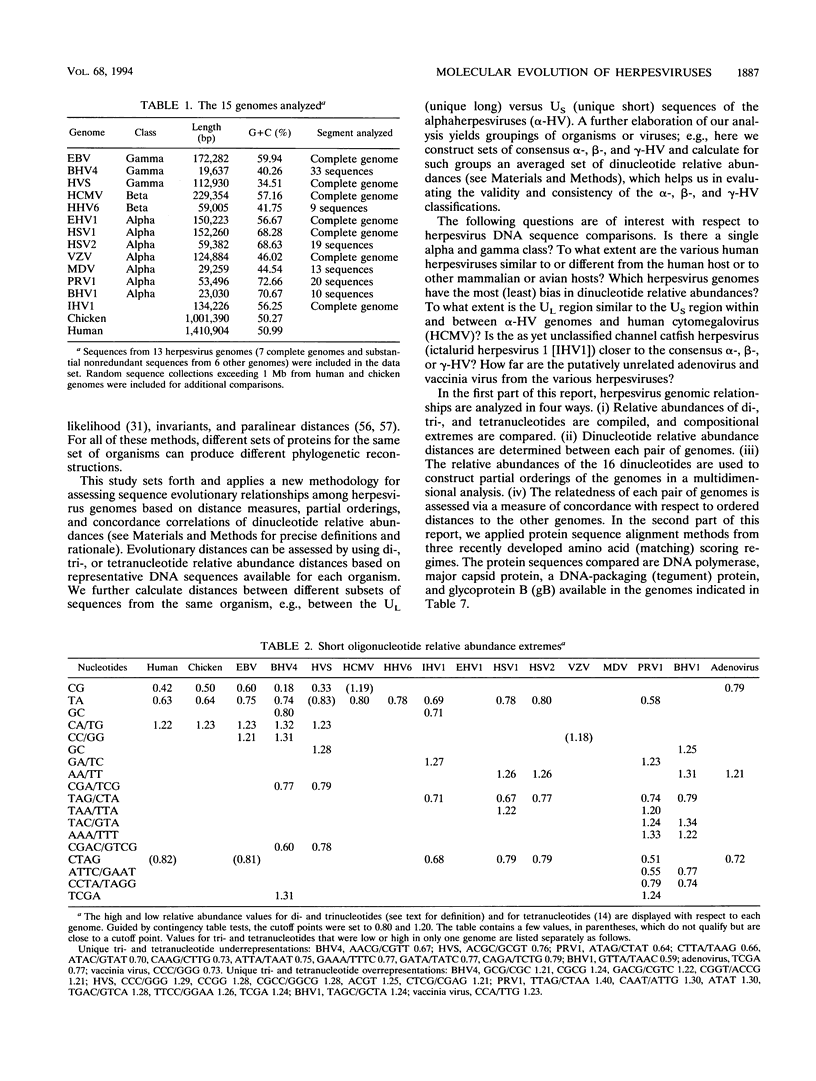
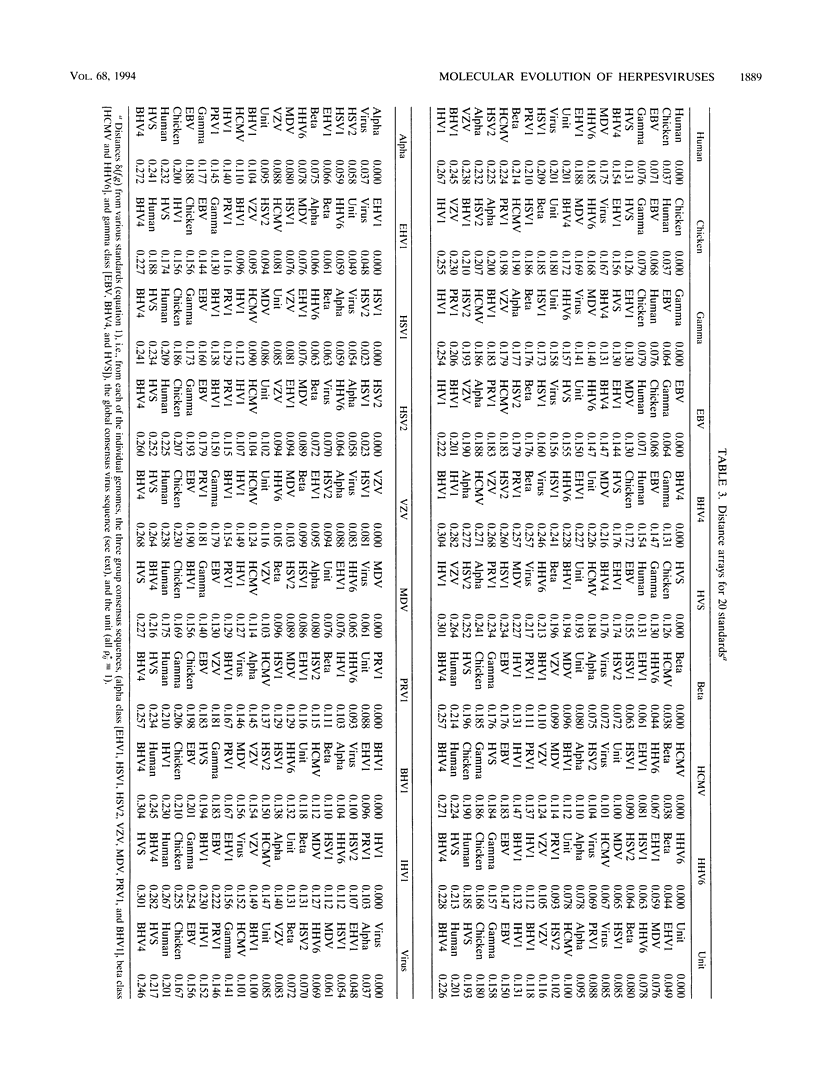
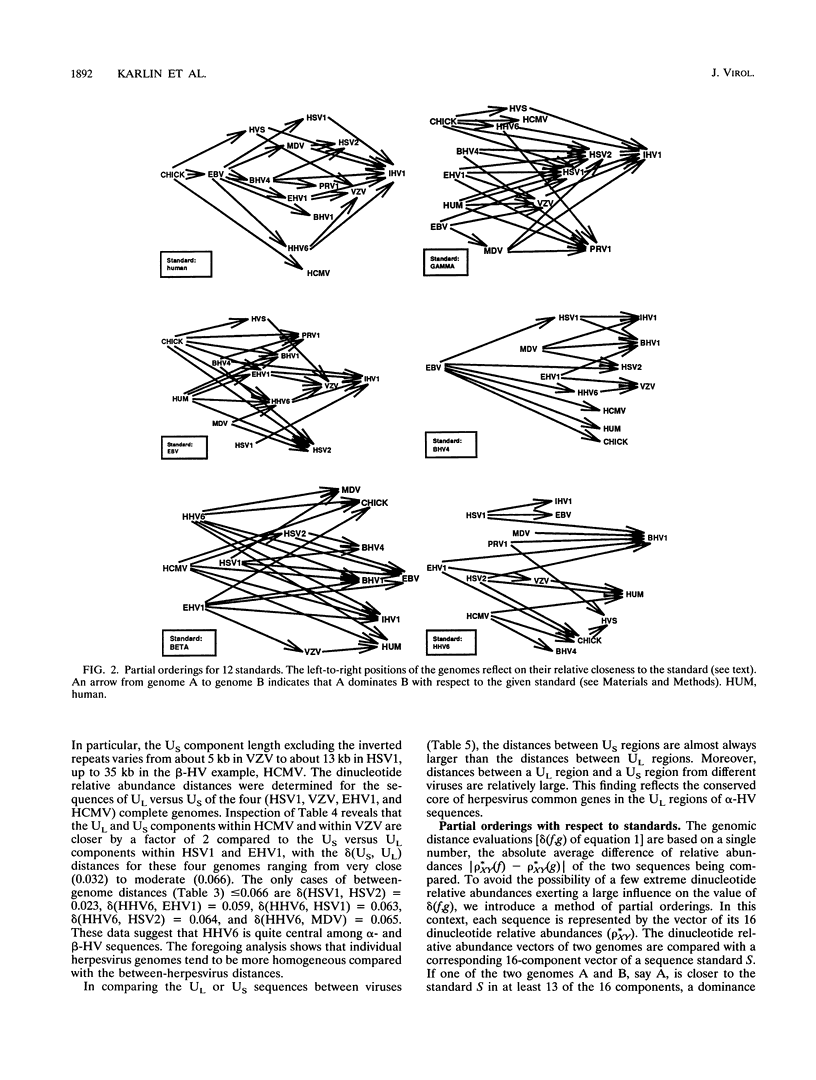
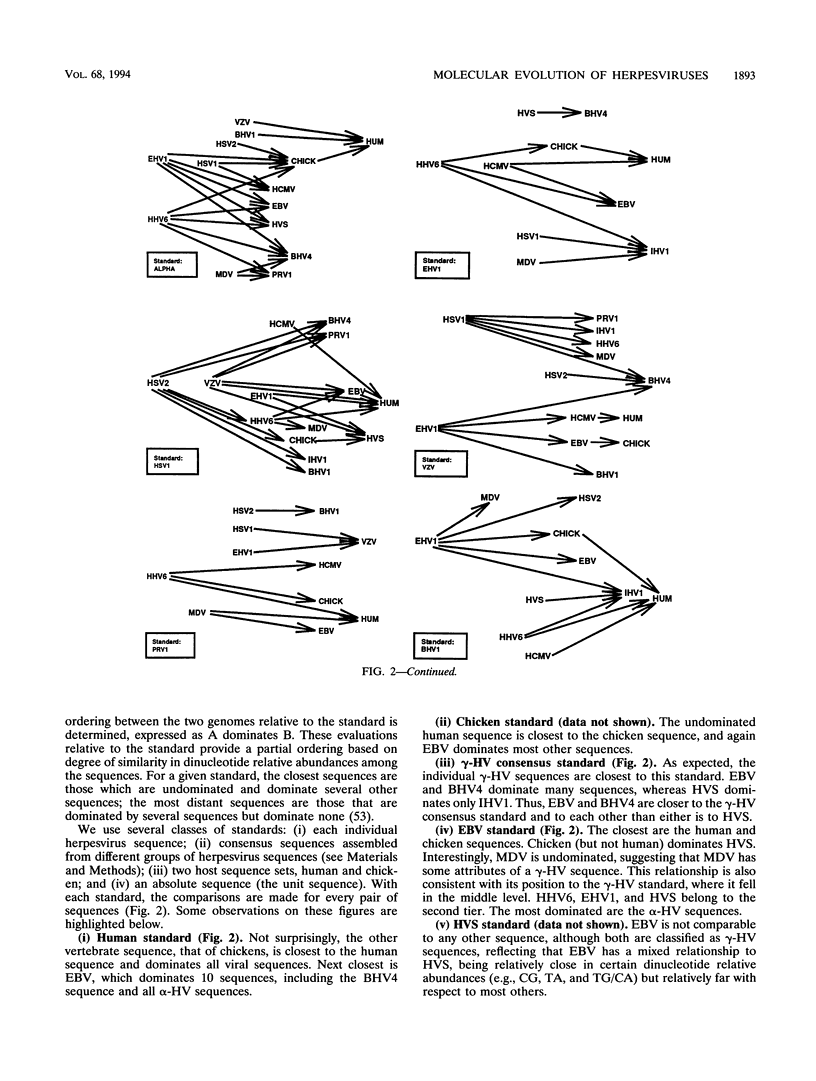
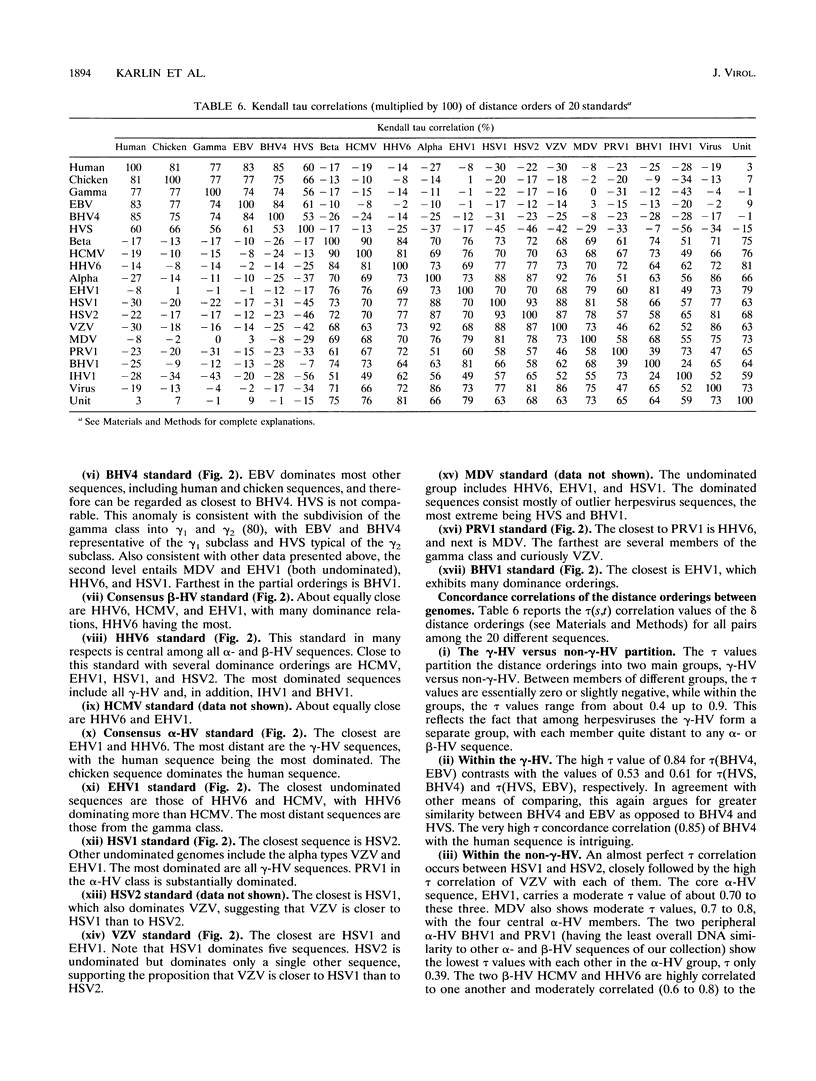
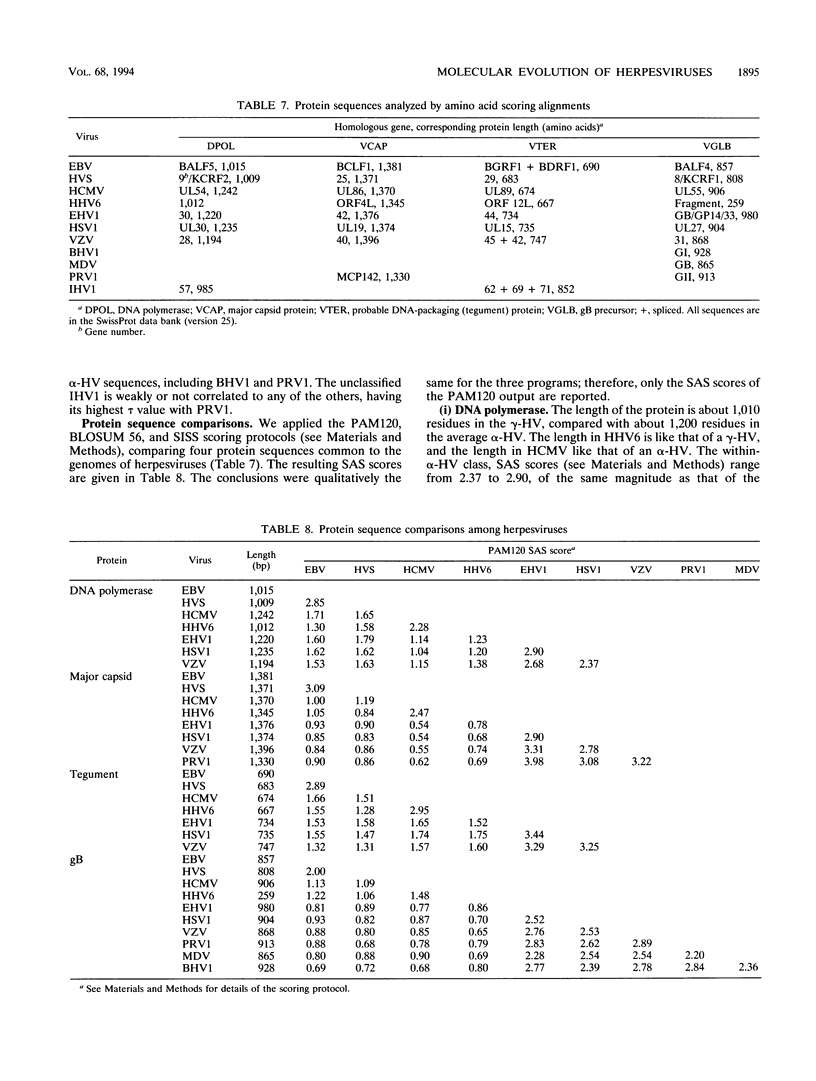
Abstract
Phylogenetic reconstruction of herpesvirus evolution is generally founded on amino acid sequence comparisons of specific proteins. These are relevant to the evolution of the specific gene (or set of genes), but the resulting phylogeny may vary depending on the particular sequence chosen for analysis (or comparison). In the first part of this report, we compare 13 herpesvirus genomes by using a new multidimensional methodology based on distance measures and partial orderings of dinucleotide relative abundances. The sequences were analyzed with respect to (i) genomic compositional extremes; (ii) total distances within and between genomes; (iii) partial orderings among genomes relative to a set of sequence standards; (iv) concordance correlations of genome distances; and (v) consistency with the alpha-, beta-, gammaherpesvirus classification. Distance assessments within individual herpesvirus genomes show each to be quite homogeneous relative to the comparisons between genomes. The gammaherpesviruses, Epstein-Barr virus (EBV), herpesvirus saimiri, and bovine herpesvirus 4 are both diverse and separate from other herpesvirus classes, whereas alpha- and betaherpesviruses overlap. The analysis revealed that the most central genome (closest to a consensus herpesvirus genome and most individual herpesvirus sequences of different classes) is that of human herpesvirus 6, suggesting that this genome is closest to a progenitor herpesvirus. The shorter DNA distances among alphaherpesviruses supports the hypothesis that the alpha class is of relatively recent ancestry. In our collection, equine herpesvirus 1 (EHV1) stands out as the most central alphaherpesvirus, suggesting it may approximate an ancestral alphaherpesvirus. Among all herpesviruses, the EBV genome is closest to human sequences. In the DNA partial orderings, the chicken sequence collection is invariably as close as or closer to all herpesvirus sequences than the human sequence collection is, which may imply that the chicken (or other avian species) is a more natural or more ancient host of herpesviruses. In the second part of this report, evolutionary relationships among the 13 herpesvirus genomes are evaluated on the basis of recent methods of amino acid alignment applied to four essential protein sequences. In this analysis, the alignment of the two betaherpesviruses (human cytomegalovirus versus human herpesvirus 6) showed lower scores compared with alignments within alphaherpesviruses (i.e., among EHV1, herpes simplex virus type 1, varicella-zoster virus, pseudorabies virus type 1 and Marek's disease virus) and within gammaherpesviruses (EBV versus herpesvirus saimiri).(ABSTRACT TRUNCATED AT 400 WORDS)
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albrecht J. C., Fleckenstein B. Structural organization of the conserved gene block of Herpesvirus saimiri coding for DNA polymerase, glycoprotein B, and major DNA binding protein. Virology. 1990 Feb;174(2):533–542. doi: 10.1016/0042-6822(90)90107-3. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Benbow R. M., Zhao J., Larson D. D. On the nature of origins of DNA replication in eukaryotes. Bioessays. 1992 Oct;14(10):661–670. doi: 10.1002/bies.950141004. [DOI] [PubMed] [Google Scholar]
- Bennett A. M., Harrington L., Kelly D. C. Nucleotide sequence analysis of genes encoding glycoproteins D and J in simian herpes B virus. J Gen Virol. 1992 Nov;73(Pt 11):2963–2967. doi: 10.1099/0022-1317-73-11-2963. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Bernardi G. Compositional constraints and genome evolution. J Mol Evol. 1986;24(1-2):1–11. doi: 10.1007/BF02099946. [DOI] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Birkenbach M., Josefsen K., Yalamanchili R., Lenoir G., Kieff E. Epstein-Barr virus-induced genes: first lymphocyte-specific G protein-coupled peptide receptors. J Virol. 1993 Apr;67(4):2209–2220. doi: 10.1128/jvi.67.4.2209-2220.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslauer K. J., Frank R., Blöcker H., Marky L. A. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bublot M., Lomonte P., Lequarre A. S., Albrecht J. C., Nicholas J., Fleckenstein B., Pastoret P. P., Thiry E. Genetic relationships between bovine herpesvirus 4 and the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. Virology. 1992 Oct;190(2):654–665. doi: 10.1016/0042-6822(92)90903-3. [DOI] [PubMed] [Google Scholar]
- Bublot M., Van Bressem M. F., Thiry E., Dubuisson J., Pastoret P. P. Bovine herpesvirus 4 genome: cloning, mapping and strain variation analysis. J Gen Virol. 1990 Jan;71(Pt 1):133–142. doi: 10.1099/0022-1317-71-1-133. [DOI] [PubMed] [Google Scholar]
- Burge C., Campbell A. M., Karlin S. Over- and under-representation of short oligonucleotides in DNA sequences. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1358–1362. doi: 10.1073/pnas.89.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., Edwards A. W. Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet. 1967 May;19(3 Pt 1):233–257. [PMC free article] [PubMed] [Google Scholar]
- Challberg M. D., Kelly T. J. Animal virus DNA replication. Annu Rev Biochem. 1989;58:671–717. doi: 10.1146/annurev.bi.58.070189.003323. [DOI] [PubMed] [Google Scholar]
- Chee M. S., Bankier A. T., Beck S., Bohni R., Brown C. M., Cerny R., Horsnell T., Hutchison C. A., 3rd, Kouzarides T., Martignetti J. A. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- Chou S., Marousek G. I. Homology of the envelope glycoprotein B of human herpesvirus-6 and cytomegalovirus. Virology. 1992 Nov;191(1):523–528. doi: 10.1016/0042-6822(92)90224-d. [DOI] [PubMed] [Google Scholar]
- Croen K. D., Ostrove J. M., Dragovic L. J., Straus S. E. Patterns of gene expression and sites of latency in human nerve ganglia are different for varicella-zoster and herpes simplex viruses. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9773–9777. doi: 10.1073/pnas.85.24.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinane A. A., Rixon F. J., Davison A. J. Characterization of the genome of equine herpesvirus 1 subtype 2. J Gen Virol. 1988 Jul;69(Pt 7):1575–1590. doi: 10.1099/0022-1317-69-7-1575. [DOI] [PubMed] [Google Scholar]
- Davison A. J., McGeoch D. J. Evolutionary comparisons of the S segments in the genomes of herpes simplex virus type 1 and varicella-zoster virus. J Gen Virol. 1986 Apr;67(Pt 4):597–611. doi: 10.1099/0022-1317-67-4-597. [DOI] [PubMed] [Google Scholar]
- Delcourt S. G., Blake R. D. Stacking energies in DNA. J Biol Chem. 1991 Aug 15;266(23):15160–15169. [PubMed] [Google Scholar]
- Earl P. L., Jones E. V., Moss B. Homology between DNA polymerases of poxviruses, herpesviruses, and adenoviruses: nucleotide sequence of the vaccinia virus DNA polymerase gene. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3659–3663. doi: 10.1073/pnas.83.11.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers B., Buhk H. J., Ludwig H. Analysis of bovine cytomegalovirus genome structure: cloning and mapping of the monomeric polyrepetitive DNA unit, and comparison of European and American strains. J Gen Virol. 1985 Jan;66(Pt 1):55–68. doi: 10.1099/0022-1317-66-1-55. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17(6):368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- Fitch W. M., Leiter J. M., Li X. Q., Palese P. Positive Darwinian evolution in human influenza A viruses. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4270–4274. doi: 10.1073/pnas.88.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammelin M., Altmüller A., Reinhardt U., Mandler J., Harley V. R., Hudson P. J., Fitch W. M., Scholtissek C. Phylogenetic analysis of nucleoproteins suggests that human influenza A viruses emerged from a 19th-century avian ancestor. Mol Biol Evol. 1990 Mar;7(2):194–200. doi: 10.1093/oxfordjournals.molbev.a040594. [DOI] [PubMed] [Google Scholar]
- Geng Y. Q., Chandran B., Josephs S. F., Wood C. Identification and characterization of a human herpesvirus 6 gene segment that trans activates the human immunodeficiency virus type 1 promoter. J Virol. 1992 Mar;66(3):1564–1570. doi: 10.1128/jvi.66.3.1564-1570.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompels U. A., Craxton M. A., Honess R. W. Conservation of gene organization in the lymphotropic herpesviruses herpesvirus Saimiri and Epstein-Barr virus. J Virol. 1988 Mar;62(3):757–767. doi: 10.1128/jvi.62.3.757-767.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. W. The endosymbiont hypothesis revisited. Int Rev Cytol. 1992;141:233–357. doi: 10.1016/s0074-7696(08)62068-9. [DOI] [PubMed] [Google Scholar]
- Griffin A. M. The nucleotide sequence of the glycoprotein gB gene of infectious laryngotracheitis virus: analysis and evolutionary relationship to the homologous gene from other herpesviruses. J Gen Virol. 1991 Feb;72(Pt 2):393–398. doi: 10.1099/0022-1317-72-2-393. [DOI] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honess R. W., Bodemer W., Cameron K. R., Niller H. H., Fleckenstein B., Randall R. E. The A+T-rich genome of Herpesvirus saimiri contains a highly conserved gene for thymidylate synthase. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3604–3608. doi: 10.1073/pnas.83.11.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honess R. W., Gompels U. A., Barrell B. G., Craxton M., Cameron K. R., Staden R., Chang Y. N., Hayward G. S. Deviations from expected frequencies of CpG dinucleotides in herpesvirus DNAs may be diagnostic of differences in the states of their latent genomes. J Gen Virol. 1989 Apr;70(Pt 4):837–855. doi: 10.1099/0022-1317-70-4-837. [DOI] [PubMed] [Google Scholar]
- Honess R. W. Herpes simplex and 'the herpes complex': diverse observations and a unifying hypothesis. The eighth Fleming lecture. J Gen Virol. 1984 Dec;65(Pt 12):2077–2107. doi: 10.1099/0022-1317-65-12-2077. [DOI] [PubMed] [Google Scholar]
- Hsu D. H., de Waal Malefyt R., Fiorentino D. F., Dang M. N., Vieira P., de Vries J., Spits H., Mosmann T. R., Moore K. W. Expression of interleukin-10 activity by Epstein-Barr virus protein BCRF1. Science. 1990 Nov 9;250(4982):830–832. doi: 10.1126/science.2173142. [DOI] [PubMed] [Google Scholar]
- Jansson A., Masucci M., Rymo L. Methylation of discrete sites within the enhancer region regulates the activity of the Epstein-Barr virus BamHI W promoter in Burkitt lymphoma lines. J Virol. 1992 Jan;66(1):62–69. doi: 10.1128/jvi.66.1.62-69.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992 Jun;8(3):275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Jones D., Isfort R., Witter R., Kost R., Kung H. J. Retroviral insertions into a herpesvirus are clustered at the junctions of the short repeat and short unique sequences. Proc Natl Acad Sci U S A. 1993 May 1;90(9):3855–3859. doi: 10.1073/pnas.90.9.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes T. H., Bhushan V. Silent nucleotide substitutions and G + C content of some mitochondrial and bacterial genes. J Mol Evol. 1986;24(1-2):39–44. doi: 10.1007/BF02099949. [DOI] [PubMed] [Google Scholar]
- Karlin S., Blaisdell B. E., Schachtel G. A. Contrasts in codon usage of latent versus productive genes of Epstein-Barr virus: data and hypotheses. J Virol. 1990 Sep;64(9):4264–4273. doi: 10.1128/jvi.64.9.4264-4273.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Burge C., Campbell A. M. Statistical analyses of counts and distributions of restriction sites in DNA sequences. Nucleic Acids Res. 1992 Mar 25;20(6):1363–1370. doi: 10.1093/nar/20.6.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Kenett R., Bonné-Tamir B. Analysis of biochemical genetic data on Jewish populations: II. Results and interpretations of heterogeneity indices and distance measures with respect to standards. Am J Hum Genet. 1979 May;31(3):341–365. [PMC free article] [PubMed] [Google Scholar]
- Klupp B. G., Kern H., Mettenleiter T. C. The virulence-determining genomic BamHI fragment 4 of pseudorabies virus contains genes corresponding to the UL15 (partial), UL18, UL19, UL20, and UL21 genes of herpes simplex virus and a putative origin of replication. Virology. 1992 Dec;191(2):900–908. doi: 10.1016/0042-6822(92)90265-q. [DOI] [PubMed] [Google Scholar]
- Kondo K., Kondo T., Okuno T., Takahashi M., Yamanishi K. Latent human herpesvirus 6 infection of human monocytes/macrophages. J Gen Virol. 1991 Jun;72(Pt 6):1401–1408. doi: 10.1099/0022-1317-72-6-1401. [DOI] [PubMed] [Google Scholar]
- Kondo K., Nagafuji H., Hata A., Tomomori C., Yamanishi K. Association of human herpesvirus 6 infection of the central nervous system with recurrence of febrile convulsions. J Infect Dis. 1993 May;167(5):1197–1200. doi: 10.1093/infdis/167.5.1197. [DOI] [PubMed] [Google Scholar]
- Lake J. A. Determining evolutionary distances from highly diverged nucleic acid sequences: operator metrics. J Mol Evol. 1987;26(1-2):59–73. doi: 10.1007/BF02111282. [DOI] [PubMed] [Google Scholar]
- Lawrence G. L., Chee M., Craxton M. A., Gompels U. A., Honess R. W., Barrell B. G. Human herpesvirus 6 is closely related to human cytomegalovirus. J Virol. 1990 Jan;64(1):287–299. doi: 10.1128/jvi.64.1.287-299.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquester G. J., Pellett P. E. Properties of the human herpesvirus 6 strain Z29 genome: G + C content, length, and presence of variable-length directly repeated terminal sequence elements. Virology. 1991 May;182(1):102–110. doi: 10.1016/0042-6822(91)90653-s. [DOI] [PubMed] [Google Scholar]
- Marchini A., Tomkinson B., Cohen J. I., Kieff E. BHRF1, the Epstein-Barr virus gene with homology to Bc12, is dispensable for B-lymphocyte transformation and virus replication. J Virol. 1991 Nov;65(11):5991–6000. doi: 10.1128/jvi.65.11.5991-6000.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. E., Nicholas J., Thomson B. J., Newman C., Honess R. W. Identification of a transactivating function mapping to the putative immediate-early locus of human herpesvirus 6. J Virol. 1991 Oct;65(10):5381–5390. doi: 10.1128/jvi.65.10.5381-5390.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Jones R., Patel Y., Nelson M. Restriction endonucleases for pulsed field mapping of bacterial genomes. Nucleic Acids Res. 1987 Aug 11;15(15):5985–6005. doi: 10.1093/nar/15.15.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D. J. Evolutionary relationships of virion glycoprotein genes in the S regions of alphaherpesvirus genomes. J Gen Virol. 1990 Oct;71(Pt 10):2361–2367. doi: 10.1099/0022-1317-71-10-2361. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J. The genomes of the human herpesviruses: contents, relationships, and evolution. Annu Rev Microbiol. 1989;43:235–265. doi: 10.1146/annurev.mi.43.100189.001315. [DOI] [PubMed] [Google Scholar]
- Minarovits J., Minarovits-Kormuta S., Ehlin-Henriksson B., Falk K., Klein G., Ernberg I. Host cell phenotype-dependent methylation patterns of Epstein-Barr virus DNA. J Gen Virol. 1991 Jul;72(Pt 7):1591–1599. doi: 10.1099/0022-1317-72-7-1591. [DOI] [PubMed] [Google Scholar]
- Neote K., DiGregorio D., Mak J. Y., Horuk R., Schall T. J. Molecular cloning, functional expression, and signaling characteristics of a C-C chemokine receptor. Cell. 1993 Feb 12;72(3):415–425. doi: 10.1016/0092-8674(93)90118-a. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Woese C. R. Ribosomal RNA: a key to phylogeny. FASEB J. 1993 Jan;7(1):113–123. doi: 10.1096/fasebj.7.1.8422957. [DOI] [PubMed] [Google Scholar]
- Quinlan M. P., Knipe D. M. Nuclear localization of herpesvirus proteins: potential role for the cellular framework. Mol Cell Biol. 1983 Mar;3(3):315–324. doi: 10.1128/mcb.3.3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B. The organization of the herpes simplex virus genomes. Annu Rev Genet. 1979;13:25–57. doi: 10.1146/annurev.ge.13.120179.000325. [DOI] [PubMed] [Google Scholar]
- Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979 Mar;16(3):481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- Ross L. J., Binns M. M. Properties and evolutionary relationships of the Marek's disease virus homologues of protein kinase, glycoprotein D and glycoprotein I of herpes simplex virus. J Gen Virol. 1991 Apr;72(Pt 4):939–947. doi: 10.1099/0022-1317-72-4-939. [DOI] [PubMed] [Google Scholar]
- Schachtel G. A., Bucher P., Mocarski E. S., Blaisdell B. E., Karlin S. Evidence for selective evolution in codon usage in conserved amino acid segments of human alphaherpesvirus proteins. J Mol Evol. 1991 Dec;33(6):483–494. doi: 10.1007/BF02102801. [DOI] [PubMed] [Google Scholar]
- Selker E. U. DNA methylation and chromatin structure: a view from below. Trends Biochem Sci. 1990 Mar;15(3):103–107. doi: 10.1016/0968-0004(90)90193-f. [DOI] [PubMed] [Google Scholar]
- Stewart C. B. The powers and pitfalls of parsimony. Nature. 1993 Feb 18;361(6413):603–607. doi: 10.1038/361603a0. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Sonoda S., Higashi K., Kondo T., Takahashi H., Takahashi M., Yamanishi K. Predominant CD4 T-lymphocyte tropism of human herpesvirus 6-related virus. J Virol. 1989 Jul;63(7):3161–3163. doi: 10.1128/jvi.63.7.3161-3163.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J., Bird A. Alternative chromatin structure at CpG islands. Cell. 1990 Mar 23;60(6):909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- Telford E. A., Studdert M. J., Agius C. T., Watson M. S., Aird H. C., Davison A. J. Equine herpesviruses 2 and 5 are gamma-herpesviruses. Virology. 1993 Aug;195(2):492–499. doi: 10.1006/viro.1993.1400. [DOI] [PubMed] [Google Scholar]
- Telford E. A., Watson M. S., McBride K., Davison A. J. The DNA sequence of equine herpesvirus-1. Virology. 1992 Jul;189(1):304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- Teo I. A., Griffin B. E., Jones M. D. Characterization of the DNA polymerase gene of human herpesvirus 6. J Virol. 1991 Sep;65(9):4670–4680. doi: 10.1128/jvi.65.9.4670-4680.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry E., Bublot M., Dubuisson J., Van Bressem M. F., Lequarre A. S., Lomonte P., Vanderplasschen A., Pastoret P. P. Molecular biology of bovine herpesvirus type 4. Vet Microbiol. 1992 Nov;33(1-4):79–92. doi: 10.1016/0378-1135(92)90037-t. [DOI] [PubMed] [Google Scholar]
- Thompson R., Honess R. W., Taylor L., Morran J., Davison A. J. Varicella-zoster virus specifies a thymidylate synthetase. J Gen Virol. 1987 May;68(Pt 5):1449–1455. doi: 10.1099/0022-1317-68-5-1449. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. DNA in profile. Trends Biochem Sci. 1991 Dec;16(12):467–470. doi: 10.1016/0968-0004(91)90181-t. [DOI] [PubMed] [Google Scholar]
- Trimble J. J., Murthy S. C., Bakker A., Grassmann R., Desrosiers R. C. A gene for dihydrofolate reductase in a herpesvirus. Science. 1988 Mar 4;239(4844):1145–1147. doi: 10.1126/science.2830673. [DOI] [PubMed] [Google Scholar]
- Wang T. S., Wong S. W., Korn D. Human DNA polymerase alpha: predicted functional domains and relationships with viral DNA polymerases. FASEB J. 1989 Jan;3(1):14–21. doi: 10.1096/fasebj.3.1.2642867. [DOI] [PubMed] [Google Scholar]


