Abstract
The domestication of crops and the development of agricultural societies not only brought about major changes in human interactions with the environment but also in plants' interactions with the diseases that challenge them. We evaluated the impact of the domestication of maize from teosinte and the widespread cultivation of maize on the historical demography of Ustilago maydis, a fungal pathogen of maize. To determine the evolutionary response of the pathogen's populations, we obtained multilocus genotypes for 1088 U. maydis diploid individuals from two teosinte subspecies in Mexico and from maize in Mexico and throughout the Americas. Results identified five major U. maydis populations: two in Mexico; two in South America; and one in the United States. The two populations in Mexico diverged from the other populations at times comparable to those for the domestication of maize at 6000–10 000 years before present. Maize domestication and agriculture enforced sweeping changes in U. maydis populations such that the standing variation in extant pathogen populations reflects evolution only since the time of the crop's domestication.
Keywords: host and pathogen coevolution, crop domestication, smut fungi, agriculture, host expansion
Abbreviations: ybp, years before present, mya, million years ago, SSR, simple sequence repeat, SMM, stepwise mutation model
1. Introduction
The domestication of plants and animals and the subsequent development of agriculture brought major changes in human societies (Smith 1995), their relationship to the natural environment and the evolution of disease. For example, the development of slash and burn agriculture in central Africa has been implicated in the evolution of more virulent forms of the malarial parasite, speciation in Anopheles mosquitoes and subsequent evolution of some forms of malarial resistance in humans (Tishkoff et al. 2001). As nascent as are archaeological studies of human disease, much less is known of agriculture's impact on plant disease dynamics. Two recent reports date speciation in plant pathogenic fungi to the times and locations of their host crop's domestication (Couch et al. 2005; Stukenbrock et al. 2007), demonstrating that the crop domestication has had tremendous impacts on the pathogen evolution over relatively short periods of time. We examined the impacts of maize domestication and subsequent cultivation throughout the Americas on the evolution of the corn smut fungus Ustilago maydis, an obligate fungal pathogen of maize.
Previously, we dated the divergence of U. maydis from its sister taxon, Ustilogo bouriquetii, at 10–25 million years ago (mya; Munkacsi et al. 2007); thus, U. maydis must have originated long before the time of maize domestication only 6000–10 000 years before present (ybp; reviewed in Staller et al. 2006). Ustilago maydis was probably a pathogen on the teosinte progenitor of maize, Zea mays ssp. parviglumis, because most Ustilago species and the members of related genera are host-specific pathogens on grasses of the Poaceae (Stoll et al. 2005), and the current host range of U. maydis is limited to Z. mays subspecies (Duran 1987; this report). We asked whether the evidence of this ancient coevolutionary history is apparent in contemporary populations or alternatively, if the dynamic response of pathogen populations to modern crop plant management has written over the historical patterns of variation as it apparently has in other systems (e.g. Roelfs & Groth 1980; Zhu et al. 2000; Kolmer et al. 2004; Stukenbrock et al. 2006). Contemporary U. maydis populations demonstrate little long-distance migration (Voth et al. 2006), yet maintain substantial variation and exhibit low levels of local inbreeding (Barnes et al. 2004), suggesting that modern practices have not homogenized corn smut populations as they have for other crop pathogens (e.g. Stukenbrock et al. 2006). The uneven impact of domestication and early agriculture across pathosystems is further suggested by the findings that the geographical origins and centre of diversity correspond for some pathogens (Banke & McDonald 2005; Stukenbrock et al. 2007), but not for others (Zaffarano et al. 2006; Brunner et al. 2007). The extent to which a pathogen's demographic history might constrain or amplify the responses to demographic and genetic changes in plant host populations is not well understood.
Domestication transformed the teosinte Z. m. ssp. parviglumis into maize (Z. m. ssp. mays) in the Balsas River valley of Mexico (Matsuoka et al. 2002; Doebley et al. 2006). As maize was subsequently moved to South America 4000–7000 ybp (Piperno & Pearsall 1998; Staller et al. 2006) and to the United States 1000–3000 ybp (Staller et al. 2006), the crop was adapted to new environments and uses. In the early part of the twentieth century, strong efforts for resistance breeding in maize to U. maydis incorporated a number of quantitative loci from a variety of genetic backgrounds (Lubberstedt et al. 1998; Baumgarten 2004; Baumgarten et al. 2007). With the native Americans' use of corn smut as a food source (Wilson 1987; Valverde et al. 1995; Iltis 2000), varying ecological conditions throughout the Americas and the quantitative nature of U. maydis resistance in maize (Lubberstedt et al. 1998; Baumgarten et al. 2007), selection on U. maydis populations has been variable over time and geography. Nonetheless, the well-documented history of maize agriculture in the Americas sets upper limits for times of U. maydis population expansion as maize is the only host of U. maydis outside of Mexico.
We investigate the null hypothesis that the domestication of maize had little measurable impact on the genetic variation that built up in the ancient coevolutionary dynamic between U. maydis and its hosts. Under the null hypothesis, we expected that the centre of genetic diversity in current U. maydis populations should be in Mexico as this is the apparent centre of origin for U. maydis and its ancestral teosinte hosts, the subspecies of Z. mays, ssp. parviglumis and ssp. mexicana (Sánchez et al. 1998). Alternatively, if the domestication of maize led to a strong genetic bottleneck of U. maydis populations, we expected that extant populations should not date older than the thousands year old history of maize in Mexico and that the diversity of populations in Mexico should not be greater than that in descendent populations. Considering more recent history, we investigate the hypothesis that modern, intensive maize agriculture practised in South America and the United States caused high rates of migration and successive bottlenecks within descendent U. maydis populations. We tested the expectation that these populations will exhibit little population substructure and lower levels of genetic diversity than that in Mexico. Pursuant to these hypotheses, we sampled U. maydis populations on maize, Z. m. ssp. parviglumis and Z. m. ssp. mexicana in Mexico and on maize in Mexico and throughout the Americas, and analysed population genetic structure using 10 simple sequence repeat (SSR) markers. We find little evidence of the ancient evolutionary history of U. maydis in Mexico, yet despite the intensive culture of modern maize, the thousands year old imprint of early maize agriculture remains evident in the genetic structure of U. maydis populations today.
2. Material and methods
(a) Ustilago maydis and sampling
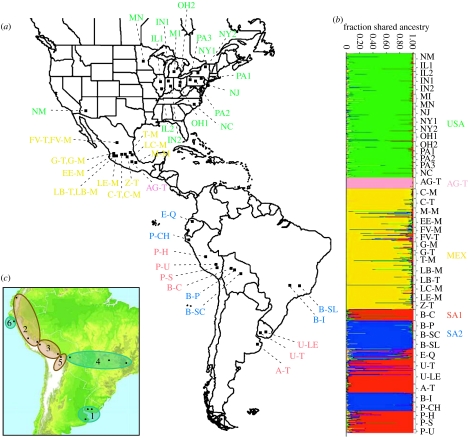
After mating of compatible haploid cells, U. maydis forms an infectious dikaryotic hypha and later develops a hypertrophy, or gall, containing diploid teliospores at the site of the localized infection. Our extensive field observations (Baumgarten et al. 2007) indicate a single generation per year in regions of annual maize cultivation with very low plant-to-plant spread within a year. Sampling was conducted between 1998 and 2002; 44 field locations were chosen to represent the geographical range of maize domestication and early cultivation, as well as variation in maize landraces and teosinte subspecies, Z. m. ssp. parviglumis and Z. m. ssp. mexicana. We sampled one gall (one diploid) per plant directly into 2 ml cryovials in the field, and sampled approximately 25 galls per field for a total of 1088 U. maydis individual galls (figure 1a; electronic supplementary material, table 1).
Figure 1.
Geographical origin and membership of U. maydis individuals in five major populations estimated using Structure (Pritchard et al. 2000) (a) Collection locations colour-coded according to the membership in five major populations. Collections in Mexico are labelled by location and host; maize (M) or teosinte (T; see table 1 in the electronic supplementary material). (b) Estimated membership of each individual to each of the five major populations. (c) Ustilago maydis subpopulations identified in South America are illustrated by elevation. High-elevation subpopulations (brown circles, more than 2300 m) and low-elevation subpopulations (green circles, less than 800 m) are shown.
(b) Scoring SSR loci
Ten SSR loci on nine different chromosomes were selected to represent a range of repeat motif lengths and varying levels of diversity appropriate to the possible time scales of this study (Munkacsi et al. 2006). The repeat motif characteristics and forward and reverse primer sequences for each locus are provided (electronic supplementary material, table 2). Total genomic DNA was extracted from approximately 10 mg of teliospores. PCR cycle conditions used to amplify fragments from 10 ng of genomic DNA using fluorescent-labelled primers and automated sequencer analysis of PCR fragment sizes (Applied Biosystems model 3100) were the same as described previously (Munkacsi et al. 2006). Fragment (allele) size ±0.5 bp was determined with Genescan Analysis software (v. 3.7, Applied Biosystems, Inc.). Background amplicons were distinguished as chromatogram peaks not larger than 25% of the tallest peak for each locus and amplifications were rerun if more than two peaks per diploid genotype were observed.
(c) Identification of populations
Genetically distinct populations were identified using a Bayesian clustering method implemented in Structure v. 2.0 (Pritchard et al. 2000). For each K number of populations modelled, individual genotypes are assigned membership in a population as a function of allele frequencies and the proportion of an individual's genotype drawn from each of the K populations. Individuals are assigned to clusters to maximize Hardy–Weinberg equilibrium, linkage equilibrium and genetic homogeneity within each cluster. For each parameter value of K, admixture and correlation between alleles were varied in triplicate runs of 100 000 iterations with a burn-in period of 30 000 iterations.
(d) Marker and population genetic analyses
We compared the observed heterozygosity and the number of alleles for each population to expectations under a neutral equilibrium model, analogous to the Ewens–Watterson test for DNA sequence variation (Ewens 1972; Watterson 1978). A deficiency or excess of heterozygosity relative to the number of alleles was evaluated against neutral expectations and a 95% confidence interval generated by permutation.
We used Arlequin (Schneider et al. 2000) to estimate the following: genetic diversity as the unbiased mean expected heterozygosity (H; Nei 1987); number of total alleles; number of rare alleles (frequency <0.05); number of private alleles (those found in only one population); and levels of genetic differentiation (RST). The RST statistic uses variation in allele sizes and incorporates the stepwise mutation model (SMM) to evaluate the proportion of the total genetic variation that is found in a subpopulation (Slatkin 1995). The significant difference of RST values from zero, at p<0.05, was determined with 1000 permutations of individuals between populations in Arlequin.
We used BayesAss+ (Wilson & Rannala 2003) to evaluate migration rates per generation as the mean proportion of one population entering another population and as the mean proportion of the population staying in that local population. Linkage equilibrium and non-overlapping generations but not Hardy–Weinberg equilibrium are assumed. We ran 3×106 iterations, discarding the first 106 iterations as burn-in, and thereafter samples were collected every 2000 iterations to infer population allele frequencies and migrant proportions. One generation per year was assumed.
We tested for the effect of population bottlenecks using Bottleneck (Cornuet & Luikart 1996) under SMM. The test examines evidence for an observed excess heterozygosity over that expected from allele numbers because with small founding populations, the number of alleles is reduced more than heterozygosity (Nei 1987). Significant deviation in observed heterozygosity from that expected was evaluated with a two-tailed test of Wilcoxon signed-rank test across loci because a deficiency of heterozygosity will occur with long-term inbreeding. We tested for linkage disequilibrium by determining the index of association (IA) using Multilocus (v. 1.2) with significance evaluated by permutation (Agapow & Burt 2001).
We conducted the k- and g-test (Reich et al. 1999) using KGTESTS (Bilgin 2007) to evaluate evidence for population expansion within each major population. Similar to mismatch distributions for DNA sequences, the k-test compares the observed distribution of allele sizes that is unimodal in an expanding population with the expected distribution of allele sizes that is multimodal in a population of constant size. The measure k, related to kurtosis, is centred on 0, and the proportion of loci that gives negative values is evaluated with a binomial test (Reich et al. 1999). The g-test uses the observed variation across loci in variance of allele sizes with the rationale that variation across loci will be greater in a population of constant size than in a newly expanding population (Reich et al. 1999). The g value is calculated as a ratio of the observed variation across loci to that expected under populations of constant size and is evaluated against a table of 5th percentile cut-off values (p<0.05) obtained by simulation for different sample sizes and numbers of loci and is robust to variation in population size (table 1 in Reich et al. 1999).
(e) Population divergence dates
We adopted the method of Zhivotovsky (2001) to obtain earlier and later bounds for divergence time between each pairwise combination of populations. Times of divergence (TD) are estimated as: , where TD is the estimate of divergence time in generations, is the average squared difference in allele size between pairs of sampled alleles over all the loci (Goldstein et al. 1995) and is the estimate of average variation in repeat number across loci in the ancestral population. The earlier date is obtained under the assumption that V0=0 and the more recent date is obtained under the assumption that V0 is the same as allelic variation among extant populations. The estimates are not affected by population growth but are affected by gene flow. Using these extremes as estimates for V0, the resulting bounds can be considered conservative estimates of divergence time. We estimated the average mutation rate over SSR loci by two methods, both assuming one generation per year. The regression for the number of repeats in an SSR locus and rate of evolution in eqn 1 of Thuillet et al. (2005) generates an estimate of rate without assuming a calibration date. A second estimate of rate was obtained using the variance in allele sizes as an estimate of 2 Nμ for USA populations (Valdes et al. 1993) and a calibration time of 2000 years for U. maydis populations in the United States.
3. Results
We obtained smut gall samples from three Z. m. ssp. parviglumis populations and three Z. m. ssp. mexicana populations in Mexico (figure 1). The Z. m. ssp. mexicana plants occurred as weeds in maize fields and galls were also collected from those maize plants. Ustilago maydis infections on teosinte plants are characterized by one to very few small galls on seeds and low infection rates (approx. 0.1% of plants). Infections on maize plants are characterized by numerous, larger galls occurring on any plant organ and at higher infection rates (approx. 3% of plants). Obtaining DNA from small gall samples representing single infection events, we evaluated a total of 1088 diploid teliospore collections (electronic supplementary material, table 1) from all plant populations for variation at 10 SSR loci (electronic supplementary material, table 2).
(a) Structure identifies five major populations
Structure assigns diploid individuals membership to genetically distinct clusters (populations) without reference to the geographical location of the collections (Pritchard et al. 2000). Modelling K=1–10 populations, we found the best support for K=5 genetically distinct clusters or ‘major populations’ (figure 1b) based on the recommended criteria that the rate of change in the log probability of the data between successive K values (ΔK; Evanno et al. 2005) was the greatest in the interval of four to five populations, and thus that it began to plateau (Pritchard et al. 2000) at K=5. Varying the degree of admixture between populations from 0.05 to 0.50 yielded little effect on the estimated log probability or ΔK values. Three independent runs for each K yielded consistent results.
The five major populations identified in Structure represent large geographical regions of Mexico, South America and the United States (figure 1a). In Mexico, we found evidence for two quite distinct populations, AG-T and MEX. The AG-T population was limited to collections from one Z. m. ssp. parviglumis population near Amates Grandes, Guerrero (n=30). The MEX population comprised all collections from the remaining 14 fields in Mexico (n=324) and included nine fields of Mexican maize landraces and five fields from the two teosinte taxa. In South America, we identified two major populations, SA1 and SA2, collected from South American maize landraces. The SA1 population comprised collections from seven maize fields (n=218) and the SA2 population comprised collections from six maize fields (n=154). SA1 and SA2 are genetically but not geographically distinct from each other (figure 1b). The fifth major population, USA, comprised collections from elite maize hybrid lines in 16 maize fields in the United States (n=362).
Using Arlequin (Schneider et al. 2000), we identified between 1 and 40 alleles and estimated expected heterozygosity (H) values between 0.04 and 0.91 per major population, across the five major populations and 10 SSR loci. Results of the Ewens–Watterson test (Ewens 1972; Watterson 1978) showed that observed heterozygosity and number of alleles fit neutral expectations within the 95% CI for all loci except locus 1.64 in the AG-T population that demonstrated a slight excess heterozygosity (electronic supplementary material, figure 1).
To quantitatively compare the levels of genetic differentiation between the five major populations, we calculated RST values across all 10 SSR loci using Arlequin and assuming SMM (Schneider et al. 2000). The RST evaluates the distribution of SSR variation within and between populations using allele sizes and assumes random mating (Slatkin 1995). High levels of genetic differentiation (RST>0.15) were found among all comparisons except that of MEX and SA1 (RST=0.07) and of SA1 and SA2 (RST=0.11); all were significantly greater than zero. The AG-T population was the most strongly differentiated one from all other populations (mean pairwise RST=0.53±0.07). Interestingly, both Structure and RST results show that the USA population is more strongly differentiated from the MEX population than are either SA1 or SA2.
To determine whether unique field populations might strongly affect the RST and Structure results for major populations, we examined RST values by field. Consistently low RST levels were found within USA (mean pairwise RST=0.01±0.02), and low to moderate RST levels were found within SA1 and MEX (mean pairwise RST=0.08±0.10 and 0.08±0.03, respectively); we concluded that local fields did not strongly affect those results. However, we did obtain higher RST levels for fields within SA2 (mean pairwise RST=0.21±0.14), motivating the investigation of substructure described below. Together, Structure and RST results demonstrate a remarkable degree of geographical structure in U. maydis populations given the very short time frame that maize has been cultivated outside of Mexico.
(b) South American populations demonstrate substructure
We next used Structure to analyse SSR variation within regions. We found little support for more than one genetic cluster (subpopulation) within the USA, MEX or AG-T populations. In contrast, analysing data for all South American collections together, K=6 subpopulations were supported and broke up SA1 and SA2 each into three subpopulations (figure 1c). SA1 consists of subpopulations 1 (U-T, U-LE, A-T), 2 (P-H, E-Q, P-U) and 3 (B-C, P-S), and SA2 consists of subpopulations 4 (B-SC, B-SL, B-I), 5 (B-P) and 6 (P-CH). No SA1 field population was grouped with SA2 field populations except E-Q, which shows evidence of mixed ancestry (figure 1b). Interestingly, substructure by landrace is not apparent while three subpopulations (1, 4 and 6) consisted of U. maydis collections from maize growing below 800 m and three subpopulations (2, 3 and 5) consisted of collections from maize growing in the Andes Mountains above 2300 m (figure 1c; see table 1 in the electronic supplementary material for landrace and elevation data).
(c) Genetic diversity is not greater in Mexico
We assumed that Mexico is the centre of origin for U. maydis and asked whether the genetic diversity of U. maydis populations is greater in Mexico than in descendent populations of South America and the United States. Across the ten SSR loci, the MEX population had approximately the same expected heterozygosity (H; Nei 1987) as the descendent USA, SA1 and SA2 populations. The AG-T population demonstrated significantly lower expected heterozygosity than any other population (table 1). The greatest numbers of total, rare and private alleles were found in the USA population (table 1), largely due to the extraordinary levels of variation at two loci (1.89, 1.206; electronic supplementary material, table 2) with 31 and 40 alleles each. Even without data for these two loci included, levels of variation observed in the USA, SA1 or SA2 populations are the same as in MEX and higher than that in AG-T. Interestingly, five loci were fixed for one allele in the AG-T population and there were no rare or private alleles, suggesting an isolated and inbreeding population.
Table 1.
For five major populations, measures of population diversity and genetic structure are given as gene diversity (H), numbers of alleles and index of association (IA) and measures of historic demographic processes are given as bottleneck and expansion test results. (*p<0.05; **p<0.01.)
| AG-T | MEX | SA1 | SA2 | USA | |
|---|---|---|---|---|---|
| no. individuals | 29 | 275 | 195 | 148 | 341 |
| Nei's Ha | 0.17* | 0.38 | 0.51 | 0.42 | 0.46 |
| no. total alleles | 16 | 59 | 84 | 55 | 123 |
| no. rare alleles | 0 | 31 | 48 | 27 | 70 |
| no. private alleles | 0 | 5 | 10 | 1 | 47 |
| IAb | 0.319* | −0.436* | 0.292* | 0.334* | −0.463* |
| bottleneckc | 3/5 n.s. | 8/9* | 9/9** | 8/9* | 9/9* |
| k-testd | 0.008* | 0.045* | 0.008** | 0.008** | 0.045* |
| g-testd | n.s. | n.s. | n.s. | n.s. | n.s. |
Unbiased mean expected heterozygosity (Nei 1987); *significant difference in pairwise comparison of AG-T with every other population.
Index of association (Agapow & Burt 2001).
Fraction of evaluated loci demonstrating observed heterozygote excess under the SMM; fixed loci were not evaluated.
k- and g-tests for expansion (Reich et al. 1999).
(d) Host subspecies contribute to genetic structure of U. maydis populations in Mexico
To evaluate the contribution of host variation to the genetic structure of U. maydis populations in Mexico, we estimated RST values among collections from maize (n=234), Z. m. ssp. mexicana (n=66) and Z. m. ssp. parviglumis (n=54). Comparing all maize (n=234) and all teosinte collections (n=120), we obtained significant, moderate levels of genetic differentiation (RST=0.14). Because the AG-T population is so distinct, we analysed all maize-infecting collections (n=234) versus all teosinte-infecting collections without the AG-T samples (n=90) and obtained a lower but still significant level of differentiation (RST=0.10). Separate comparisons of U. maydis populations on maize with those on Z. m. ssp. mexicana or Z. m. ssp. parviglumis (without AG-T) yielded similar and significant values (RST=0.10 and 0.08, respectively). These results show low to moderate levels of differentiation of U. maydis populations by host subspecies, perhaps surprising, since U. maydis collections were obtained from both Z. m. ssp. mexicana and maize growing in the same fields, and since Z. m. ssp. parviglumis is the direct ancestor of maize.
(e) Little migration occurs between major populations
Cognizant of continuing trade of maize in the Americas and the potential for long-distance wind-borne smut spore dispersal, we used BayesAss+ (Wilson & Rannala 2003) to estimate bidirectional migration rates among the five major populations. Results show little evidence of ongoing gene flow among the major populations because each retains a mean of 99% of its own population per generation, with the exception of SA1. SA1 retains approximately 95% of its own population and acquires approximately 4% of the SA2 population per generation (table 2). These results suggest that neither wind-borne spores nor contemporary agricultural trade have lead to significant levels of long-distance migration between major U. maydis populations.
Table 2.
Estimates of migration rates between five major populations as the mean of the posterior probabilities of migrants (m) per year (95% CI). (Rates of migration from populations listed across the top into populations listed along the side of the table. The proportion of individuals derived from the source population is shown on the diagonal.)
| into | from | ||||
|---|---|---|---|---|---|
| AG-T | MEX | SA1 | SA2 | USA | |
| AG-T | 99 (96.4–100) | 0.3 (0–1.6) | 0.3 (0–1.7) | 0.3 (0–1.6) | 0.2 (0–1.7) |
| MEX | 0 (0–0.2) | 99 (98.5–100) | 0 (0–0.3) | 0.3 (0–1.1) | 0.2 (0–0.6) |
| SA1 | 0 (0–0.4) | 0.2 (0–0.9) | 95.3 (93.0–97.4) | 4.2 (2.2–6.4) | 0 (0–0.9) |
| SA2 | 0 (0–0.4) | 0.1 (0–0.6) | 0.3 (0–1.5) | 99.2 (97.7–100) | 0.3 (0–1.1) |
| USA | 0 (0–0.1) | 0 (0–0.2) | 0 (0–0.2) | 0 (0–0.2) | 99.9 (99.6–100) |
(f) Historical demographic processes affected extant population structure
Bottleneck uses a Bayesian framework to evaluate the evidence for founder effects in descendent populations as observed excess heterozygosity over that expected for the number of alleles at each locus. Bottleneck removes data for loci fixed for one allele (Cornuet & Luikart 1996). Evidence for bottlenecks was strong for eight to nine out of nine loci in each of the major populations except AG-T (table 1). In AG-T, five out of the 10 loci were fixed for one allele, and the evidence for bottlenecks was not significant at the remaining five loci.
We evaluated disequilibrium association of alleles across loci using the measure IA (Agapow & Burt 2001) and found evidence of significant disequilibrium within all major populations (table 1). However, varied causes underlie apparent disequilibrium. The IA statistic is probably significant in the AG-T and MEX populations owing to low heterozygosity, sampling across subpopulations in SA1 and SA2 and insufficient time for random association of alleles in the USA population.
We next used the k- and g-tests to examine the evidence for population expansion (Reich et al. 1999). The k-test (variation within loci) returned significant results for population expansion for all five major populations (table 1). The g values (variation among loci) were large and not significant for any of the major populations (table 1). As a ratio, the g-test lacks power to distinguish from the null hypothesis; however, the non-significant g-test results could also result from uneven effects of bottlenecks and expansion across each major population. The bottleneck and expansion k-test results give evidence for recent bottlenecks and subsequent expansion in Mexico, and in descendent populations in the United States and South America.
Together, evidence for low migration and strong subdivision between large geographical regions and recent population bottlenecks within those regions suggest that descendent populations of U. maydis have been largely isolated since the establishment on their maize host populations.
(g) Corn smut populations are as old as maize agriculture and not much older
We used two approaches to estimate SSR mutation rates, , one using a calibration date and the other based on regression analysis in plants Thuillet et al. 2005. With either method, we found that the rates for loci 1.89 and 1.206 in USA populations were 5- to 10-fold higher (1.2×10−3 and 9.2×10−4 mutations per year, respectively) than that estimated for the remaining eight SSR loci. After removing data for 1.89 and 1.206 and assuming SMM, we obtained =1.55×10−4 steps per year using regression eqn 1 of Thuillet et al. (2005), and =1.89×10−4 steps per year using SSR variance in USA populations as an estimate of 2 Nμ (Valdes et al. 1993) and a calibration of 2000 years for the USA population. Both are within the range of SSR rates estimated for other organisms (Estoup et al. 2002). We used the latter estimate of SSR rate as it involved results and assumptions from our study system.
We estimated divergence times between major populations using the pairwise distance method of Zhivotovsky (2001). The assumption V0=0 should generate dates approaching the ancient origin of U. maydis (10–25 mya) if ancestral variation is still represented in extant populations. Results show the most recent dates at ca 1000–2000 ybp in comparisons that included the descendent populations, USA, SA1 and SA2, while the earliest dates at ca 9000–10 000 ybp were obtained in comparisons of AG-T with other populations (table 3). Using the lower rates of SSR evolution (1.55×10−4), we obtained using Thuillet et al. (2005) gives dates not earlier than 12 000 ybp for comparisons including AG-T. Upper bounds obtained for the MEX–USA divergence range up to 6400 ybp and are earlier than expected from the 1000–3000 ybp archaeological dates for maize in the United States (reviewed in Staller et al. 2006). All dates of population divergence are on the order expected for maize domestication in Mexico and cultivation in the Americas and none reflect the ancient origin U. maydis millions of years ago.
Table 3.
Bounds for pairwise divergence times (as ybp) estimated between five major U. maydis populations calculated with an average per locus mutation rate of 1.89×10−4 steps per year and assuming one generation per year.
| MEX | SA1 | SA2 | USA | |
|---|---|---|---|---|
| AG-T | 5200–9100 | 5500–12 200 | 8500–12 000 | 7500–11 100 |
| MEX | 1200–9500 | 1300–6400 | 2700–7800 | |
| SA1 | 1300–9100 | 2800–10 800 | ||
| SA2 | 3400–8200 | |||
| USA |
4. Discussion
We show that the domestication of maize in Mexico and later expansion of maize throughout the Americas had a dramatic and lasting impact on the evolution of corn smut populations. Although U. maydis interacted with its grass hosts over millions of years (Munkacsi et al. 2007), extant U. maydis populations date not much older than the time of maize domestication at 6000–10 000 ybp (reviewed in Staller et al. 2006). Furthermore, the results for dates of divergence, population structure and historical demographic processes show a history of U. maydis populations closely associated with that of maize domestication and early cultivation throughout the Americas.
Our results do not provide support for the null hypothesis that maize domestication had little impact on the genetic structure of U. maydis populations. The earliest population divergence times dated to only ca 10 000 ybp and were found in comparisons that included the Z. m. ssp. parviglumis-infecting AG-T population. While such recent dates might be obtained if U. maydis moved to the Z. mays subspecies from an alternate host soon after domestication, hosts other than the Z. mays subspecies are not described for U. maydis (Duran 1987). Homoplasy is unlikely to account for the recent dates we obtain. We calculated an SSR evolutionary rate of the order of 10−4 and dates we obtained are of the order of 103 years. To obtain dates closer to the origin of the species U. maydis, which are of the order of 107 years, the rate of evolution at the SSR loci would have to be approximately 10−8 steps per year, much too slow for most SSR loci (Estoup et al. 2002). Most importantly, the dates obtained with SSR loci for the USA and MEX populations are similar to those we obtained using sequence for the dsRNA virus in U. maydis (Voth et al. 2006). We infer that the distinct AG-T population is very likely an older, isolated and inbreeding population. With the recent decline and fragmentation of natural teosinte populations (Sánchez et al. 1998; Ruiz et al. 2001), other small, unique teosinte-infecting U. maydis populations must exist, and like AG-T could support little representation of ancient U. maydis variation. The population substructure by host subspecies demonstrated here could either represent adaptation to host differences since the time of domestication or a relict of ancestral variation, yet neither scenario pushes dates for the establishment of extant smut populations in Mexico to a time much older than the domestication.
The impact of maize domestication on U. maydis populations is further illustrated by the finding that MEX populations harbour no greater genetic diversity than do more recently founded, descendent USA, SA1 and SA2 populations, and similar to these descendents show evidence of population bottlenecks and recent expansion. Other pathosystems also display little correspondence of a species' geographical origin and diversity, implying that the early agricultural practices may often redistribute pathogen variation (Stukenbrock et al. 2006; Zaffarano et al. 2006). For example, Mexico is the centre of genetic diversity for Phytophthora infestans, late blight of potato (Goodwin 1997; Grunwald & Flier 2005) while genealogical results place the origin in the Andes with potatoes (Gomez-Alpizar et al. 2007). The contrast between results for diversity and origin points out that standing genetic diversity is strongly affected by recent demographic events and illustrates the challenge of relating the ancient history of crop pathogens to current coevolutionary dynamics. In the case of U. maydis, biological evidence that all extant hosts of U. maydis originated in Mexico places the ancient origin of U. maydis there (Munkacsi et al. 2007) whereas our results for demographic history demonstrate that domestication and early agriculture strongly affected both the overall levels of genetic variation and its geographical distribution.
We hypothesized that the intensity of modern agricultural practices and trade would largely obscure the impact of early maize domestication and cultivation in USA and South America. Nonetheless, against a background of low ongoing gene flow between the major populations, we inferred that the pathogen closely tracked maize as native peoples moved the new crop plant to South America (Piperno & Pearsall 1998; Staller et al. 2006) and more recently to the United States (Staller et al. 2006). In South America, two major populations, SA1 and SA2, each break up into three subpopulations, yet dates of divergence from MEX were estimated at only 1000–7800 ybp. If the SA1 and SA2 populations correspond to the two separate introductions of maize into South America (Frietas et al. 2003), then subpopulations within SA1 and SA2 must have independently adapted to high- and low-elevation regions over a relatively short time. For example, both subpopulations 4 and 5 belong to SA2, but lowland subpopulation 4 stretches from Brazil west to Bolivia and is geographically adjacent to the highland subpopulation 5 in the Bolivian Andes. Strikingly, subpopulations 2 (SA1), 3 (SA1) and 5 (SA2) range down the Andes but originated from within the two major populations. The geographical structure demonstrated in South American U. maydis subpopulations mirrors the patchwork nature of maize introductions (Piperno & Pearsall 1998; Frietas et al. 2003; Iriarte et al. 2004; Staller et al. 2006), and the apparent, rapid demographic shifts of U. maydis populations strongly parallel the intense selection and diversified use of maize.
In contrast to the quite varied structure of U. maydis populations in South America, the USA populations we sampled demonstrate the characteristics of recently founded and expanding populations, similar to other pathogen populations (Carbone & Kohn 2001; Brown & Hovmoller 2002; Banke & McDonald 2005; Stukenbrock et al. 2006; Brunner et al. 2007). However, our results provide a contrast to the human-mediated long-distance dispersal of fungal pathogens (citations above) and instead evidence for dates of divergence, strong differentiation from the MEX population and the presence of unique alleles suggest that USA populations were established at times coincident with the introduction of maize. At 2200–6400 ybp, the dates of divergence we obtained for USA from MEX were broader than we might have expected, but are similar to those of Voth et al. (2006) for the dsRNA of U. maydis (up to 3700 ybp). Interestingly, smut co-occurs with maize in archaeological remains from southwestern United States at 1500 ybp (Reinhard 2006) and smut spores co-occur with Iroquoian maize pollen at 500–800 ybp in a Canadian lake's sediments (McAndrews & Turton 2007). Considering that the MEX population also shows evidence of recent bottlenecks, we can also infer that ancestral USA populations were founded from a source more diverse than those in the extant MEX population. Variation at the b mating-type locus supports that inference as we readily found 20 b types in the United States and South America but not more than 13 in Mexico (G. May 2001, unpublished data). Together, the results for divergence dates and genetic structure provide evidence that extant U. maydis populations were founded from relatively few but diverse source populations and represent the historic imprints left behind by the expansion of maize agriculture into the United States.
We show that maize domestication and early cultivation enforced dramatic changes in the structure of U. maydis populations but without speciation demonstrated for other pathogens (Couch et al. 2005; Stukenbrock et al. 2007). What then determines the evolutionary response of pathogens to the human impact on host populations and environment? We propose a model for further investigation that the environmental disruption of emerging agricultural systems alters spatial structure of wild and domesticated plant populations such that acquired pathogens are likely to evolve virulence and specialize on the crop. The times and mechanisms by which crops acquire pathogens vary tremendously: at the centre of crop origin from the crop's progenitor species (Stukenbrock et al. 2007; this report) or other wild plants (Couch et al. 2005), or as a crop is moved into contact with wild pathogen populations (Zaffarano et al. 2006). Rather than the acquisition of pathogens per se, the most radical impact of altered spatial structure should be that gene flow between pathogen populations on the wild and crop plants will decrease, and breeding within the domesticated host population will increase (Garrett & Mundt 1999; Zhu et al. 2000). Further increases in the area of crop planting will result in expanding pathogen populations on domesticated hosts relative to those on wild hosts. Consequently, unlike crop plants themselves (Thuillet et al. 2005; Vigouroux et al. 2005), genetic diversity in crop pathogen populations will rebound from bottlenecks and support higher levels of genetic diversity than will the populations in surrounding wild plant populations. In addition to these demographic effects, agricultural practices may reduce resistance in host populations (Cohen 2000; Tishkoff et al. 2001) or reduce competition (Cohen 2000) and accelerate the evolution of specialized pathogens. For U. maydis, we speculate that historical demographic processes and human cultural practices for both the host (Eyre-Walker et al. 1998; Fukunaga et al. 2005; Wright et al. 2005; Staller et al. 2006) and the pathogen (Wilson 1987; Valverde et al. 1995; Iltis 2000) produced spatially structured populations by host and geography but that gene flow was sufficient to maintain virulence polymorphism (Thrall & Burdon 2003) and impede speciation of the maize-infecting U. maydis. More recent maize breeding in the twentieth century incorporated varied maize genetic backgrounds and numerous resistance loci of quantitative effect against U. maydis infection (Lubberstedt et al. 1998; Wisser et al. 2006; Baumgarten et al. 2007) and will have contributed to a relatively slow virulence evolution (McDonald & Linde 2002; Carbone & Kohn 2004). Our results demonstrate an immediate impact of crop domestication on pathogen demography and provide a model for the impact of historical events on the ongoing coevolutionary processes among hosts and pathogens.
Acknowledgments
Funding for this research was provided by the National Science Foundation Grant (DMS-0083468) to G.M., and research grants from the Minnesota Center for Community Genetics to A.B.M. We are grateful for U. maydis collections from Charles Barnes, Marc Cubeta, James Groth, Nick Lauter, Luis Narro, Jean Pan, Jesús Sánchez, Peter Tiffin and Peter Voth. We thank Sharon Jansa and Keith Barker for their valuable input on analyses, and Nick Lauter, Robert Zink and two anonymous reviewers for their comments on earlier versions of the manuscript. We thank past and present members of the May Lab for their enthusiastic discussions. The work partially fulfils requirements of the PhD by A.B.M. in the Plant Biological Sciences Graduate Program at the University of Minnesota; A.B.M. is indebted to his Thesis Committee (Peter Tiffin, George Weiblen, David McLaughlin, Sue Wick) for their contributions.
Supplementary Material
Sample collection information
References
- Agapow P.M, Burt A. Indices of multilocus linkage disequilibrium. Mol. Ecol. Notes. 2001;1:101–102. doi:10.1046/j.1471-8278.2000.00014.x [Google Scholar]
- Banke S, McDonald B.A. Migration patterns among global populations of the pathogenic fungus Mycosphaerella graminicola. Mol. Ecol. 2005;14:1881–1896. doi: 10.1111/j.1365-294X.2005.02536.x. doi:10.1111/j.1365-294X.2005.02536.x [DOI] [PubMed] [Google Scholar]
- Barnes C.W, Szabo L.J, May G, Groth J.V. Inbreeding levels of two Ustilago maydis populations. Mycologia. 2004;96:1236–1244. [PubMed] [Google Scholar]
- Baumgarten, A. M. 2004 Positions of U. maydis QTLs, NBS-LRR genes, and defense genes within the maize genome. In Graduate program in plant biological sciences St Paul, MN: University of Minnesota.
- Baumgarten A.M, Suresh J, May G, Phillips R.L. Mapping QTLs contributing to Ustilago maydis resistance in specific plant tissues of maize. Theor. Appl. Genet. 2007;114:1229–1238. doi: 10.1007/s00122-007-0513-5. doi:10.1007/s00122-007-0513-5 [DOI] [PubMed] [Google Scholar]
- Bilgin R. Kgtests: a simple Excel macro program to detect signatures of population expansion using microsatellites. Mol. Ecol. Notes. 2007;7:416–417. doi:10.1111/j.1471-8286.2006.01671.x [Google Scholar]
- Brown J.K.M, Hovmoller M.S. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science. 2002;297:537–541. doi: 10.1126/science.1072678. doi:10.1126/science.1072678 [DOI] [PubMed] [Google Scholar]
- Brunner P.C, Schurch S, McDonald B.A. The origin and colonization history of the barley scald pathogen Rhynchosporium secalis. J. Evol. Biol. 2007;20:1311–1321. doi: 10.1111/j.1420-9101.2007.01347.x. doi:10.1111/j.1420-9101.2007.01347.x [DOI] [PubMed] [Google Scholar]
- Carbone I, Kohn L. A microbial population–species interface: nested cladistic and coalescent inference with multilocus data. Mol. Ecol. 2001;10:947–964. doi: 10.1046/j.1365-294x.2001.01244.x. doi:10.1046/j.1365-294X.2001.01244.x [DOI] [PubMed] [Google Scholar]
- Carbone, I. & Kohn, L. 2004 Inferring process from pattern in fungal population genetics. In Applied mycology and biotechnology, vol. 4 (eds D. K. Arora & G. G. Khachatourians), pp. 29–58. Amsterdam, The Netherlands: Elsevier.
- Cohen M.L. Changing patterns of infectious disease. Nature. 2000;406:762–767. doi: 10.1038/35021206. doi:10.1038/35021206 [DOI] [PubMed] [Google Scholar]
- Cornuet J.M, Luikart G. Description and power analysis for two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch B.C, Fudal I, Lebrun M.-H, Tharreau D, Valent B, Kim P.v, Notteghem J.-L, Kohn L.M. Origins of host-specific populations of the blast pathogen Magnaporthe oryzae in crop domestication with subsequent expansion of pandemic clones on rice and weeds of rice. Genetics. 2005;170:613–630. doi: 10.1534/genetics.105.041780. doi:10.1534/genetics.105.041780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J.F, Gaut B.S, Smith B.D. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. doi:10.1016/j.cell.2006.12.006 [DOI] [PubMed] [Google Scholar]
- Duran R. Washington State University; Pullman, WA: 1987. Ustilaginales of Mexico: taxonomy, symptomatology, spore germination, and basidial cytology. [Google Scholar]
- Estoup A, Jarne P, Cornuet J.-M. Homoplasy and mutation model at microsatellite and their consequences for population genetic analyses. Mol. Ecol. 2002;11:1591–1604. doi: 10.1046/j.1365-294x.2002.01576.x. doi:10.1046/j.1365-294X.2002.01576.x [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut G, Goudet J. Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. doi:10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Ewens W.J. The sampling theory of selectively neutral alleles. Theor. Popul. Biol. 1972;3:87–112. doi: 10.1016/0040-5809(72)90035-4. doi:10.1016/0040-5809(72)90035-4 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Gaut R.L, Hinton H, Feldman D.L, Gaut B. Investigation of the bottleneck leading to the domestication of maize. Proc. Natl Acad. Sci. USA. 1998;95:441–446. doi: 10.1073/pnas.95.8.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frietas F.O, Bendel G, Allaby R.G, Brown T.A. DNA from primitive maize landraces and archaeological remains: implications for the domestication of maize and its expansion into South America. J. Archaeol. Sci. 2003;30:901–908. doi:10.1016/S0305-4403(02)00269-8 [Google Scholar]
- Fukunaga K, Hill J, Vigouroux Y, Matsuoka Y, Sanchez J, Liu J, Buckler E.S, Doebley J. Genetic diversity and population structure of teosinte. Genetics. 2005;93:382–390. doi: 10.1534/genetics.104.031393. doi:10.1534/genetics.104.031393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett K.A, Mundt C.C. Epidemiology in mixed host populations. Phytopathology. 1999;89:984–990. doi: 10.1094/PHYTO.1999.89.11.984. doi:10.1094/PHYTO.1999.89.11.984 [DOI] [PubMed] [Google Scholar]
- Goldstein D.B, Linares A.R, Cavelli-Sforza L.L, Feldman M.W. Genetic absolute dating based on microsatellites and the origin of modern humans. Proc. Natl Acad. Sci. USA. 1995;92:6723–6727. doi: 10.1073/pnas.92.15.6723. doi:10.1073/pnas.92.15.6723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Alpizar L, Carbone I, Ristaino J.B. An Andean origin of Phytophthora infestans inferred from mitochondrial and nuclear gene genealogies. Proc. Natl Acad. Sci. USA. 2007;104:3306–3311. doi: 10.1073/pnas.0611479104. doi:10.1073/pnas.0611479104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S.B. The population genetics of Phytophthora. Phytopathology. 1997;87:462–473. doi: 10.1094/PHYTO.1997.87.4.462. doi:10.1094/PHYTO.1997.87.4.462 [DOI] [PubMed] [Google Scholar]
- Grunwald N.J, Flier W.G. The biology of Phytophthora infestans at its center of origin. Annu. Rev. Phytopathol. 2005;43:171–190. doi: 10.1146/annurev.phyto.43.040204.135906. doi:10.1146/annurev.phyto.43.040204.135906 [DOI] [PubMed] [Google Scholar]
- Iltis H. Homeotic sexual translocations and the origins of maize (Zea mays, Poacaea): a new look at an old problem. Econ. Bot. 2000;54:7–42. [Google Scholar]
- Iriarte J, Holst I, Marozzi O, Listopad C, Alonso E, Riderknecht A, Montan J. Evidence for cultivar adoption and emerging complexity during the middle Holocen in La Plata basin. Nature. 2004;432:614–617. doi: 10.1038/nature02983. doi:10.1038/nature02983 [DOI] [PubMed] [Google Scholar]
- Kolmer J.A, Long D.L, Hughes M.E. Physicologic specialization of Puccinia triticina on wheat in the United States. Plant Dis. 2004;8:1079–1084. doi: 10.1094/PDIS.2004.88.10.1079. doi:10.1094/PDIS.2004.88.10.1079 [DOI] [PubMed] [Google Scholar]
- Lubberstedt T, Klein D, Melchinger A. Comparative QTL mapping of resistance to Ustilago maydis across four populations of European flint-maize. Theor. Appl. Genet. 1998;97:1321–1330. doi:10.1007/s001220051025 [Google Scholar]
- Matsuoka Y, Vigouroux Y, Goodman M.M, Sanchez J, Buckler E, Doebley J. A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl Acad. Sci. USA. 2002;99:6080–6084. doi: 10.1073/pnas.052125199. doi:10.1073/pnas.052125199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAndrews J.H, Turton C. Canada geese dispersed cultigen pollen grains from prehistoric Iroquoian fields to Crawford Lake, Ontario. Palynology. 2007;31:9–18. [Google Scholar]
- McDonald B.A, Linde C. Pathogen population genetics, evolutionary potential and durable resistance. Annu. Rev. Phytopathol. 2002;40:349–379. doi: 10.1146/annurev.phyto.40.120501.101443. doi:10.1146/annurev.phyto.40.120501.101443 [DOI] [PubMed] [Google Scholar]
- Munkacsi A.B, Kawakami S, Pan J.J, Lee K, Stoxen S, Hang J, May G. Genome-wide asessment of tandem repeat markers for biogeographical analyses of the corn smut fungus, Ustilago maydis. Mol. Ecol. Notes. 2006;6:221–223. doi:10.1111/j.1471-8286.2005.01199.x [Google Scholar]
- Munkacsi A.B, Stoxen S, May G. Domestication of maize, sorghum, and sugarcane did not drive the domestication of their smut pathogens. Evolution. 2007;61:388–403. doi: 10.1111/j.1558-5646.2007.00036.x. doi:10.1111/j.1558-5646.2007.00036.x [DOI] [PubMed] [Google Scholar]
- Nei M. Columbia University Press; New York, NY: 1987. Molecular evolutionary genetics. [Google Scholar]
- Piperno D.R, Pearsall D.M. Academic Press; San Diego, CA: 1998. The origins of agriculture in the lowland neotropics. [Google Scholar]
- Pritchard J.K, Stephens P, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D.E, Feldman M.W, Goldstein D.B. Statistical properties of two tests that use multilocus data sets to detect population expansions. Mol. Biol. Evol. 1999;16:453–466. [Google Scholar]
- Reinhard K.J. A coprological view of ancestral Pueblo cannibalism. Am. Sci. 2006;94:254–261. doi:10.1511/2006.3.254 [Google Scholar]
- Roelfs A.P, Groth J.V. A comparison of virulence phenotypes in wheat stem rust populations reproducing sexually and asexually. Phytopathology. 1980;70:855–862. [Google Scholar]
- Ruiz J.A.C, Sanchez J.J.G, Aguilar M.S. Potential geographical distribution of teosinte in Mexico: a GIS approach. Maydica. 2001;46:105–110. [Google Scholar]
- Sánchez, G. J. J., Kato, Y. T. A., Aguilar, S. M., Hernandez, C. J. M., Lopez, A. R. & Ruiz, J. A. C. 1998 Distribución y caracterización del teosintle. Libro Técnico no. 2 INIFAP-CIRPAC. Ed. Conexión Gráfica, Guadalajara, Jalisco, México.
- Schneider S, Roessli D, Excoffier L. Genetics and Biometry Laboratory, University of Geneva; Geneva, Switzerland: 2000. Arlequin. [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B.D. Scientific American Library; New York, NY: 1995. The emergence of agriculture. [Google Scholar]
- Staller J, Tykot R, Benz B.F. Elsevier; New York, NY: 2006. Histories of maize: multidisciplinary approaches to the prehistory, linguistics, biogeography, domestication, and evolution of maize. [Google Scholar]
- Stoll M, Begerow D, Oberwinkler F. Molecular phylogeny of Ustilago, Sporisorium, and related taxa based on combined analyses of rDNA sequences. Mycol. Res. 2005;109:342–356. doi: 10.1017/s0953756204002229. doi:10.1017/S0953756204002229 [DOI] [PubMed] [Google Scholar]
- Stukenbrock E.H, Banke S, McDonald B.A. Global migration patterns in the fungal wheat pathogen, Phaeosphaeria nodorum. Mol. Ecol. 2006;15:2895–2904. doi: 10.1111/j.1365-294X.2006.02986.x. doi:10.1111/j.1365-294X.2006.02986.x [DOI] [PubMed] [Google Scholar]
- Stukenbrock E.H, Banke S, Javan-Nikkhah M, McDonald B.A. Origin and domestication of the fungal wheat pathogen Mycosphaerella graminicola via sympatric speciation. Mol. Biol. Evol. 2007;24:398–411. doi: 10.1093/molbev/msl169. doi:10.1093/molbev/msl169 [DOI] [PubMed] [Google Scholar]
- Thrall P.H, Burdon J.J. Evolution of virulence in a plant host–pathogen metapopulation. Science. 2003;299:1735–1737. doi: 10.1126/science.1080070. doi:10.1126/science.1080070 [DOI] [PubMed] [Google Scholar]
- Thuillet A.-C, Bataillon T, Poirier S, Santoni S, David J.L. Estimation of long-term effective population sizes through the history of durum wheat using microsatellite data. Genetics. 2005;169:1589–1599. doi: 10.1534/genetics.104.029553. doi:10.1534/genetics.104.029553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff S.A, et al. Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science. 2001;293:455–462. doi: 10.1126/science.1061573. doi:10.1126/science.1061573 [DOI] [PubMed] [Google Scholar]
- Valdes A.M, Slatkin M, Freimer N.B. Allele frequencies at microsatellite loci: the stepwise mutation model revisited. Genetics. 1993;133:737–749. doi: 10.1093/genetics/133.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde M.E, Paredes-Lopez O, Pataky J.K, Guevara-Lara F. Huitlacoche (Ustilago maydis) as a food source—biology, composition, and production. Crit. Rev. Food Sci. Nutr. 1995;35:191–229. doi: 10.1080/10408399509527699. [DOI] [PubMed] [Google Scholar]
- Vigouroux Y, Mitchell S, Matsuoka Y, Hamblin M, Kresovich S, Smith J.S.C, Jaqueth J, Smith O.S, Doebley J. An analysis of genetic diversity across the maize genome using microsatellites. Genetics. 2005;169:1617–1630. doi: 10.1534/genetics.104.032086. doi:10.1534/genetics.104.032086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth P.D, Mairura L, Lockhart B.E, May G. Phylogeography of Ustilago maydis virus H1 in the USA and Mexico. J. Gen. Virol. 2006;87:3433–3441. doi: 10.1099/vir.0.82149-0. doi:10.1099/vir.0.82149-0 [DOI] [PubMed] [Google Scholar]
- Watterson G.A. The homozygosity test of neutrality. Genetics. 1978;88:405–417. doi: 10.1093/genetics/88.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. Minnesota Historical Society Press; Saint Paul, MN: 1987. Buffalo bird woman's garden: agriculture of the Hidatsa Indians. [Google Scholar]
- Wilson G.A, Rannala B. Bayesian inference of recent migration rates using multilocus genotypes. Genetics. 2003;163:1177–1191. doi: 10.1093/genetics/163.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisser R.J, Balint-Kurti P.J, Nelson R.J. The genetic architecture of disease resistance in maize: a synthesis of published studies. Phytopathology. 2006;96:120–129. doi: 10.1094/PHYTO-96-0120. doi:10.1094/PHYTO-96-0120 [DOI] [PubMed] [Google Scholar]
- Wright S.I, Bi I.V, Schroeder S.G, Yamasaki M, Doebley J.F, McMullen M.D, Gaut B.S. The effects of artificial selection on the maize genome. Science. 2005;308:1310–1314. doi: 10.1126/science.1107891. doi:10.1126/science.1107891 [DOI] [PubMed] [Google Scholar]
- Zaffarano P.L, McDonald B.A, Zala M, Linde C.C. Global hierarchical gene diversity analysis suggests the fertile crescent is not the center of origin of the barley scald pathogen Rhynchosporium secalis. Phytopathology. 2006;96:941–950. doi: 10.1094/PHYTO-96-0941. doi:10.1094/PHYTO-96-0941 [DOI] [PubMed] [Google Scholar]
- Zhivotovsky L.A. Estimating divergence time with the use of microsatellite genetic distances: impacts of population growth and gene flow. Mol. Biol. Evol. 2001;18:700–709. doi: 10.1093/oxfordjournals.molbev.a003852. [DOI] [PubMed] [Google Scholar]
- Zhu Y, et al. Genetic diversity and disease control in rice. Nature. 2000;406:719–722. doi: 10.1038/35021046. doi:10.1038/35021046 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample collection information