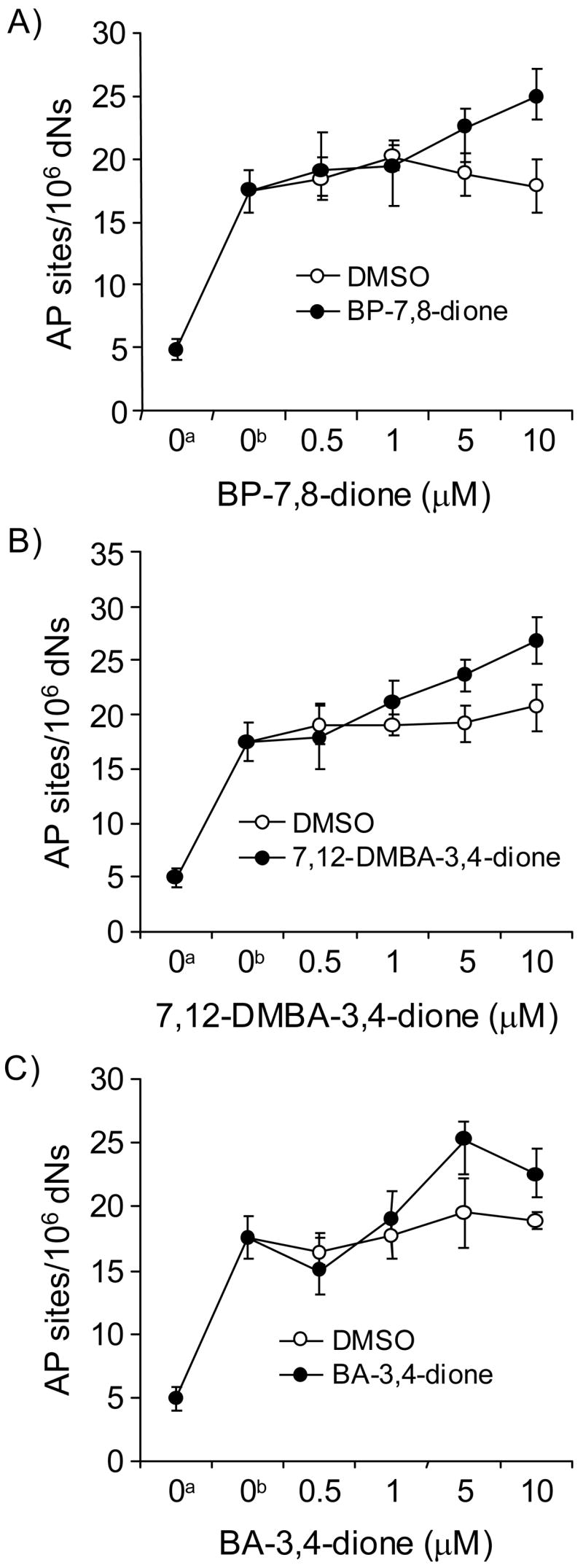
Detection of AP sites in PAH o-quinone-treated calf thymus DNA produced from heat depurination and depyrimidination for 2 h at 70 °C. (A) BP-7,8-dione, (B) 7,12-DMBA-3,4-dione and (C) BA-3,4-dione. See the Materials and methods for details. a, Basal level of AP sites in methoxyamine-treated calf thymus DNA; b, Heat-treated methoxyamine-treated calf thymus DNA (70 °C). A Wilcoxon two-sample exact test shows a marginally significant effect of temperature, i.e., more adducts were generated at 70 °C than 4 °C (p = 0.05). With the temperature set at 70 °C, Kruskal-Wallis tests show that BP-7,8-dione and 7,12-DMBA-3,4-dione had a significant effect on the quantity of adducts generated (p = 0.0084 and 0.0112, respectively) while BA-3,4-dione had no effect (p = 0.1304). Also, with the temperature set at 70 °C, linear regression analysis showed that the concentration of two quinones (BP-7,8-dione and 7,12-DMBA-3,4-dione) had a highly significant positive effect on the amount of depurinating adducts detected (p <0.0001 for both quinones; a positive effect indicates that the amount of the adducts increases as the concentration of each quinone increases).
Formation of AP sites in calf thymus DNA treated with PAH o-quinones under redox-cycling conditions
The formation of AP sites was detected in calf thymus DNA treated with PAH o-quinones, NADPH and Cu(II) using the ARP assay. A concentration dependence of formation of AP sites was observed with all three PAH o-quinones (Figure 2). At 5 – 10 μM PAH o-quinone, the amounts AP sites increased to 100 ~ 300 AP sites per 106 dNs. The rank order of AP site formation generated in calf thymus DNA was BA-3,4-dione > 7,12-DMBA-3,4-dione > BP-7,8-dione. This rank order shows a close relationship with the ability of each PAH o-quinone to consume NADPH and molecular oxygen as described before (15). The addition of sodium azide to the mixture abolished the AP site formation during PAH o-quinone redox-cycling.
Figure 2.
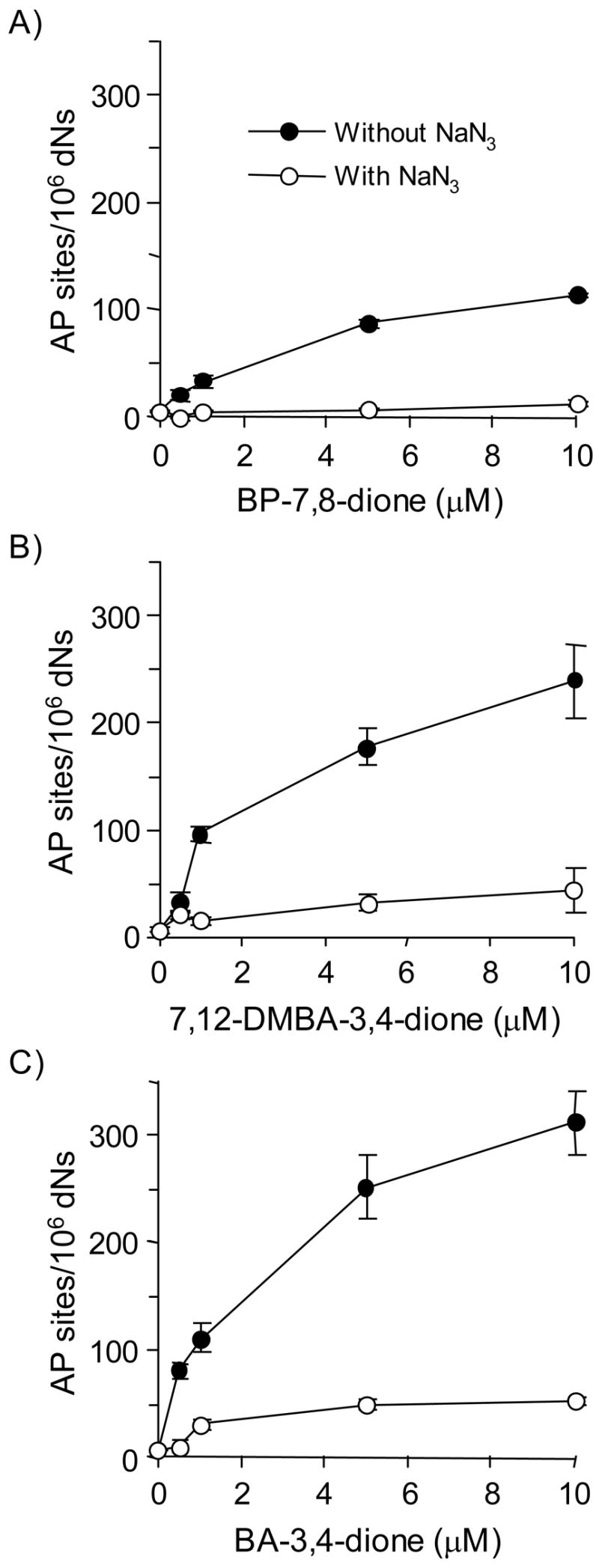
Detection of directly formed AP sites in PAH o-quinone-treated calf thymus DNA under redox-cycling conditions. (A) BP-7,8-dione, (B) 7,12-DMBA-3,4-dione and (C) BA-3,4-dione. See the Materials and methods for details. Kruskal-Wallis tests show that all three quinones had a significant effect on the AP sites generated (p = 0.0094, 0.0367 and 0.0093 for BP-7,8-dione, BA-3,4-dione and 7,12-DMBA-3,4-dione, respectively). Linear regression analysis showed that the concentration of all three quinones had a significant effect on the amount of AP sites (p <0.0001 for all three quinones). In each case, sodium azide blocked the formation of AP sites with p value < 0.0001 at all concentrations of PAH o-quinone tested.
Detection of 8-oxo-dGuo lesions by the coupled ARP-hOGG1 assay
Previously, we have shown that submicromolar amounts of PAH o-quinones plus Cu(II) and NADPH produced significant levels of 8-oxo-dGuo (15). We next quantified these lesions by coupling the ARP assay to the BER enzyme hOGG1. In this assay methoxyamine-treated calf thymus DNA was incubated with PAH o-quinones under redox-cycling conditions. Additional AP sites generated were protected with methoxyamine and the DNA sample was the treated with hOGG1 to remove 8-oxo-guanine. The abasic sites revealed by this treatment were then quantified with the ARP.
The formation of 8-oxo-dGuo by three PAH o-quinones was again concentration-dependent and the amounts of lesions formed at 5 μM PAH o-quinone was > 500 AP sites per 106 dNs. The formation of these lesions was abolished by sodium azide (Figure 3).
Figure 3.
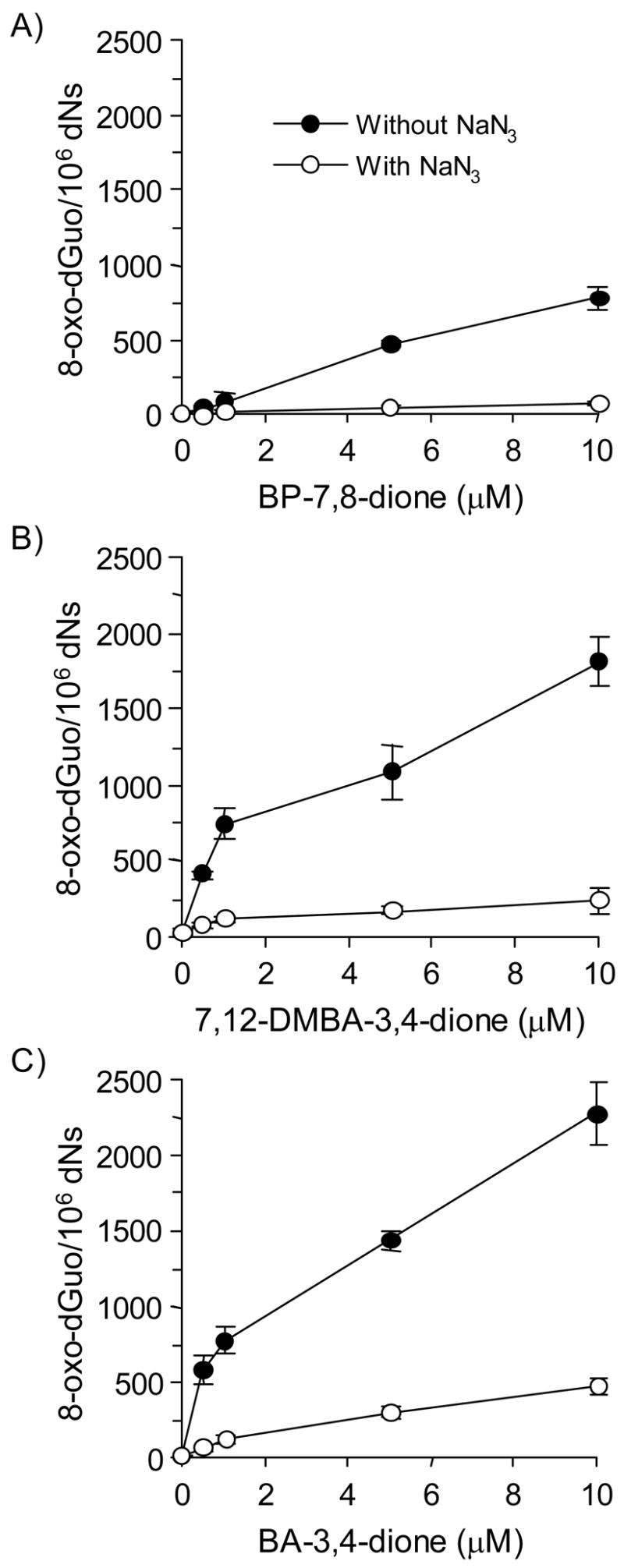
Detection of AP sites in calf thymus DNA treated with PAH o-quinones in the presence of NADPH and CuCl2 revealed by enzymatic base excision by hOGG1. (A) BP-7,8-dione, (B) 7,12-DMBA-3,4-dione and (C) BA-3,4-dione. See the Materials and methods for details. Kruskal-Wallis tests showed that all three quinones had a significant effect on 8-oxo-dGuo generated (p = 0.0304, 0.0094 and 0.0094 for BP-7,8-dione, BA-3,4-dione and 7,12-DMBA-3,4-dione, respectively). Linear regression analysis showed that the concentration of all three quinones had a significant effect on the amount of 8-oxo-dGuo detected (p <0.0001 for all three quinones). In each case, sodium azide blocked the formation of 8-oxo-dGuo with p < 0.0001 at all concentrations of PAH o-quinone tested.
The detection of 8-oxo-dGuo by the coupled ARP-hOGG1 assay was validated using an HPLC-ECD method to analyze the same DNA samples. In these experiments methoxyamine-treated calf thymus DNA was treated with PAH o-quinone in the presence of NADPH and Cu(II) and divided into two portions. One portion was used for the ARP assay coupled to hOGG1 and other portion was used for HPLC-ECD for the detection of 8-oxo-dGuo. The amounts of 8-oxo-dGuo detected with the ARP assay coupled to hOGG1 were in good agreement with those detected by HPLC-ECD (Figure 4). This shows that the protocol for the coupled ARP-hOGG1 assay provides a good estimate of 8-oxo-dGuo present in calf thymus DNA treated with PAH o-quinone and can be used to compare the abundance of 8-oxo-dGuo with other oxidative lesions.
Figure 4.
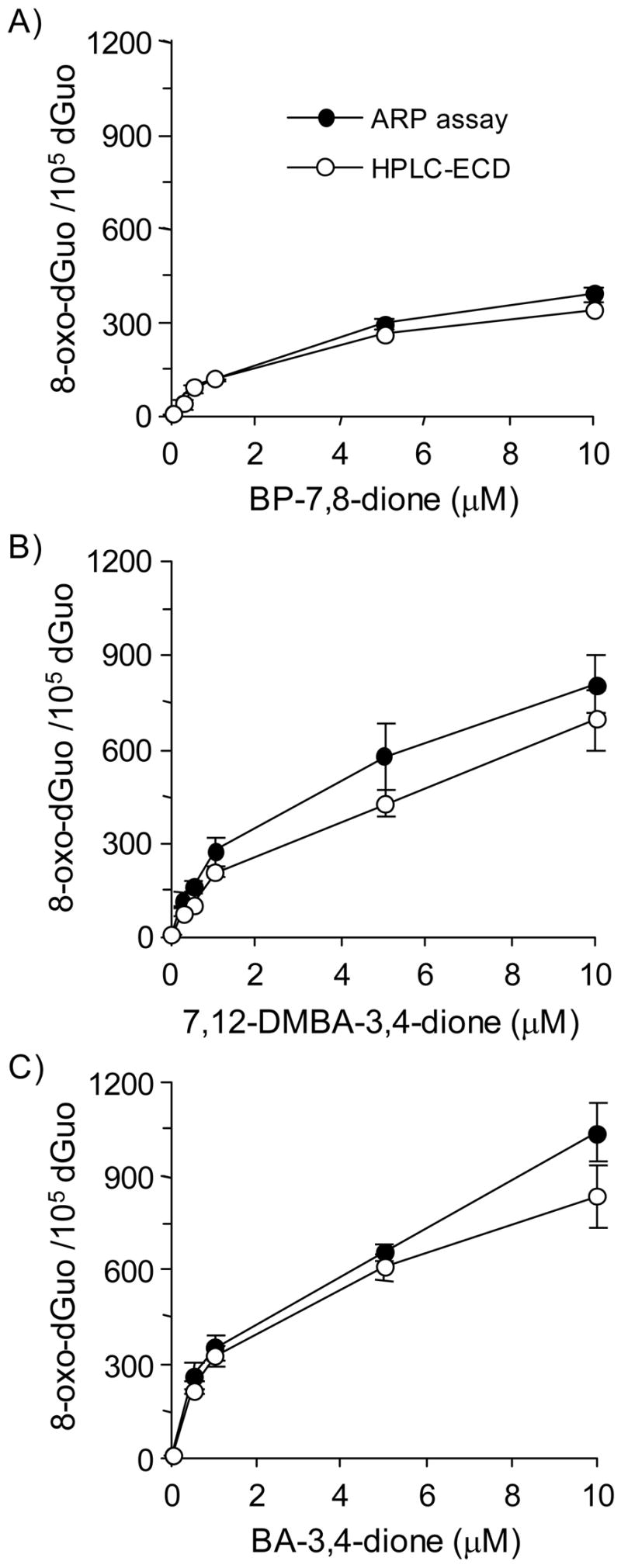
Relationship between ARP and HPLC-ECD methods for the detection of 8-oxo-dGuo. (A) BP-7,8-dione, (B) 7,12-DMBA-3,4-dione and (C) BA-3,4-dione. See the Materials and methods for details.
Formation of oxidized pyrimidine lesions detected by coupled ARP-endonuclease III assay
We next quantified oxidized pyrimidine lesions by coupling the ARP assay to the BER enzyme endonuclease III. In this assay, methoxyamine-treated calf thymus DNA was incubated with PAH o-quinones under redox-cycling conditions. Additional AP sites generated were protected with methoxyamine and the DNA sample was then treated with Endo III to remove oxidized pyrimidines. The abasic sites revealed by this treatment were then quantified with ARP.
The formation of oxidized pyrimidine lesions were again concentration-dependent with all three PAH o-quinones tested. At the concentration of 5 – 10 μM PAH o-quinone the lesions ranged from 100 ~ 300 per 106 dNs. The formation of their lesions was abolished by sodium azide (Figure 5).
Figure 5.
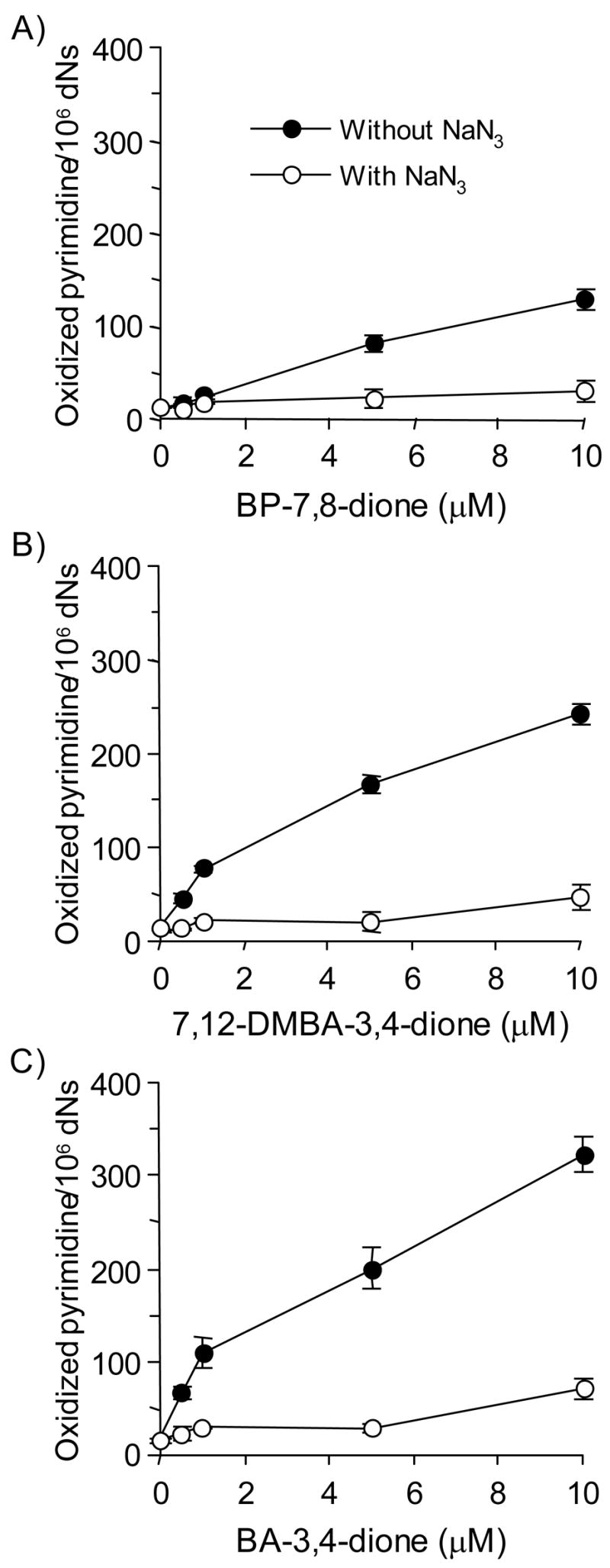
Detection of AP sites in calf thymus DNA treated with PAH o-quinones in the presence of NADPH and CuCl2 revealed by enzymatic base excision by Endo III. (A) BP-7,8-dione, (B) 7,12-DMBA-3,4-dione and (C) BA-3,4-dione. See the Materials and methods for details. Kruskal-Wallis tests showed that all three quinones had a significant effect on the amount of oxidized pyrimidines formed (p = 0.0299, 0.0094 and 0.0094 for BP-7,8-dione, BA-3,4-dione and 7,12-DMBA-3,4-dione, respectively). In each case, sodium azide blocked the formation of oxidized pyrimidines with p < 0.0001 at all concentrations of PAH o-quinone tested.
Relative abundance of aldehydic sites in calf thymus DNA treated with PAH o-quinones
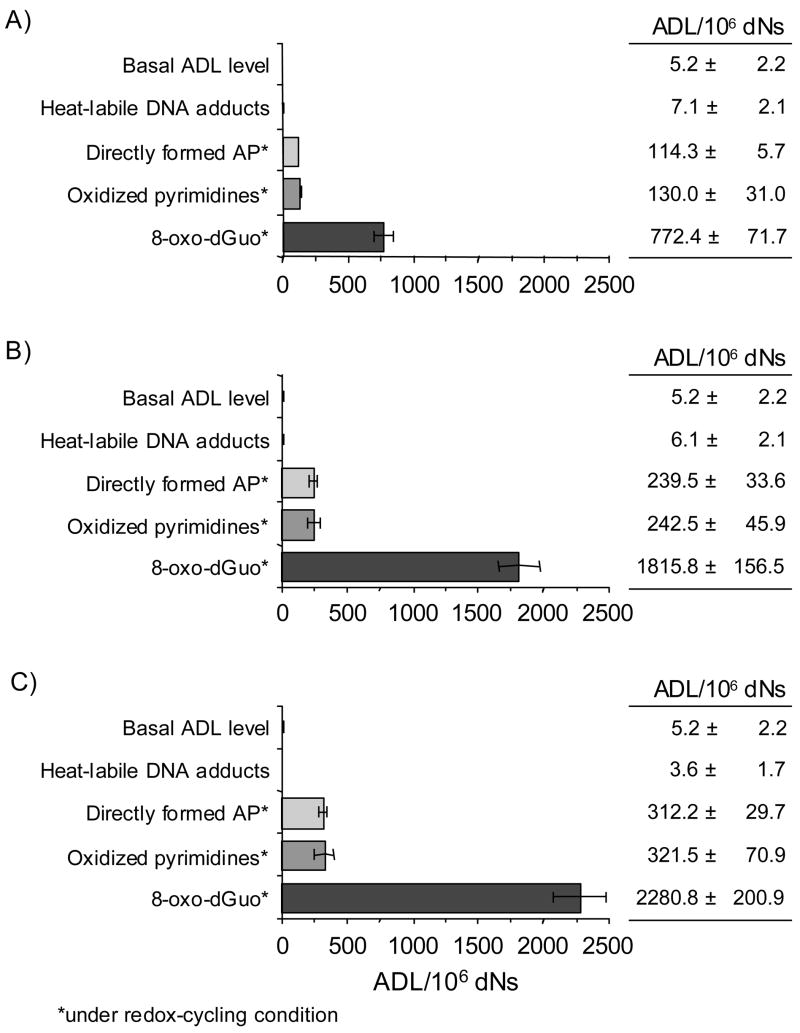
For each of the three quinones tested the relative abundance of heat-labile DNA adducts (depurination/depyrimidination), AP sites detected from ROS attack, oxidized pyrimidines and 8-oxo-dGuo were found to be the very different (Figure 6). The rank order of AP sites in PAH o-quinone treated DNA was found to be 8-oxo-dGuo > oxidized pyrimidines = directly formed AP sites ≫ heat-labile DNA adducts. These data show that the major lesions generated by PAH o-quinone are oxidative lesions caused by ROS and are not the PAH o-quinone N7-guanine depurinating adducts.
Figure 6.
PAH o-quinones can produce 100 times higher 8-oxo-dGuo than the heat-labile unstable DNA depurinating adducts in vitro. The experiments were conducted with 10 μM concentration of three PAH o-quinones in the presence or absence of NADPH and Cu(II). (A) BP-7,8-dione, (B) 7,12-DMBA-3,4-dione, (C) BA-3,4-dione. AP sites formed during redox-cycling were significantly different from the basal level of AP sites seen for all three quinones (p = 0.0101, 0.0297 and 0.0101 for BP-7,8-dione, BA-3,4-dione and 7,12-DMBA-3,4-dione, respectively). Oxidized pyrimidines generated during redox-cycling were significantly different from the basal level of AP sites observed with all three quinones (p = 0.0101 for all three quinones). 8-oxo-dGuo lesions formed during redox-cycling was significantly different from the basal level of AP sites seen for all three quinones (p = 0.0101 for all three quinones).
Comparison of the formation of AP sites, oxidized pyrimidines and 8-oxo-dGuo under different BP-7,8-dione redox-cycling conditions
The formation of AP sites, oxidized pyrimidines and 8-oxo-dGuo were measured using redox-cycling conditions in which either Cu(II) or Fe(III) were used as the transition metal (Figure 7). The formation of the three types of oxidative lesions in calf thymus DNA increased with BP-7,8-dione in a concentration-dependent manner using both metal ions. When BP-7,8-dione was used at its highest concentration (10 μM) in the presence of Cu(II)-mediated redox-cycling, the levels of the lesions generated were 164.0 AP sites per 106 dNs, 118 oxidized pyrimidines per 106 dNs and 1129.0 8-oxo-dGuo per 106 dNs, respectively. The efficiency of Fe(III)-mediated BP-7,8-dione redox-cycling to cause these lesions in calf thymus DNA was much lower than that of the Cu(II)-mediated redox-cycling. The frequency of the lesions were 48.7 AP sites per 106 dNs, 41.2 oxidized pyrimidines per 106 dNs and 386.0 8-oxo-dGuo per 106 dNs, respectively.
Figure 7.
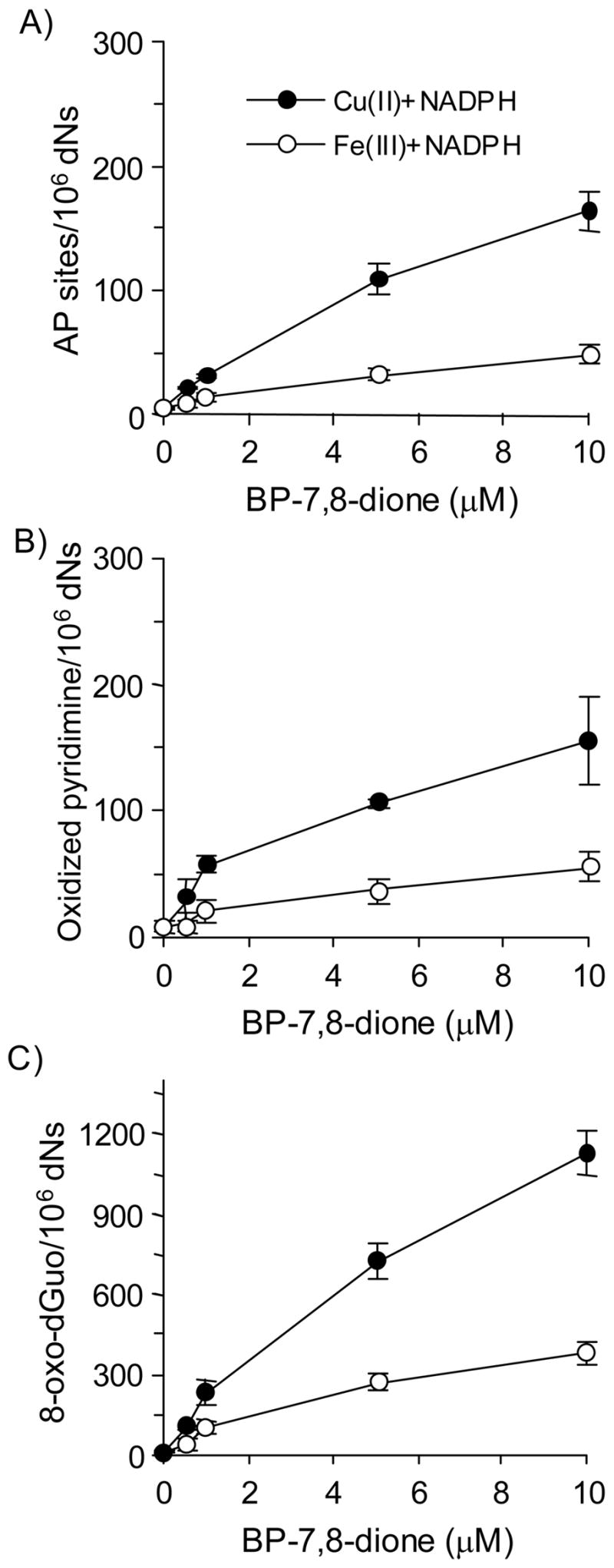
Detection of AP sites, oxidized pyrimidines and 8-oxo-dGuo in calf thymus DNA treated with BP-7,8-dione under Cu(II)- and Fe(III)-mediated redox-cycling conditions. (A) Directly formed AP sites, (B) Oxidized pyrimidines (by Endo III-coupled assay) and (C) 8-oxo-dGuo (by hOGG1-coupled assay). See the Materials and methods for details. For both systems, the difference in the amount of three lesions was concentration dependent. For Cu(II) plus NADPH system, the p-value for the interaction between lesions and the linear component of quinone concentration was <0.0001. For Fe(III) plus NADPH system, the p-value for the interaction between lesions and the linear component of quinone concentration was <0.0001.
Experiments were repeated using hydrogen peroxide in the presence of Cu(II) and Fe(III) without BP-7,8-dione (Figure 8). The levels of AP sites, oxidized pyrimidines and 8-oxo-dGuo by hydrogen peroxide detected increased linearly with the concentration of hydrogen peroxide (0 – 400 μM). Interestingly, 100 μM H2O2 produced a level of lesions equal to that seen with 5 μM PAH o-quinone. Similar to BP-7,8-dione, the rank order of three oxidative lesions caused by hydrogen peroxide was 8-oxo-dGuo > oxidized pyrimidines = AP sites. These results suggest that the mechanism to DNA lesions in calf thymus DNA is the same irrespective of whether a PAH o-quinone or H2O2 is added with the transition metal.
Figure 8.
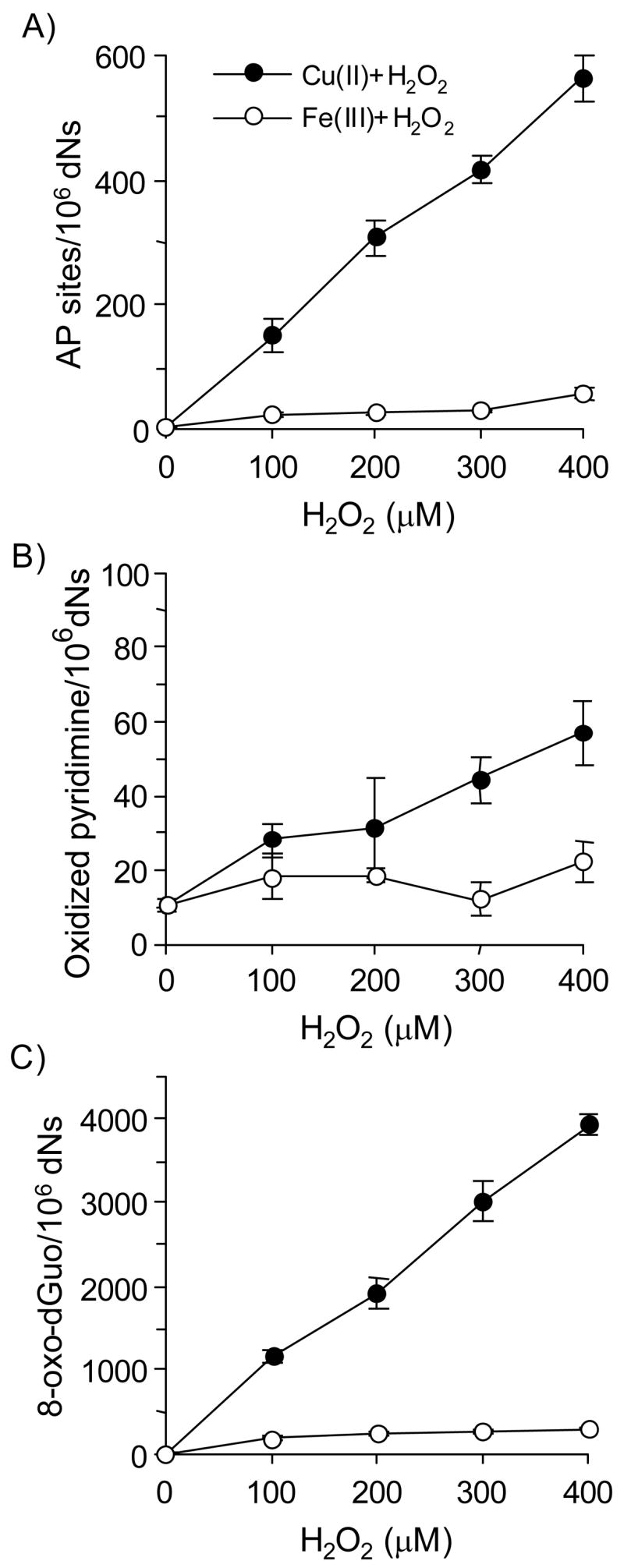
Detection of AP sites, oxidized pyrimidines and 8-oxo-dGuo in calf thymus DNA treated with hydrogen peroxide under Cu(II)- and Fe(III)-mediated redox-cycling conditions. (A) Directly formed AP sites, (B) Oxidized pyrimidines (by Endo III-coupled assay) and (C) 8-oxo-dGuo (by hOGG1-coupled assay). See the Materials and methods for details. For both redox-cycling conditions, the difference in the amount of all three lesions was significant; 8-oxo-dGuo gave the most lesions, while oxidized pyrimidines gave the fewest lesions. However, the differences were dependent upon hydrogen peroxide concentration. The p-value for the linear interaction between lesions and hydrogen peroxide concentration was <0.0001 for both Cu(II) and Fe(III) systems.
Effects of ROS scavengers on the formation of AP sites, oxidized pyrimidines and 8-oxo-dGuo
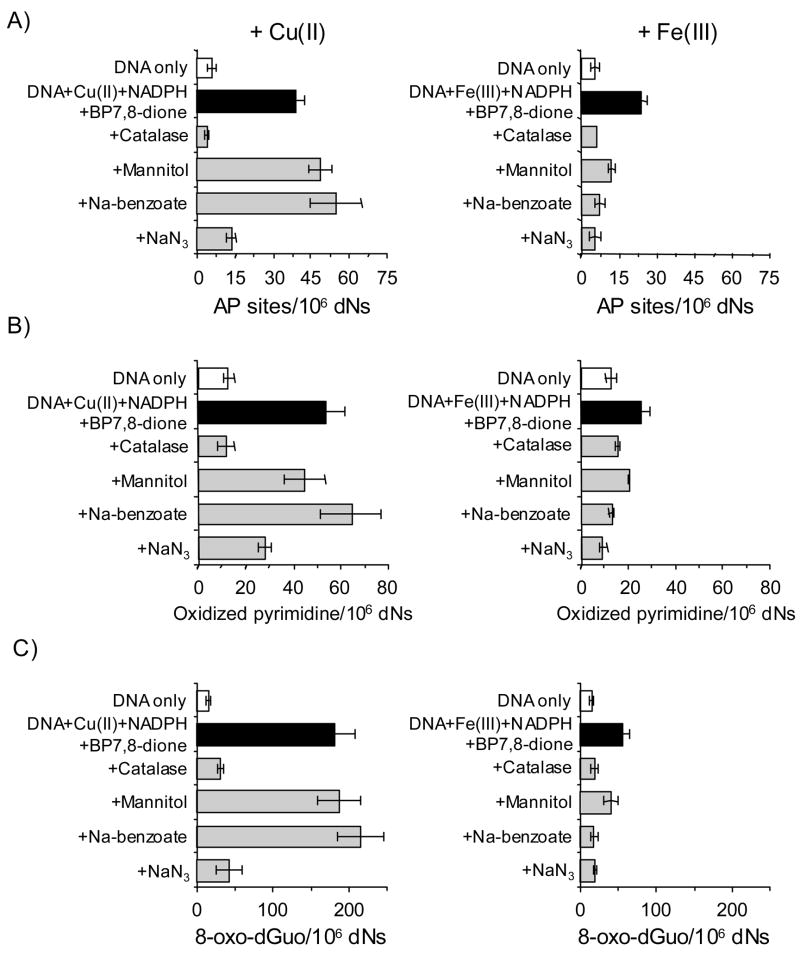
To identify which ROS is responsible for the formation of AP sites, oxidized pyrimidines and 8-oxo-dGuo in the presence of Cu(II)- and Fe(III)-mediated PAH o-quinone redox-cycling conditions, respectively, three types of ROS scavenging agents were used. Although each of the oxidative lesions detected in calf thymus DNA using the ARP assay was chemically different, the effects of scavengers observed under each redox redox cycling condition showed the same pattern (Figure 9). In the Cu(II)-mediated BP-7,8-dione redox-cycling conditions catalase eliminated the formation of the AP sites, oxidized pyrimidines and 8-oxo-dGuo, showing the dependence on hydrogen peroxide, but hydroxyl radical scavengers such as 0.1 M mannitol and 0.1 M sodium benzoate did not suppress the formation of these lesions. By contrast, sodium azide, a singlet oxygen and hydroxyl radical scavenger, abolished the formation of AP sites, oxidized pyrimidines and 8-oxo-dGuo, suggesting that the 1O2 was the responsible oxidant under these conditions.
Figure 9.
The effects of ROS scavenging agents on the formation of direct AP sites, oxidized pyrimidines and 8-oxo-dGuo in calf thymus DNA treated with 1 μM BP-7,8-dione under Cu(II)-and Fe(III)-mediated redox-cycling conditions. (A) Directly formed AP sites, (B) Oxidized pyrimidines (by Endo III-coupled assay) and (C) 8-oxo-dGuo (by hOGG1-coupled assay). The concentration of each scavenger was as follows. Catalase was added as 800 U/mL. Mannitol, sodium benzoate and sodium azide were added to achieve a final concentration of 0.1 M. In the data presented, the effects of each scavenger on the levels of directly formed AP site, oxidized pyrimidines (by Endo III-coupled assay) and 8-oxo-dGuo (by hOGG1-coupled assay) were compared with the levels of each control group (complete system), respectively. Kruskal-Wallis tests were conducted to determine which scavengers altered the amount of each lesions in the Cu(II) plus NADPH or Fe(III) plus NADPH redox-cycling conditions. With the Cu(II) plus NADPH system, catalase reduced the amount of all three adducts significantly (p = 0.0495 for each lesion type); mannitol produced significantly more AP sites (p = 0.0495) but had no significant effect on oxidized pyrimidines (p = 0.1266) or 8-oxo-dGuo ( p = 0.8273); sodium benzoate produced significantly more AP sites (p = 0.0495) but had no significant effect on oxidized pyrimidines or 8-oxo-dGuo (p = 0.2754 and 0.1266, respectively); sodium azide reduced the amounts of all three adducts significantly (p = 0.0495 for each lesion type). Inspection of the data shows that the effects of mannitol and sodium benzoate were statistically significant but their effects were marginal when compared with sodium azide.
With the Fe(III) plus NADPH system, catalase reduced the amount of all three adducts significantly (p = 0.0495 for each adduct); mannitol reduced the amount of 8-oxo-dGuo and AP sites significantly (p = 0.0495) but not oxidized pyrimidines (p = 0.1266); Both sodium benzoate and sodium azide reduced the amount of all three adducts significantly (p = 0.0495 for each lesion type).
At the same time, the experiments were repeated under the Fe(III)-mediated redox-cycling conditions. However, the observed results under these conditions showed a different pattern compared to that seen with Cu(II)-mediated redox-cycling conditions. In the Fe(III)-mediated BP-7,8-dione redox-cycling conditions hydroxyl radical scavengers attenuated all the lesions detected. The ability of hydroxyl radical scavengers to work under the Fe(III)-mediated redox-cycling conditions when the same scavengers failed to inhibit the their formation under Cu(II)-mediated redox-cycling conditions indicates that the main oxidant in the Fe(III)-mediated BP-7,8-dione redox-cycling is not singlet oxygen but •OH radical.
Discussion
PAH o-quinones are reactive and redox-active PAH metabolites produced by human AKRs in vitro and in lung cells (6,7,29). These quinones can form abasic sites by forming unstable N7-depurinating covalent DNA adducts or ROS-mediated DNA lesions (8,9,12,15). This study addresses the relative abundance of these lesions using the ARP assay.
AP sites generated by thermal depurination of PAH o-quinone-treated calf thymus DNA in vitro could be quantified using the ARP assay. The amounts of these adducts were relatively low (< 7 adducts per 106 dNs) even at high concentrations of PAH o-quinone (10 μM). Others have shown that pyrene-4,5-dione and fluroanthene-2,3-dione did not cause significant AP sites following thermal depurination (30), thus PAH o-quinone derived depurinating adducts appear to be a minor route to DNA adduct formation.
By contrast the formation of AP sites in PAH o-quinone-treated calf thymus DNA in the presence of Cu(II) and NADPH was substantial (> 100 adducts per 106 dNs). Under these conditions the amounts of AP sites detected varied according to the PAH o-quinone used, yielding a rank order of BA-3,4-dione > 7,12-DMBA-3,4-dione > BP-7,8-dione which is reminiscent of their rank order they consume NADPH and molecular oxygen (15). The suppression of these AP sites by ROS scavengers further implicated ROS in their formation.
A selection of ROS scavenging agents implicated singlet oxygen and not hydroxyl radical as the oxidant responsible. AP sites could arise from singlet oxygen by nonspecific hydrogen atom abstraction from the C-1′, C-2′ and C-4′ positions of deoxyribose (31). Abstraction at C-1′ leads to mere hydrolysis of the N-glycosidic bond to yield an AP site whereas abstraction at C-4′ leads to a Creigee rearrangement and the production of a base-propenal (32). Abstraction at the C-2′ position results in a ring-opened form of deoxyribose (33). Additionally, 8-oxo-dGuo which is formed under these conditions is more reactive than dGuo and can undergo secondary oxidation leading to destabilization of the N-glycosidic bond, and production of the aldehydic sites. Each of the resultant AP sites can be detected by ARP probe because they all generate an aldehydic group at the ROS-targeted carbon. The ARP is unable to distinguish between these lesions.
A combination of human oxoguanine glycosylase (hOGG1) with the ARP assay made it possible to detect 8-oxo-dGuo as AP sites in PAH o-quinone-treated calf thymus DNA. This BER enzyme recognizes 8-oxo-Gua resulting in AP sites in DNA and subsequently causes strand breaks through β-elimination. As expected, AP sites detected in this assay were observed with all three PAH o-quinones and required the presence of Cu(II) and NADPH (Figure 3); they were not suppressed by a hydroxyl radical scavenger but were completely abolished by sodium azide, suggesting the involvement of singlet oxygen as the immediate oxidant.
The hOGG1 coupled ARP assay was validated using HPLC-ECD by analyzing the level of 8-oxo-dGuo present in the same DNA pellets. To obtain accurate values for 8-oxo-dGuo present in calf-thymus DNA treated with PAH o-quinones significant precautions were taken to avoid adventitious oxidation of dGuo (see materials and methods). A comparison of both methods gave close agreement, indicating that the hOGG1 coupled ARP assay was a useful method for the detection of 8-oxo-dGuo. AP sites revealed by hOGG1 treatment may not only originate from 8-oxo-dGuo. hOGG1 also recognizes 2,6-diamino-4-hydroxy-5-formamidopyrimidine (Fapy-Gua) (34). Moreover, Fapy-Gua can be formed in dsDNA by singlet oxygen (35,36). The close agreement between the two methods for 8-oxo-dGuo determination suggests that Fapy-Gua was not produced significantly during the redox-cycling of PAH o-quinones.
The ARP assay detected additional AP sites in PAH o-quinone-treated calf thymus DNA when Escherichia coli endonuclease III (Endo III) was included (Figure 6). The AP sites revealed by Endo III treatment had the same requirement of Cu(II) and NADPH as observed for the formation of AP sites and 8-oxo-dGuo. The formation of these AP sites by all three PAH o-quinones and their suppression by sodium azide and not by hydroxyl radical scavengers implicates singlet oxygen in their formation. Endo III can recognize various oxidized thymines and cytosines (37). Among, these are the mutagenic lesions of 5,6-dihydroxy-5,6-dihydrothymine (thymine glycol), 5-hydroxycytosine (5-OH-C), 5-hydroxyuracil (5-OH-U) and 5,6-dihydroxy-5,6-dihydrouracil (uracil glycol) (38,39). However, studies show that these lesions are not produced by singlet oxygen at neutral pH (40). Therefore it is likely that Endo III may recognize secondary oxidation products of these lesions that are singlet oxygen dependent. These oxidized pyrimidines were formed in a much lower amount than 8-oxo-dGuo but at a level that was slightly higher or equal to the AP sites detected with PAH o-quinones under redox-cycling conditions in the absence of base-excision repair enzyme (Figure 4).
These data suggest that singlet oxygen generated during the redox-cycling of PAH o-quinones can cause a distinctive profile of oxidative damage in calf-thymus DNA, were 8-oxo-dGuo > oxidized pyrimidines = abasic sites. Similar results have been observed by others with singlet oxygen. Will et al (41) showed that the photosentizer R019-8022 yielded higher levels of 8-oxo-dGuo than single strand breaks, AP sites or oxidized pyrimidines in AS52 Chinese hamster ovary cells. Similarly, Pflaum et al (42) have observed a 10-fold higher formation of formamidopyrimidine-DNA glycosylase (FPG) sensitive base modification than either single strand breaks or AP sites in L1210 mouse leukemia cells exposed to visible light.
Our data show the following rank-order of oxidative DNA lesions 8-oxo-dGuo > oxidized pyrimidines = abasic sites. The question arises as to whether this rank order can explain the mutational pattern observed in the p53 tumor suppressor gene observed with PAH o-quinones under redox cycling conditions. Our published studies showed the following mutational pattern: G to T transversions (46% of 63 base substitutions), followed by C to T transitions (27% of 63 base substitutions), followed by T to C transitions (7.9% of 63% base substitutions) (24). The formation of 8-oxo-dGuo if unrepaired will lead to base-mispairing with A so that during replication a G to T transverison will occur. dATP is also preferentially incorporated opposite an abasic site providing another route to G to T transversions (43). 5-Hydroxy-cytosine, 5-hydroxyuracil and uracil glycol, which are oxidatively deaminated cytosines, induce C to T transitions (38), and thymine glycol gives rise to T to C transitions (39). Thus there is a reasonable correlation between the point mutations seen in p53 with PAH o-quinones under redox-cycling conditions with oxidative DNA lesions observed in naked DNA. The lack of mutation observed in p53 when it is reacted with PAH o-quinones in the absence of redox-cycling (24) is also consistent with the low level of depurination seen under these conditions.
The addition of different transition metals Cu(II) or Fe(III) to the redox-cycling conditions for BP-7,8-dione resulted in differences in the amount of 8-oxo-dGuo, oxidized pyrimidine and AP site formation, but the rank order of these lesions remained the same. Substitution of BP-7,8-dione with hydrogen peroxide showed a similar pattern of oxidative lesions suggesting that hydrogen peroxide was a common intermediate. In the presence of Fe(III) and hydrogen peroxide Fenton-like chemistry is expected:
| (Equation 1) |
| (Equation 2) |
From these equations, 2 moles of hydroxyl radical are produced from the consumption of 3 moles of hydrogen peroxide (44,45). Therefore, a plausible mechanism of PAH o-quinone-mediated oxidative damage under Fe(III)/Fe(II) redox-cycling is presumed to be the formation of •OH. However, the amount of lesions observed with Cu(II) and Fe(III) are different.
The formation of AP sites, oxidized pyrimidines and 8-oxo-dGuo by PAH o-quinones under the two redox cycling conditions (e.g. Cu(II) or Fe(III)) was completely blocked by catalase confirming that hydrogen peroxide was indispensable for the formation of the lesions. By contrast, •OH scavengers such as mannitol and sodium benzoate gave different outcomes. The formation of these lesions using Fe(III)-mediated redox-cycling was consistently attenuated by •OH scavengers but those formed by Cu(II)-mediated redox-cycling conditions were not suppressed suggesting that the responsible oxidant in both conditions was different. Sodium azide a •OH and 1O2 scavenger identified the oxidant responsible. In Fe(III)-mediated redox-cycling conditions, sodium azide gave the same result as the •OH scavengers and indicated that the dominant oxidant was •OH. In Cu(II)-mediated redox-cycling conditions, the lesions were inhibited by sodium azide and not by the •OH scavengers suggesting that singlet oxygen was the dominant oxidant.
It is proposed that the oxidants produced by PAH o-quinones in the two redox-cycling conditions produce their lesions by different mechanisms. In the Fe(III)-mediated redox-cycling system hydroxyl radical produced in Fenton-like chemistry produces a guanine radical (G8OH) by reaction at the C8 position of guanine and induces 8-oxo-dGuo formation by subsequent oxidation of the guanine radical (46). In the Cu(II)-mediated redox-cycling system singlet oxygen is produced by the binding of Cu(II) to DNA and the eventual formation of a Cu(I)OOH complex. The resultant singlet oxygen produces 8-oxo-Gua via a 4,8-endoperoxide of guanine (15).
Our results show that PAH o-quinones produced by AKRs generate aldehydic sites in DNA which can be accounted for by the formation of three types of oxidative lesions: 8-oxo-dGuo > oxidized pyrimidines = abasic sites. These lesions provide a good correlation with the mutation pattern seen in the p53 tumor suppressor gene when treated with PAH o-quinones under redox-cycling conditions. These lesions arise due to either singlet oxygen or hydroxyl radical formation, which is determined by the transition metal used. These lesions are more abundant than those that arise due to the formation of PAH o-quinone N7-depurinating adducts. These studies add weight to the hypothesis that PAH-induced ROS formation by AKR generated PAH o-quinones may contribute to PAH mutagenesis and carcinogenesis.
Acknowledgments
This work was supported by grants from NIH (P01-CA-092537) and (R01-CA-39504) awarded to TMP. The PAH o-quinones used for the study was supplied by Dr. Ronald Harvey at The Ben May Institute for Cancer Research. We thank Xingmei Wang for the statistical analysis.
Footnotes
Abbreviations: ADL, aldehydic DNA lesion; AKR, aldo-keto-reductase; (±)-anti-BP-7,8-diol-9,10-epoxide, (±)-anti-7,8-dihydroxy-9α,10α–epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene; AP sites, apurinic and apyrimidinic sites (abasic sites); ARP, aldehyde reactive probe; BP-7,8-dione, benzo[a]pyrene-7,8-dione; BA-3,4-dione, benz[a]anthracene-3,4-dione; BER, base excision repair; P450, cytochrome P450; Desferal®, deferoxamine mesylate; dGuo, 2′-deoxyguanosine; 7,12-DMBA-3,4-dione, 7,12-dimethylbenz[a]anthracene-3,4-dione; Endo III, Escherichia coli endonuclease III; Fapy-Gua, 2,6-diamino-4-hydroxy-5-formamidopyrimidine; hOGG1, human 8-oxoguanine glycosylase; HRP, horse radish peroxidase; M1-dGuo, 3-(2′-deoxy-β-D-erythro-pentofuranosyl)pyrimido[1,2-a]purin-10(3H)one; NADPH, β-nicotinamide adenine dinucleotide phosphate (reduced); NP-1,2-dione, naphthalene-1,2-dione; 8-oxo-dGuo, 8-oxo-7,8-dihydro-2′-deoxyguanosine; PA-1,2-dione, phenanthrene-1,2-dione; PAH, polycyclic aromatic hydrocarbon; PDE I, phosphodiesterase I; ROS, reactive oxygen species.
References
- 1.Xue W, Warshawsky D. Metabolic activation of polycyclic and heterocyclic aromatic hydrocarbons and DNA damage. Toxicol Appl Pharmacol. 2005;206:73–93. doi: 10.1016/j.taap.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Rothman N, Poirier MC, Baser ME, Hansen JA, Gentile C, Bowman ED, Strickland PT. Formation of polycyclic aromatic hydrocarbon-DNA adducts in peripheral white blood cells during consumption of charcoal-broiled beef. Carcinogenesis. 1990;11:1241–1243. doi: 10.1093/carcin/11.7.1241. [DOI] [PubMed] [Google Scholar]
- 3.Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 4.Gelboin HV. Benzo[a]pyrene metabolism, activation and carcinogenesis: role and regulation of mixed function oxidases and related enzymes. Physiol Rev. 1980;60:1107–1166. doi: 10.1152/physrev.1980.60.4.1107. [DOI] [PubMed] [Google Scholar]
- 5.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons. G H A Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 6.Palackal NT, Burczynski ME, Harvey RG, Penning TM. Metabolic activation of polycyclic aromatic hydrocarbon trans-dihydrodiols by ubiquitously expressed aldehyde reductase (AKR1A1) Chem Biol Interact 130–. 2001;132:815–824. doi: 10.1016/s0009-2797(00)00237-4. [DOI] [PubMed] [Google Scholar]
- 7.Jiang H, Shen YM, Quinn AM, Penning TM. Competing roles of cytochrome P450 1A1/1B1 and aldo-keto reductase 1A1 in the metabolic activation of (+/−)-7,8-dihydroxy-7,8-dihydro-benzo[a]pyrene in human bronchoalveolar cell extracts. Chem Res Toxicol. 2005;18:365–374. doi: 10.1021/tx0497245. [DOI] [PubMed] [Google Scholar]
- 8.Flowers L, Harvey RG, Penning TM. Examination of polycyclic aromatic hydrocarbon o-quinones produced by dihydrodiol dehydrogenase as substrates for redox-cycling in rat liver. Biochem (Life Sci Adv) 1992;11:49–58. [Google Scholar]
- 9.Flowers L, Ohnishi ST, Penning TM. DNA strand scission by polycyclic aromatic hydrocarbon o-quinones: role of reactive oxygen species, Cu(II)/Cu(I) redox cycling and o-semiquinone anion radicals. Biochemistry. 1997;36:8640–8648. doi: 10.1021/bi970367p. [DOI] [PubMed] [Google Scholar]
- 10.Balu N, Padgett WT, Lambert GR, Swank AE, Richard AM, Nesnow S. Identification and characterization of novel stable deoxyguanosine and deoxyadenosine adducts of benzo[a]pyrene-7,8-quinone from reactions at physiological pH. Chem Res Toxicol. 2004;17:827–838. doi: 10.1021/tx034207s. [DOI] [PubMed] [Google Scholar]
- 11.Shou M, Harvey RG, Penning TM. Reactivity of benzo[a]pyrene-7,8-dione with DNA. Evidence for the formation of deoxyguanosine adducts. Carcinogenesis. 1993;14:475–482. doi: 10.1093/carcin/14.3.475. [DOI] [PubMed] [Google Scholar]
- 12.McCoull KD, Rindgen D, Blair IA, Penning TM. Synthesis and characterization of polycyclic aromatic hydrocarbon o-quinone depurinating N7-guanine adducts. Chem Res Toxicol. 1999;12:237–246. doi: 10.1021/tx980182z. [DOI] [PubMed] [Google Scholar]
- 13.Ohnishi S, Kawanishi S. Double base lesions of DNA by a metabolite of carcinongenic benzo[a]pyrene. Biochem Biophys Res Commun. 2002;290:778–782. doi: 10.1006/bbrc.2001.6249. [DOI] [PubMed] [Google Scholar]
- 14.Seike K, Murata M, Oikawa S, Hiraku Y, Hirakawa K, Kawanishi S. Oxidative DNA damage induced by benz[a]anthracene metabolites via redox cycles of quinone and unique non-quinone. Chem Res Toxicol. 2003;16:1470–1476. doi: 10.1021/tx034103h. [DOI] [PubMed] [Google Scholar]
- 15.Park JH, Sridhar GR, Szewczuk LM, Troxel AB, Harvey RG, Penning TM. Formation of 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxo-dGuo) by PAH o-quinones: involvement of reactive oxygen species and copper(II)/copper(I) redox cycling. Chem Res Toxicol. 2005;18:1026–1037. doi: 10.1021/tx050001a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong YC, Sangaiah R, Nakamura J, Pachkowski BF, Ranasinghe A, Gold A, Ball LM, Swenberg JA. Analysis of M1G-dR in DNA by aldehyde reactive probe labeling and liquid chromatography tandem mass spectrometry. Chem Res Toxicol. 2005;18:51–60. doi: 10.1021/tx049853l. [DOI] [PubMed] [Google Scholar]
- 17.Douki T, Odine F, Caillat S, Favier A, Cadet J. Predominance of the 1,N2-propano-2′-deoxyguanosine adduct among 4-hydroxy-2-nonenal-induced DNA lesions. Free Radic Biol Med. 2004;37:62–70. doi: 10.1016/j.freeradbiomed.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 18.Lee SH, Oe T, Blair IA. 4,5-Epoxy-2(E)-decenal-induced formation of 1,N(6)-etheno-2′-deoxyadenosine and 1,N(2)-etheno-2′-deoxyguanosine adducts. Chem Res Toxicol. 2002;15:300–304. doi: 10.1021/tx010147j. [DOI] [PubMed] [Google Scholar]
- 19.Frenkel K, Wei L, Wei H. 7,12-dimethylbenz[a]anthracene induces oxidative DNA modification in vivo. Free Radic Biol Med. 1995;19:373–380. doi: 10.1016/0891-5849(95)00046-z. [DOI] [PubMed] [Google Scholar]
- 20.Leadon SA, Stampfer MR, Bartley J. Production of oxidative DNA damage during the metabolic activation of benzo[a]pyrene in human mammary epithelial cells correlates with cell killing. Proc Natl Acad Sci USA. 1988;85:4365–4368. doi: 10.1073/pnas.85.12.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leadon SA, Sumerel J, Minton TA, Tischler A. Coal tar residues produce both DNA adducts and oxidative DNA damage in human mammary epithelial cells. Carcinogenesis. 1995;16:3021–3026. doi: 10.1093/carcin/16.12.3021. [DOI] [PubMed] [Google Scholar]
- 22.Tang DW, Chang KW, Chi CW, Liu TY. Hydroxychavicol modulates benzo[a]pyrene-induced genotoxicity through induction of dihydrodiol dehydrogenase. Toxicol Lett. 2004;152:235–243. doi: 10.1016/j.toxlet.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Chen HJC, Chiu WL. Association between cigarette smoking and urinary excretion of 1,N2-Ethenoguanine measured by isotope dilution liquid chromatography-electrospray ionization/tandem mass spectrometry. Chem Res Toxicol. 2005;18:1593–1599. doi: 10.1021/tx050145p. [DOI] [PubMed] [Google Scholar]
- 24.Yu D, Berlin JA, Penning TM, Field J. Reactive oxygen species generated by PAH o-quinones cause change-in-function mutations in p53. Chem Res Toxicol. 2002;15:832–842. doi: 10.1021/tx010177m. [DOI] [PubMed] [Google Scholar]
- 25.Harvey RG, Dai Q, Ran C, Penning TM. Synthesis of the o-quinones and other oxidized metabolites of polycyclic aromatic hydrocarbons implicated in carcinogenesis. J Org Chem. 2004;69:2024–2032. doi: 10.1021/jo030348n. [DOI] [PubMed] [Google Scholar]
- 26.Fu PP, Cortez C, Sukumaran KB, Harvey RG. Synthesis of isomeric phenols of benz[a]anthracene from benz[a]anthracene. J Org Chem. 1979;44:4265–4271. [Google Scholar]
- 27.Nakamura J, Walker VE, Upton PB, Chiang SY, Kow YW, Swenberg JA. Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions. Cancer Res. 1998;58:222–225. [PubMed] [Google Scholar]
- 28.Nakamura J, Swenberg JA. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–2526. [PubMed] [Google Scholar]
- 29.Jiang H, Vudathala DK, Blair IA, Penning TM. Competing roles of aldo-keto reductase 1A1 and cytochrome P450-1B1 in benzo[a]pyrene-7,8-diol activation in human bronchoalveolar H358 cells: Role of AKRs in P450-1B1 Induction. Chem Res Toxicol. 2006;19:68–78. doi: 10.1021/tx0502488. [DOI] [PubMed] [Google Scholar]
- 30.Zielinska-Park J, Nakamura J, Swenberg JA, Aitken MD. Aldehydic DNA lesions in calf thymus DNA and Hela S3 cells produced by bacterial quinone metabolites of fluranthene and pyrene. Carcinogenesis. 2004;25:1727–1733. doi: 10.1093/carcin/bgh174. [DOI] [PubMed] [Google Scholar]
- 31.Povirk LF, Steighner RJ. Oxidized apurinic/apyrimidinic sites formed in DNA by oxidative mutagens. Mutat Res. 1989;214:13–22. doi: 10.1016/0027-5107(89)90193-0. [DOI] [PubMed] [Google Scholar]
- 32.Casadevall M, Fresco PC, Kortenkamp A. Chromium (VI)-mediated DNA damage: oxidative pathway resulting in the formation of DNA breaks and abasic sites. Chem Biol Interact. 1999;123:117–132. doi: 10.1016/s0009-2797(99)00128-3. [DOI] [PubMed] [Google Scholar]
- 33.Greenberg MM, Weledji YN, Kroeger KM, Kim J. In vitro replication and repair of DNA containing a C2′-oxidized abasic sites. Biochemistry. 2004;43:15217–15222. doi: 10.1021/bi048360c. [DOI] [PubMed] [Google Scholar]
- 34.Karahalil B, Girard PM, Boiteux S, Dizdaroglu M. Substrate specificity of the Ogg1 protein of Saccharomyces cerevisiae: excision of guanine lesions produced in DNA by ionizing radiation- or hydrogen peroxide/metal ion-generated free radicals. Nucleic Acids Res. 1998;26:1228–1232. doi: 10.1093/nar/26.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boiteux S, Gajewski E, Laval J, Dizdaroglu M. Substrate specificity of the Escherichia coli Fpg protein formamidopyrimidine-DNA glycosylase: excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry. 1992;31:106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- 36.Steenken S. Purine bases, nucleosides, and nucleotides: Aqueous solution redox chemistry and transformation reactions of their radical cations and e- and OH adducts. Chem Rev. 1989;89:503–520. [Google Scholar]
- 37.Gasparutto D, Bourdat AG, D’Ham C, Duarte V, Romieu A, Cadet J. Repair and replication of oxidized DNA bases using modified oligodeoxyribonucleotides. Biochimie. 2000;82:19–24. doi: 10.1016/s0300-9084(00)00347-3. [DOI] [PubMed] [Google Scholar]
- 38.Kreutzer DA, Essigmann JM. Oxidized, deaminated cytosines are a source of C to T transitions in vivo. Proc Natl Acad Sci USA. 1998;95:3578–3582. doi: 10.1073/pnas.95.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Basu AK, Loechler EL, Leadon SA, Essigmann JM. Genetic effects of thymine glycol: site-specific mutagenesis and molecular modeling studies. Proc Natl Acad Sci USA. 1989;86:7677–7681. doi: 10.1073/pnas.86.20.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ravanat JL, Mascio PD, Martinez GR, Medeiros MHG, Cadet J. Singlet oxygen induces oxidation of cellular DNA. J Biol Chem. 2000;275:40601–40604. doi: 10.1074/jbc.M006681200. [DOI] [PubMed] [Google Scholar]
- 41.Will O, Gocke E, Eckert I, Schulz I, Pflaum M, Mahler HC, Epe B. Oxidative DNA damage and mutations induced by a polar photosensitizer, Ro19-8022. Mutat Res. 1999;435:89–101. doi: 10.1016/s0921-8777(99)00039-7. [DOI] [PubMed] [Google Scholar]
- 42.Pflaum M, Boiteux S, Epe B. Visible light generates oxidative DNA base modifications in high excess of strand breaks in mammalian cells. Carcinogenesis. 1994;15:297–300. doi: 10.1093/carcin/15.2.297. [DOI] [PubMed] [Google Scholar]
- 43.Sagher D, Strauss B. Abasic sites from cytosine as termination signals for DNA synthesis. Nucleic Acids Res. 1985;13:4285–4298. doi: 10.1093/nar/13.12.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Josephson RA, Silverman HS, Lakatta EG, Stern MD, Zweier JL. Study of the mechanisms of hydrogen peroxide and hydroxyl free radical-induced cellular injury and calcium overload in cardiac myocytes. J Biol Chem. 1991;266:2354–2361. [PubMed] [Google Scholar]
- 45.Lee YH, Lee CH, Yoon JY. High temperature dependence of 2,4-dichlorophenoxyacetic acid degradation by Fe3+/H(2)O(2) system. Chemosphere. 2003;51:963–971. doi: 10.1016/S0045-6535(03)00043-2. [DOI] [PubMed] [Google Scholar]
- 46.Dizdaroglu M. Formation of an 8-hydroxyguanine moiety in deoxyribonucleic acid on gamma-irradiation in aqueous solution. Biochemistry. 1985;24:4476–4481. doi: 10.1021/bi00337a032. [DOI] [PubMed] [Google Scholar]