Abstract
Human deoxyribonucleoside kinases are required for the pharmacological activity of several clinically important anticancer and antiviral nucleoside analogs. Human deoxycytidine kinase and thymidine kinase 1 are described as cytosolic enzymes in the literature, whereas human deoxyguanosine kinase and thymidine kinase 2 are believed to be located in the mitochondria. We expressed the four human deoxyribonucleoside kinases as fusion proteins with the green fluorescent protein to study their intracellular locations in vivo. Our data showed that the human deoxycytidine kinase is located in the cell nucleus and the human deoxyguanosine kinase is located in the mitochondria. The fusion proteins between green fluorescent protein and thymidine kinases 1 and 2 were both predominantly located in the cytosol. Site-directed mutagenesis of a putative nuclear targeting signal, identified in the primary structure of deoxycytidine kinase, completely abolished nuclear import of the protein. Reconstitution of a deoxycytidine kinase-deficient cell line with the wild-type nuclear or the mutant cytosolic enzymes both restored sensitivity toward anticancer nucleoside analogs. This paper reports that a deoxyribonucleoside kinase is located in the cell nucleus and we discuss the implications for deoxyribonucleotide synthesis and phosphorylation of nucleoside analogs.
DNA replication and repair require synthesis of deoxyribonucleotides. During the S-phase of the cell cycle, deoxyribonucleotides are mainly synthesized de novo in the cytosol via reduction of ribonucleotides by the cell cycle-regulated ribonucleotide reductase (1, 2). Deoxyribonucleotides can also be synthesized via phosphorylation of deoxyribonucleosides by deoxyribonucleoside kinases, both in dividing and resting cells (1). Biochemical studies show that there are four distinct human deoxyribonucleoside kinases (3). Deoxycytidine kinase (dCK), deoxyguanosine kinase (dGK), and thymidine kinase 2 (TK2) are constitutively expressed throughout the cell cycle and are closely related members of an enzyme family (3, 4). The fourth deoxyribonucleoside kinase, thymidine kinase 1 (TK1), is not sequence-related to the other enzymes and is strictly S-phase-regulated (3). Like ribonucleotide reductase, dCK and TK1 are described in the literature as cytosolic enzymes (3, 5, 6). dGK and TK2 are both believed to be located in the mitochondria (5–7). The physiological importance of the constitutively expressed deoxyribonucleoside kinases are suggested to be in providing nondividing cells with deoxyribonucleotides for DNA repair and mitochondrial DNA replication.
We have recently cloned the cDNAs that encode human dGK and TK2 (4, 8). The cDNAs that encode all four known human deoxyribonucleoside kinases are thereby cloned. Deoxyribonucleoside kinases are pharmacologically important because they phosphorylate several nucleoside analogs. dCK has been carefully characterized because it phosphorylates clinically important anticancer and antiviral nucleoside analogs in addition to deoxycytidine, deoxyadenosine, and deoxyguanosine. The nucleoside analogs phosphorylated by dCK include 1-β-d-arabinofuranosylcytosine (araC), 9-β-d-arabinofuranosyladenine, and 2-chloro-2′-deoxyadenosine (CdA), which are commonly used in the treatment of hematological malignancies (9), and 2′,2′-difluorodeoxycytidine (dFdC), which is active against several solid malignant tumors (10). The nucleoside analogs are inactive pro-drugs that are dependant on intracellular phosphorylation by dCK for their pharmacological activities.
The mitochondrial deoxyribonucleoside kinases dGK and TK2 phosphorylate several nucleoside analogs in vitro (7, 11). Little is known, however, about the contribution of the mitochondrial deoxyribonucleoside kinases for deoxyribonucleotide synthesis or for the pharmacological effects of nucleoside analogs. The N-terminal part of the predicted primary structure of human dGK contains a sequence motif similar to a mitochondrial import signal (8). We were, however, unable to find a similar motif in the predicted amino acid sequence of TK2 (4). To determine the true subcellular location of the human deoxyribonucleoside kinases, we decided to express the enzymes as fusion proteins with the green fluorescent protein (GFP) from Aequorea victoria in mammalian cells. The GFP is used as a tool to study protein targeting to both the mitochondria and the nucleus in vivo (12, 13). The main finding of these experiments was that the fusion protein between dCK and GFP was not located in the cytosol as expected, but located almost exclusively in the cell nucleus. We further identified a nuclear import signal in the primary structure of human dCK and showed that the signal was required for nuclear import of the protein.
EXPERIMENTAL PROCEDURES
Construction of Plasmids.
We used the pEGFP-N1 plasmid vector (CLONTECH) to express the human deoxyribonucleoside kinases as fusion proteins with the GFP in mammalian cells. The plasmid encodes the red-shifted S65T mutant of GFP (14). It also contains a neomycin resistance gene for selection of stable transfected cells. We designed oligonucleotide primers flanking the ORF sequences of the four human deoxyribonucleoside kinases dCK, dGK, TK1, and TK2 (4, 8, 15, 16). Restriction enzyme sites for EcoRI and SalI were inserted in the 5′ part of the primers for cloning of the cDNA fragments into the EcoRI–SalI site of the pEGFP-N1 plasmid vector. The oligonucleotides were used in a PCR with plasmid cDNA clones of the deoxyribonucleoside kinases as templates. The PCR products were cloned into the pGEM-T plasmid vector (Promega). These plasmids were subsequently restriction-digested with EcoRI and SalI (Promega), and the cDNA fragments were ligated into the pEGFP-N1 plasmid. Site-directed mutagenesis of the dCK–GFP fusion was done with an extended oligonucleotide in the 5′ region (5′-TCGAATTCATGGCCACCCC GCCCAACGGAAGCTGCCC). All plasmids were propagated in the DH5α Escherichia coli strain and the plasmids were purified using the midi-prep kit (Qiagen, Chatsworth, CA). The plasmid vectors containing the fusions between the cDNA encoding the human deoxyribonucleoside kinases and the cDNA encoding GFP were checked by DNA sequence determinations (Automatic Laser Fluorescence sequencer, Pharmacia).
Culture and Transfection of Cell Lines.
The dCK-deficient CHO cell line (17) was a gift from W. Plunkett. The human melanoma cell line G361 and the human osteosarcoma cell line were gifts from J. Balzarini (Rega Institute, Belgium). The CHO cells were cultured in McCoy 5A modified medium and the human cell lines were cultured in DMEM. The cell culture medium was supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 0.1 mg/ml streptomycin. All cell lines were transfected using LipofectAmine (GIBCO/BRL). Plasmid DNA (1 μg) and 9 μl of liposomes dissolved in OPTI-MEM medium (GIBCO/BRL) were used for each transfection according to the GIBCO protocol.
Detection of GFP with Fluorescence Microscopy.
The culture plates with transfected cells were washed in PBS 24–48 h after transfection. Coverslips were placed directly onto the culture plates. Microscopy was performed on a Nikon Optiphot microscope with epifluorescence illumination. GFP fluorescence was observed in the living cells with a Nikon B1-A fluorescein isothiocyanate filter cube (470–490 nm excitation filter, 510 nm emission filter) and a ×40 Nikon Fluor 40/1.30 oil-immersion objective lens. Cells were photographed with Kodak Ektachrome ASA 400 film.
Stable Transfection of Cell Lines and Clonogenicity Assays.
dCK-deficient CHO cells were transfected with plasmids encoding the GFP, dCK–GFP, and mutdCK–GFP, as described above. The cells were subcultured 1:5 3 days after transfection and 0.9 mg/ml Geneticin (GIBCO/BRL) was added to the cell culture medium. The cells were cultured in the presence of Geneticin for 3 weeks. More than 90% of the cells showed green fluorescence. The stably transfected cells were plated in 25 cm2 flasks at ≈104 cells/flask. araC (Sigma), CdA (Sigma), or dFdC (Eli Lilly) were added to the indicated concentrations. After 3 days, the number of green fluorescent cell colonies in 1.0 cm2 with >15 cells was determined by fluorescence microscopy as described above.
RESULTS
Intracellular Locations of Human Deoxyribonucleoside Kinases Fused to GFP.
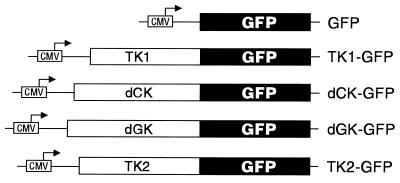
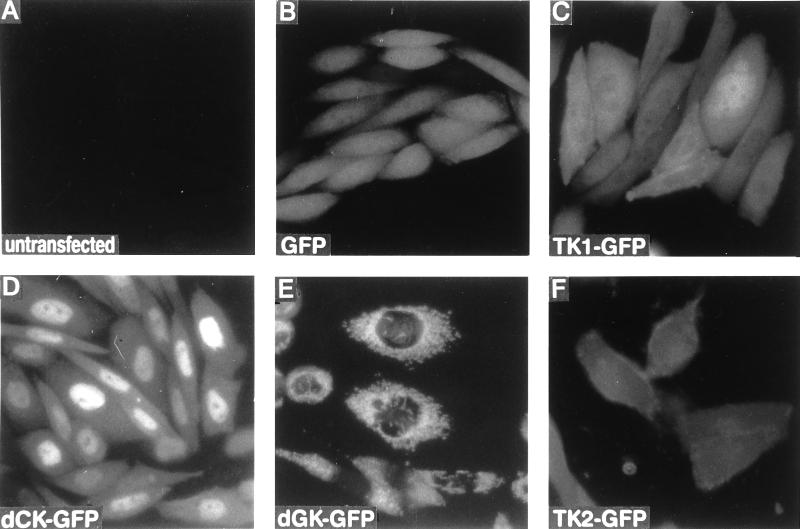
We constructed plasmid vectors to express the human deoxyribonucleoside kinases as fusion proteins to the GFP (Fig. 1). The deoxyribonucleoside kinases were inserted at the N-terminal part of the GFP, because mitochondrial targeting requires an intact N-terminal sequence. The plasmids were transiently transfected into CHO cells and the fluorescent fusion proteins detected by fluorescence microscopy (Fig. 2). Untransfected cells showed no fluorescence. The cells transfected with the plasmid encoding GFP showed, as expected, fluorescence in both the cytosol and the nucleus of the cells. The 27-kDa molecular mass of the monomeric GFP allows it to passively diffuse between the cytosol and nucleus. The fusion between TK1 and GFP showed predominant fluorescence in the cytosol with low levels in the cell nucleus. The dCK–GFP fusion showed strong fluorescence in the nucleus and weak fluorescence in the cytosol. dGK-GFP showed an irregular fluorescent pattern around a nonfluorescent nucleus, identical to the pattern described for mitochondrial location of the GFP (12, 13). TK2–GFP showed mainly cytosolic fluorescence with a fluorescent border outlining the cell membrane.
Figure 1.
Plasmids constructed to express the four human deoxyribonucleoside kinases as fusion proteins with the GFP. CMV, cytomegalovirus promoter.
Figure 2.
Fluorescence microscopy images of CHO cells transfected with the plasmids encoding the human deoxyribonucleoside kinases as fusion proteins with the GFP. (A) Untransfected cells. (B) GFP. (C) TK1–GFP. (D) dCK–GFP. (E) dGK–GFP. (F) TK2–GFP.
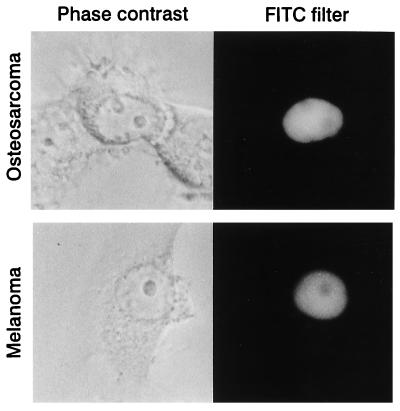
To verify the unexpected finding that the dCK–GFP fusion protein was imported into the nucleus, we decided to test if it occurred in other than CHO cells. We transfected a human osteosarcoma and a human melanoma cell line with the dCK–GFP plasmid. Both cell lines showed strong fluorescence in the cell nucleus with no fluorescence in the cytosol (Fig. 3). The nucleoli were visible and nonfluorescent in all the transfected cell lines, indicating that dCK–GFP was located inside the cell nucleus and not associated only with the nuclear membrane. The TK1–GFP, dGK–GFP, and TK2–GFP plasmids were also transfected into the osteosarcoma and melanoma cell lines with similar results as described above (data not shown).
Figure 3.
Phase contrast and fluorescence microscopy images of human osteosarcoma and human melanoma cell lines transfected with the dCK–GFP plasmid. Both cell lines showed strong GFP fluorescence in the nucleus with no fluorescence in the cytosol.
Identification and Mutagenesis of the dCK Nuclear Import Signal.
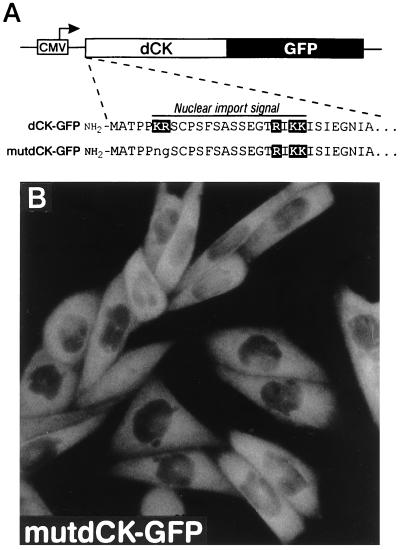
Proteins imported into the cell’s nucleus contain a signal for nuclear import (18). Nuclear targeting signals usually consist of either one or two clusters of positively charged amino acid residues. A bipartite signal motif comprising 2 basic amino acids, a spacer region of any 10 or 11 amino acids, and a basic cluster in which 3 of the next 5 amino acids must be basic, is present in many nuclear proteins (19). An identical motif was identified in the N-terminal region of dCK at amino acids 6–23 (Fig. 4A). We decided to mutate the putative nuclear targeting signal of human dCK and to determine the intracellular location of the mutant protein (mutdCK–GFP). The two N-terminally located arginine residues were changed to noncharged amino acids by site-directed mutagenesis (Fig. 4A). CHO cells transfected with the mutdCK–GFP plasmid showed strong fluorescence in the cytosol but no fluorescence in the nucleus (Fig. 4B). In summary, we conclude that the human dCK contains a nuclear targeting signal and that the bipartite motif in the N-terminal region is required for the nuclear import of dCK.
Figure 4.
Identification and mutagenesis of the bipartite nuclear import signal of human dCK. (A) The N-terminal sequence of the wild-type and mutated human dCK. Positively charged amino acid residues are shown in black boxes. The putative nuclear import signal is located as indicated. The mutated amino acid residues are shown in lower case letters. (B) CHO cells transfected with the plasmid encoding the mutant dCK fused to GFP.
Nucleoside Analog Sensitivity of Cells Expressing dCK in the Nucleus and the Cytosol.
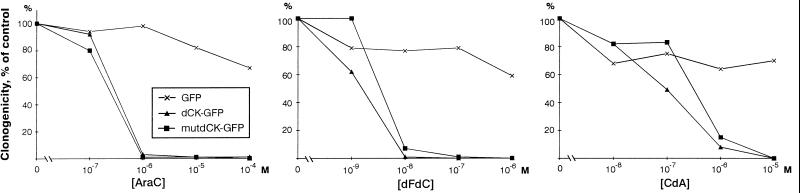
We decided to determine whether the nuclear location of human dCK was important for the cytotoxic effect of nucleoside analogs. We stably transfected dCK-deficient CHO cells with plasmids that encoded either the GFP, the wild-type nuclear dCK–GFP, or the cytosolic mutant of dCK–GFP. A clonogenicity assay was used to determine the sensitivity of the three cell lines for the nucleoside analogs araC, dFdC, and CdA (Fig. 5). The dCK-deficient CHO cells transfected with only GFP were not sensitive to the nucleoside analogs within the tested concentration range. The cells expressing dCK–GFP in the nucleus were sensitive to all three tested nucleoside analogs. The cell line expressing the mutant cytosolic dCK–GFP was also sensitive to the nucleoside analogs, at similar concentrations as the cells expressing the wild-type dCK–GFP.
Figure 5.
Sensitivity of dCK-deficient CHO cells expressing either GFP, dCK–GFP, or the mutant cytosolic dCK–GFP for the nucleoside analogs AraC, CdA, and dFdC.
DISCUSSION
The main finding of this report is that human “cytosolic” dCK is located in the cell nucleus. This implies that phosphorylation of deoxyribonucleosides and nucleoside analogs by dCK to the corresponding nucleoside monophosphates occurs in the nucleus. The deoxyribonucleoside monophosphates must, however, be further phosphorylated to deoxyribonucleoside triphosphates to be incorporated into DNA. The pharmacological activities of most nucleoside analogs are also dependant on phosphorylation to the triphosphate forms. Human cells contain several nucleoside monosphosphate kinases with different substrate specificities (1). Adenylate kinases are located both in the cytosol and the mitochondria (20), but the subcellular locations of the other nucleoside monosphate kinases have not been determined. It is, therefore, presently not known if the cell nucleus contains all enzymes required for the salvage of deoxycytidine, or if dCMP is further phosphorylated in the cytosol. The third phosphorylation step is catalyzed by nucleoside diphosphate kinases (1). Several human nucleoside diphosphate kinases, encoded by the nm23 gene family, are cloned and characterized (21). Nucleoside diphosphate kinase type A and B are both mainly located in the cytosol, but there are reports that these enzymes are present in the nucleus (22, 23). The nucleoside diphosphate kinases also phosphorylate deoxyribonucleotides synthesized de novo, because reduction of ribonucleotides occurs at the diphosphate level in mammalian cells. Among the enzymes of the de novo pathway, only ribonucleotide reductase and thymidylate synthase have been investigated for their subcellular locations. Ribonucleotide reductase was suggested to be located in the cell nucleus (24), but subsequent biochemical and immunocytochemical studies demonstrate that the enzyme is exclusively located in the cytosol (2, 25). A study with two monoclonal anti-thymidylate synthase antibodies shows that thymidylate synthase is located in the cytosol (26). A recent report on immunogold electron microscopy with a polyclonal antibody, however, indicates that the enzyme is present both in the cytosol and the nucleus (27). Although suggested to be located in the nucleus, neither thymidylate synthase nor the nucleoside diphosphate kinases contain known nuclear import signal sequences.
Enzymes involved in both the de novo synthesis of deoxyribonucleotides and DNA replication have been postulated to form a nuclear multienzyme complex. This enzyme complex, known as the “replitase,” was reported to contain ribonucleotide reductase, and to channel de novo synthesised deoxyribonucleotides to DNA replication (24). The replitase has been used to explain the apparent poor incorporation of salvaged dCTP as compared with de novo synthesized dCTP into DNA (28). The existence of a nuclear replitase enzyme complex in human cells is, however, very doubtful. De novo synthesis of deoxyribonucleotides requires ribonucleotide reductase and there is compelling evidence that this enzyme is located exclusively in the cytosol as discussed above (2, 25). In addition, recent isotope labeling experiments clearly show that dCTP synthesized de novo rapidly equilibrates with dCTP produced by phosphorylation of deoxycytidine (29). Deoxyribonucleotides from both pathways are incorporated into DNA with equal efficiency during the S-phase of the cell cycle (29). There are accordingly no apparent differences in utilization of deoxyribonucleotides synthesised de novo or by salvage of deoxyribonucleosides, in spite of the different subcellular locations of the enzymes involved. We showed in this report that dCK-negative cell lines reconstituted with either wild-type nuclear dCK or mutant cytosolic dCK were equally sensitive toward dCK phosphorylated nucleoside analogs. If dCK phosphorylated deoxyribonucleotides were compartmentalized within the cell nucleus because of the nuclear location of dCK, the cells that expressed wild-type nuclear dCK would be expected to be more sensitive to the nucleoside analogs than the cells that expressed dCK in the cytosol. Our data suggest that nucleotides are freely distributed between the cytosol and the nucleus. This is in accordance with studies demonstrating that the nuclear membrane contains protein pores that facilitate diffusion of metabolites across the membrane (18). Because there is no evidence that the nucleus and the cytosol have separate deoxyribonucleotide pools, we cannot presently explain, from a physiological point of view, why dCK is located in the nucleus.
The evidence we provide in this report, that human dCK is located in the cell nucleus, is based on the nuclear import of the artificial fusion protein of dCK and GFP. We believe that the native dCK protein without GFP also is located in the cell nucleus, because a nuclear import signal was identified in the dCK protein. The fusion protein of TK1 and GFP was mainly located in the cytosol in accordance with immunocytochemical studies (30). Low levels of TK1–GFP fluorescence was, however, also detected in the cell nucleus. It is possible that the fusion of TK1 with GFP disrupts the multimerization of the four TK1 subunits, which would lower the molecular mass of the protein to allow it to enter the nucleus without a nuclear import signal (31). TK2 is, in the literature, generally described as a mitochondrial enzyme, although TK2 activity has been reported to be present in the cytosol (3, 32). A mitochondrial location of TK2 is supported by experiments that show exclusive salvage of thymidine to mitochondrial DNA in TK1-deficient cell lines (6). We have recently cloned a cDNA that encodes an enzyme with similar biochemical properties as described for native TK2 (4). In contrast to the mitochondrial human dGK (8), we were not able to identify a mitochondrial import signal in the predicted primary structure of TK2, and the TK2–GFP fusion protein was not targeted to the mitochondria. A possible explanation of the discrepancy may be that we have cloned and studied an alternatively spliced cDNA isoform of human TK2 that lacks the N-terminal mitochondrial targeting sequence. We are currently generating antibodies toward recombinant TK2 to determine the true intracellular location of the native protein.
In contrast to the nucleotide-permeable nuclear membrane, the inner mitochondrial membrane is impermeable to most hydrophilic and charged molecules. No carrier protein for transport of deoxyribonucleotides between the mitochondrial matrix and the cytosol has, so far, been identified in human cells, and there is evidence that the mitochondrial deoxyribonucleotide pools are physically separated from the cytosolic pools (6, 33). Several nucleoside analogs interfere with mitochondrial DNA replication, which results in severe delayed adverse effects (34). Phosphorylation of these nucleoside analogs by mitochondrial deoxyribonucleoside kinases and subsequent inhibition of the mitochondrial DNA polymerase γ are implicated as determinants in the pathogenesis of the nucleoside analog toxicity. Nucleoside analogs phosphorylated by dCK do, surprisingly, also damage mitochondrial DNA (35). The data reported in this paper will serve as the basis for the construction of artificial human deoxyribonucleoside kinases that are targeted to either the cytosol or the mitochondria. These genetically engineered enzymes will be used as tools to study the importance of mitochondrial compartmentalization of the human deoxyribonucleoside kinases for both therapeutic and toxic effects of nucleoside analogs.
Acknowledgments
This work was supported by grants from the Swedish Medical Research Council, the Biomedical Research Programme and the Human Capital and Mobility Programme of the European Community, the Harald and Greta Jeansson Foundation, and the Robert Lundberg Foundation.
ABBREVIATIONS
- dCK
deoxycytidine kinase
- dGK
deoxyguanosine kinase
- TK1
thymidine kinase 1
- TK2
thymidine kinase 2
- GFP
green fluorescent protein
- araC
1-β-d-arabinofuranosylcytosine
- CdA
2-chloro-2′-deoxyadenosine
- dFdC
2′,2′-difluorodeoxycytidine
- CHO
Chinese hamster ovary
References
- 1.Reichard P. Annu Rev Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- 2.Engström Y, Rozell B. EMBO J. 1988;7:1615–1620. doi: 10.1002/j.1460-2075.1988.tb02987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnér E S J, Eriksson S. Pharmacol Ther. 1995;67:155–186. doi: 10.1016/0163-7258(95)00015-9. [DOI] [PubMed] [Google Scholar]
- 4.Johansson M, Karlsson A. J Biol Chem. 1997;272:8454–8458. doi: 10.1074/jbc.272.13.8454. [DOI] [PubMed] [Google Scholar]
- 5.Gower W R, Jr, Carr M C, Ives D H. J Biol Chem. 1979;254:2180–2183. [PubMed] [Google Scholar]
- 6.Berk A J, Clayton D A. J Biol Chem. 1973;248:2722–2729. [PubMed] [Google Scholar]
- 7.Wang L, Karlsson A, Arnér E S J, Eriksson S. J Biol Chem. 1993;268:22847–22852. [PubMed] [Google Scholar]
- 8.Johansson M, Karlsson A. Proc Natl Acad Sci USA. 1996;93:7258–7262. doi: 10.1073/pnas.93.14.7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tallman M S, Hakimian D. Blood. 1995;86:2463–2474. [PubMed] [Google Scholar]
- 10.Hui Y F, Reitz J. Am J Health-Syst Pharm. 1997;54:162–170. doi: 10.1093/ajhp/54.2.162. [DOI] [PubMed] [Google Scholar]
- 11.Munch-Petersen B, Cloos L, Tyrsted G, Eriksson S. J Biol Chem. 1991;266:9032–9038. [PubMed] [Google Scholar]
- 12.Rizzuto R, Brini M, Pizzo P, Murgia M, Pozzan T. Curr Biol. 1995;5:635–642. doi: 10.1016/s0960-9822(95)00128-x. [DOI] [PubMed] [Google Scholar]
- 13.Yano M, Kanazawa M, Terada K, Namchai C, Yamaizumi M, Hanson B, Hoogenraad N, Mori M. J Biol Chem. 1997;272:8459–8465. doi: 10.1074/jbc.272.13.8459. [DOI] [PubMed] [Google Scholar]
- 14.Heim R, Cubitt A B, Tsien R Y. Nature (London) 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 15.Bradshaw H D, Jr, Deininger P L. Mol Cell Biol. 1984;4:2316–2320. doi: 10.1128/mcb.4.11.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chottiner E G, Shewach D S, Datta N S, Ashcraft E, Gribbin D, Ginsburg D, Fox I H, Mitchell B S. Proc Natl Acad Sci USA. 1991;88:1531–1535. doi: 10.1073/pnas.88.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris B A, Saunders P P, Plunkett W. Mol Pharmacol. 1981;20:200–205. [PubMed] [Google Scholar]
- 18.Nigg E A. Nature (London) 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 19.Dingwall C, Laskey R A. Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 20.Walker E J, Dow J W. Biochem J. 1982;203:361–369. doi: 10.1042/bj2030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De La Rosa A, Williams R L, Steeg P S. BioEssays. 1995;17:53–62. doi: 10.1002/bies.950170111. [DOI] [PubMed] [Google Scholar]
- 22.Rosengard A M, Krutzsch H C, Shearn A, Biggs J R, Barker E, Margulies I M K, King C R, Liotta L A, Steeg P S. Nature (London) 1989;342:177–180. doi: 10.1038/342177a0. [DOI] [PubMed] [Google Scholar]
- 23.Kraeft S-K, Traincart F, Mesnildrey S, Bourdais J, Véron M, Chen L B. Exp Cell Res. 1996;227:63–69. doi: 10.1006/excr.1996.0250. [DOI] [PubMed] [Google Scholar]
- 24.Reddy G P V, Pardee A B. Proc Natl Acad Sci USA. 1980;77:3312–3316. doi: 10.1073/pnas.77.6.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leeds J M, Slabaugh M B, Mathews C K. Mol Cell Biol. 1985;5:3443–3450. doi: 10.1128/mcb.5.12.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston P G, Liang C-M, Henry S, Chabner B A, Allegra C J. Cancer Res. 1991;51:6668–6676. [PubMed] [Google Scholar]
- 27.Samsonoff W A, Reston J, McKee M, O′Connor B, Galivan J, Maley G, Maley F. J Biol Chem. 1997;272:13281–13285. doi: 10.1074/jbc.272.20.13281. [DOI] [PubMed] [Google Scholar]
- 28.Xu Y-Z, Huang P, Plunkett W. J Biol Chem. 1995;270:631–637. [PubMed] [Google Scholar]
- 29.Bianchi V, Borella S, Rampazzo C, Ferraro P, Calderazzo F, Bianchi L C, Skog S, Reichard P. J Biol Chem. 1997;272:16118–16124. doi: 10.1074/jbc.272.26.16118. [DOI] [PubMed] [Google Scholar]
- 30.He Q, Skog S, Wang N, Eriksson S, Tribukait B. Eur J Cell Biol. 1996;70:117–124. [PubMed] [Google Scholar]
- 31.Sherley J L, Kelly T J. J Biol Chem. 1988;263:375–382. [PubMed] [Google Scholar]
- 32.Söderlund J C F, Arnér E S J. Adv Exp Med Biol. 1995;370:201–204. doi: 10.1007/978-1-4615-2584-4_43. [DOI] [PubMed] [Google Scholar]
- 33.Bestwick R K, Moffett G L, Mathews C K. J Biol Chem. 1982;257:9300–9304. [PubMed] [Google Scholar]
- 34.Lewis W, Dalakas M C. Nat Med. 1995;1:417–422. doi: 10.1038/nm0595-417. [DOI] [PubMed] [Google Scholar]
- 35.Chen C-H, Cheng Y-C. J Biol Chem. 1992;267:2856–2859. [PubMed] [Google Scholar]