Summary
Drosophila Dscam encodes 38,016 distinct axon guidance receptors through the mutually exclusive alternative splicing of 95 variable exons. Importantly, known mechanisms that ensure the mutually exclusive splicing of pairs of exons cannot explain this phenomenon in Dscam. I have identified two classes of conserved elements in the Dscam exon 6 cluster, which contains 48 alternative exons—the docking site, located in the intron downstream of constitutive exon 5, and the selector sequences, which are located upstream of each exon 6 variant. Strikingly, each selector sequence is complementary to a portion of the docking site, and this pairing juxtaposes one, and only one, alternative exon to the upstream constitutive exon. The mutually exclusive nature of the docking site:selector sequence interactions suggests that the formation of these competing RNA structures is a central component of the mechanism guaranteeing that only one exon 6 variant is included in each Dscam mRNA.
Introduction
Alternative splicing is a widespread mechanism for regulating gene expression and increasing protein diversity in eukaryotes (Black, 2000; Graveley, 2001). Most metazoan genes encode pre-mRNAs that undergo alternative splicing. For example, recent microarray analyses suggest that approximately 40% and 74% of Drosophila and human genes, respectively, encode alternatively spliced pre-mRNAs (Johnson et al., 2003; Stolc et al., 2004). Moreover, while most genes are thought to generate only two alternatively spliced mRNA isoforms, many genes can encode a much greater repertoire of mRNA variants (Black, 2000; Graveley, 2001). The most dramatic example is the Drosophila Dscam gene, which can potentially generate over 38,000 mRNA isoforms via alternative splicing (Schmucker et al., 2000).
Alternative splicing can occur through intron retention, alternative 5′ or 3′ splice site selection, the inclusion or exclusion of cassette exons, the selection of alternative 3′ terminal exons, and even alternative trans-splicing (Maniatis and Tasic, 2002). On a genome-wide level, the most frequent alternative splicing events involve cassette exons—exons that are either included or excluded from an mRNA in their entirety (Thanaraj et al., 2004). Moreover, approximately 10% of cassette exons exist in pairs in which only one of the two exons is included in the mRNA (Thanaraj et al., 2004). In fact, thousands of genes exist that contain such an arrangement.
Several mechanisms have been discovered that are used to ensure that the splicing of pairs of alternative exons is strictly mutually exclusive. First, the splice sites in the intron separating the two alternative exons can be spatially arranged such that when splicing factors recognize one splice site, they prevent the binding of splicing factors to the other splice site through steric hindrance. Specifically, the binding of U1 snRNP to the 5′ splice site in the intron separating the two mutually exclusive exons will prevent the binding of U2 snRNP to the branchpoint in the same intron. Conversely, the binding of U2 snRNP to the branchpoint will prevent the binding of U1 snRNP to the 5′ splice site. In this way, the spliceosome can recognize only one of the two mutually exclusive exons. This mechanism has been shown to occur in the α-tropomyosin (Smith and Nadal-Ginard, 1989) and α-actinin (Southby et al., 1999) genes.
A related mechanism could operate if the intron separating the two alternative exons is too small to be efficiently spliced. For example, in Drosophila, introns smaller than 59 nucleotides cannot be removed by the spliceosome (Kennedy and Berget, 1997). Thus, the presence of introns smaller than this lower limit would prevent the alternative exons from being spliced together. As a result, only one of the two exons would be included in the pre-mRNA.
A third mechanism involves a unique arrangement of splice sites that are recognized by the major and minor spliceosomes. Most introns are removed by the major spliceosome, which consists of U1, U2, U4, U6, and U5 snRNPs. However, a small number of introns are removed by the minor spliceosome, which consists of the U11, U12, U4atac, U6atac, and U5 snRNPs (Patel and Steitz, 2003). The splice sites that are recognized by these two spliceosomes are distinct. Moreover, neither spliceosome can remove an intron containing a mixture of major and minor splice sites (Sharp and Burge, 1997). Thus, one way in which alternative splicing can be mutually exclusive is to use a combination of splice sites recognized by the two spliceosomes. For instance, if the 5′ splice site of the upstream constitutive exon and the 3′ splice sites of the two alternative exons are major spliceosome splice sites and the 5′ splice sites of the alternative exons and the 3′ splice site of the downstream constitutive exon are minor spliceo-some splice sites, the splicing of the alternative exons is guaranteed to be mutually exclusive. The human stress-activated protein kinase (JNK 1) gene contains an alternatively spliced region with this type of organization (Letunic et al., 2002).
Finally, the splicing of two cassette exons may not be truly mutually exclusive, but rather may simply appear to be so. This could occur if the two alternative exons are not a multiple of three nucleotides. If neither or both exons are included, premature termination codons will be introduced into the mRNA and those isoforms will be subject to degradation by the non-sense-mediated decay pathway (Jones et al., 2001). As a result, mRNAs containing one and only one exon will be stable and the splicing of such pre-mRNAs will appear to be mutually exclusive, though in reality it is not.
The Drosophila melanogaster Dscam gene contains 95 mutually exclusive alternative exons arranged into four clusters. The exon 4, 6, 9, and 17 clusters contain 12, 48, 33, and 2 exons, respectively (Figure 1A). Despite the fact that each alternative exon is flanked by splice sites, the alternative exons have not been observed to be spliced together. Thus, a powerful mechanism(s) must exist to ensure that multiple alternative exons are not included in the Dscam mRNAs. Interestingly, none of the known mechanisms can explain the mutually exclusive splicing of the Dscam exon 4, 6, and 9 clusters. First, although the steric hindrance of snRNP binding could prevent adjacent alternative exons from being spliced together, it could not do so for nonadjacent exons. Second, all of the introns separating the alternative exons are larger, and in many cases significantly larger, than the lower limit for intron size. Third, the dual spliceosome mechanism is not involved for Dscam because all of the splice sites in the gene conform to the major spliceosome consensus sequences. Finally, the NMD mechanism cannot be sufficient either. In the exon 4 cluster, all 12 exons are a multiple of 3. As a result, regardless of the number of exons included, no change in reading frame would occur. In contrast, for the exon 6 and 9 clusters, the alternative exons are not divisible by 3. However, the NMD mechanism appears unlikely because transcripts containing one exon, four exons, seven exons, etc. should be insensitive to NMD. However, mRNAs containing multiples of 3 exons have not been detected. Thus, the mechanism involved in the mutually exclusive splicing of Dscam must be novel.
Figure 1. The D. melanogaster Dscam Gene and Insect Phylogeny.
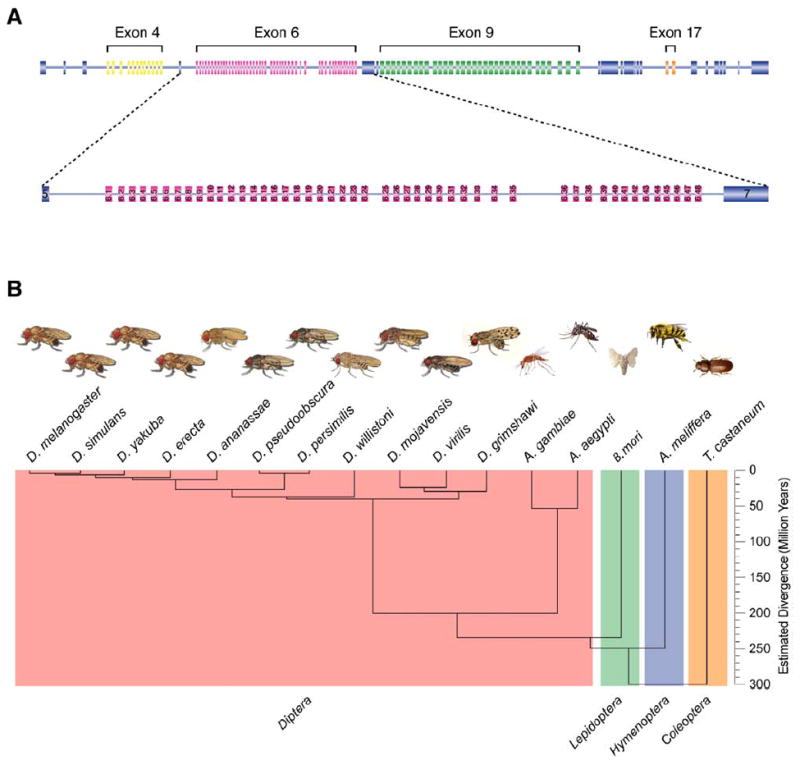
(A) Organization of the D. melanogaster Dscam gene. Dscam contains 115 exons, 95 of which are alternatively spliced. The exon 4, 6, and 9 clusters contain 12, 48, and 33 alternative exons, respectively, that each encode variable immunoglobulin domains. The exon 17 cluster contains two exons that encode alternate versions of the transmembrane domain. The exons within each cluster are alternatively spliced in a mutually exclusive manner. The exon 6 cluster is enlarged to depict its organization.
(B) Dendrogram of the phylogenetic relationship among the insects used in this study. The tree represents the estimated evolutionary distance of each organism in millions of years. These organisms represent four major orders of Insecta—Diptera, Lepidoptera, Hymenoptera, and Coleoptera—which are each shaded in different colors.
I have used comparative genomics to identify two classes of conserved sequence elements that appear to play a role in the mutually exclusive splicing of the Dscam exon 6 cluster. The first element is the docking site and is located in the intron between constitutive exon 5 and the first exon 6 variant. The second type of element is called the selector sequence, one of which is located upstream of each exon 6 variant. This extraordinarily complex alternative splicing event appears to involve mutually exclusive RNA base-pairing interactions between the docking site and one of the 48 selector sequences. Thus, an elegant and novel mechanism appears to play a critical role in the mutually exclusive splicing of the exon 6 cluster of the insect Dscam genes.
Results
Comparison of the Dscam Gene from 16 Insect Species
Comparative sequence analysis was used to identify RNA sequence elements that could potentially be involved in the regulation of Dscam alternative splicing. The sequences of the Dscam genes of 16 different insects were extracted from GenBank as either preassembled genes or as individual sequence reads from the trace archives that were subsequently assembled into a contig covering the gene. The organisms analyzed consisted of 13 Dipteran species, including 11 Drosophila species (D. melanogaster, D. simulans, D. yakuba, D. erecta, D. ananassae, D. pseudoobscura, D. persimilis, D. willistoni, D. mojavensis, D. virilis, and D. grimshawi) and two mosquito species (Anopheles gambiae [malaria mosquito] and Aedes aegypti [yellow fever mosquito]), the Lepidopteran Bombyx mori (silk worm), the Hymenopteran Apis mellifera (honeybee), and the Coleopteran Tribolium castaneum (red flour beetle; Figure 1B). Together these organisms encompass four major taxanomic groups of insects that last shared a common ancestor at least 300 million years ago (Powell, 1997). As described previously for five of these species (Graveley et al., 2004), the Dscam genes of each organism can each potentially generate tens of thousands of isoforms by alternative splicing, though the exact number of alternative exons differs in most species.
The Docking Site
Multiple sequence alignment of the Dscam genes from these 16 species revealed a number of conserved intronic elements, the majority of which were located in the exon 6 cluster. The most highly conserved element in the entire Dscam gene, which is greater than 60,000 bp in D. melanogaster, is located in the intron between the constitutive exon 5 and exon 6.1 and will be referred to as the docking site. The docking site is a 66 nt sequence element in D. melanogaster that is 90%–100% identical in the 10 other Drosophila species examined (see Figure S1 in the Supplemental Data available with this article online). Moreover, the central 24 nt of the docking site is nearly invariant in all 16 species (Figure 2). The only exceptions are D. willistoni, which contains a 2 nt insertion at position 34 of the docking site (Figure S1), and A. mellifera, which contains two T to C transitions at positions 22 and 28 (Figure 2). A docking site consensus sequence can be derived from the first 37 nt of the alignment shown in Figure 2.
Figure 2. The Docking Site.
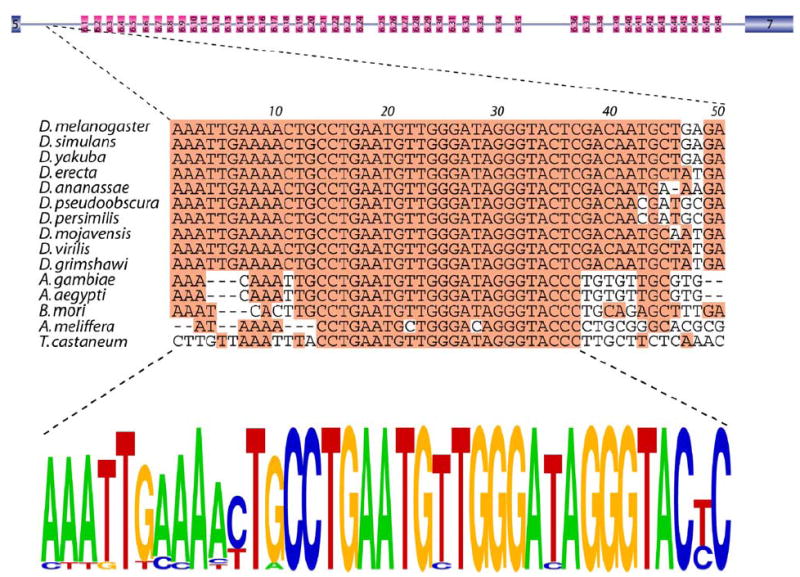
The nucleotide sequence alignment of the docking sites of 15 insects. The most common nucleotide at each position is shaded. The docking site consensus is represented as a pictogram (bottom). The height of each letter represents the frequency of each nucleotide at that position.
The Selector Sequences
The second class of conserved elements that were identified will be referred to as selector sequences. The initial selector sequences were identified as relatively conserved sequences in the introns upstream of some of the exon 6 variants. Examples of several of these elements are shown in Figure 3. Some of these elements are related to one another. For instance, the selector sequences upstream of exons 6.5, 6.19, and 6.43 all contain the sequence CAGGCAG, while the selector sequences upstream of exons 6.28, 6.36, and 6.44 contain sequences that deviate from CAGGCAG by only one nucleotide. However, this is not universally true since the exon 6.12 selector sequence does not contain this motif. By searching the remaining exon 6 cluster for sequences that are similar to but not identical to the initially identified selector sequences, a potential selector sequence was identified upstream of each exon 6 variant that was also similar in other Drosophila species. An alignment of the selector sequences located upstream of all 48 D. melanogaster exon 6 variants, together with some flanking sequence, revealed that all of the selector sequences overlap with one another to a certain extent (Figure 4A). This alignment was used to generate a consensus selector sequence (Figure 4B). Importantly, this consensus sequence does not resemble any known splicing regulatory elements or splice site sequences.
Figure 3. Conservation of Selector Sequences.
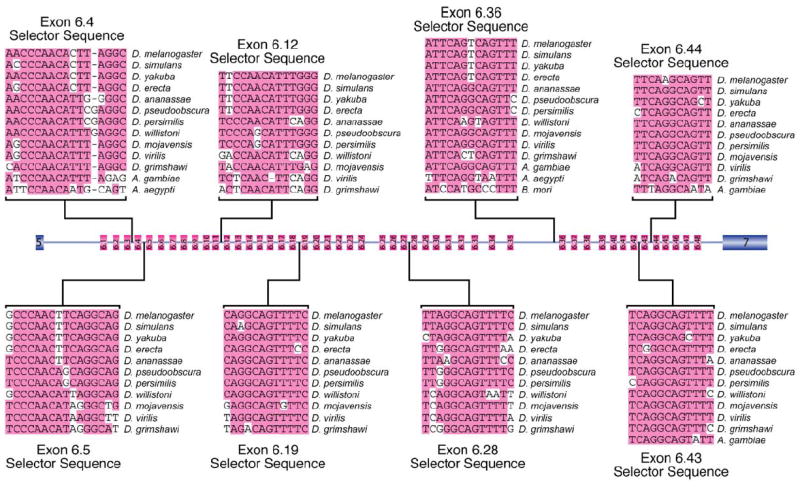
Alignment of eight of the selector sequences and their locations with the exon 6 cluster are depicted. The most common nucleotides at each position are shaded.
Figure 4. The D. melanogaster Selector Sequence Consensus.
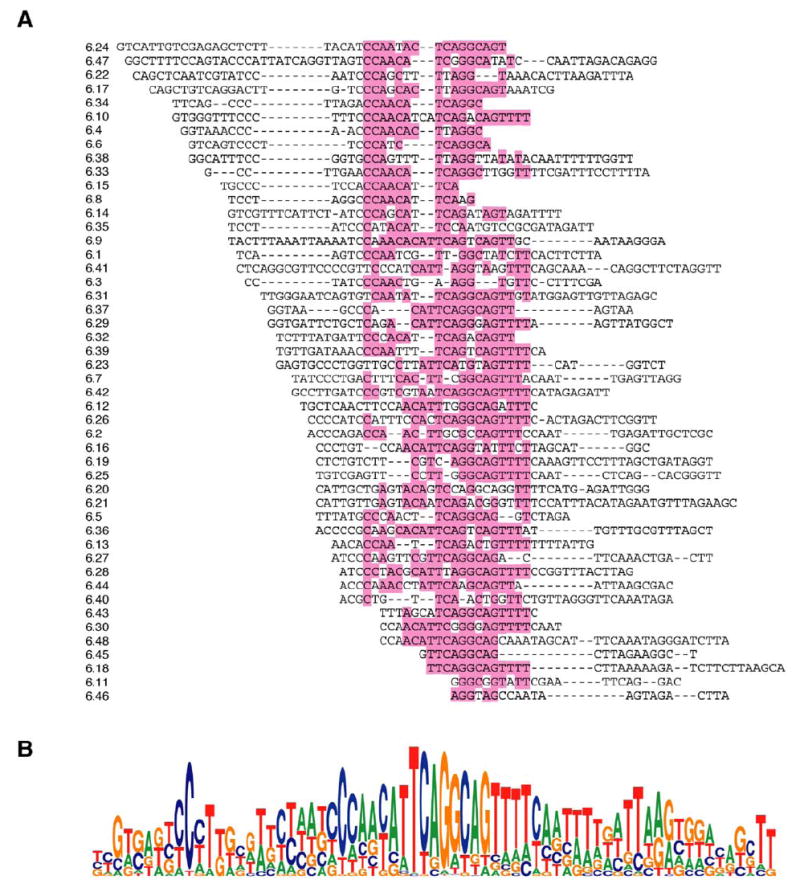
(A) The 48 selector sequences and flanking sequence were aligned together. The most frequent nucleotides in the central portion of the alignment are highlighted.
(B) The alignment was used to generate a selector sequence consensus.
Docking Site:Selector Sequence Interactions
Strikingly, the central 28 nt of the consensus selector sequence is complementary to the docking site consensus sequence (Figure 5). Moreover, all 48 predicted D. melanogaster selector sequences are complementary to the docking site (See Figure 6A for two examples and Figure S2 for all 48 structures). Because the selector sequences all overlap with one another to some extent, the docking site is predicted to interact with only one selector sequence at a time. Thus, the docking site: selector sequence interactions would simultaneously juxtapose exon 5 with the exon 6 variant that is to be included and could explain how the alternative splicing of these 48 exons is mutually exclusive.
Figure 5. The Docking Site and Selector Sequences Consensus Are Complementary.
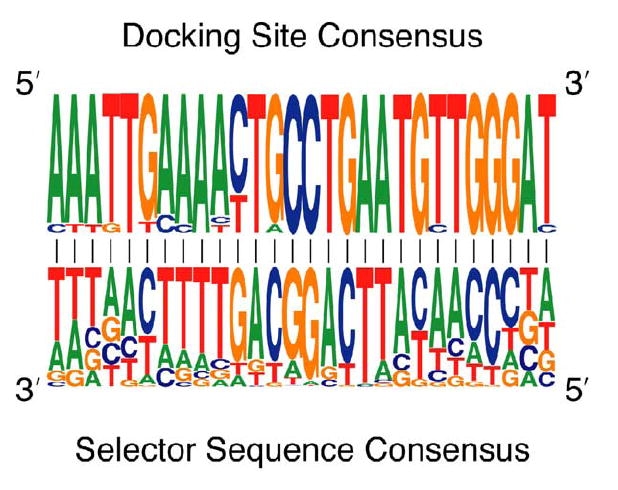
The docking site consensus sequence is complementary to the central 28 nucleotides of the selector sequence consensus. The most frequent nucleotide at each position of the selector sequence is complementary to the docking site.
Figure 6. Conservation of the Docking Site:Selector Sequence Secondary Structures.
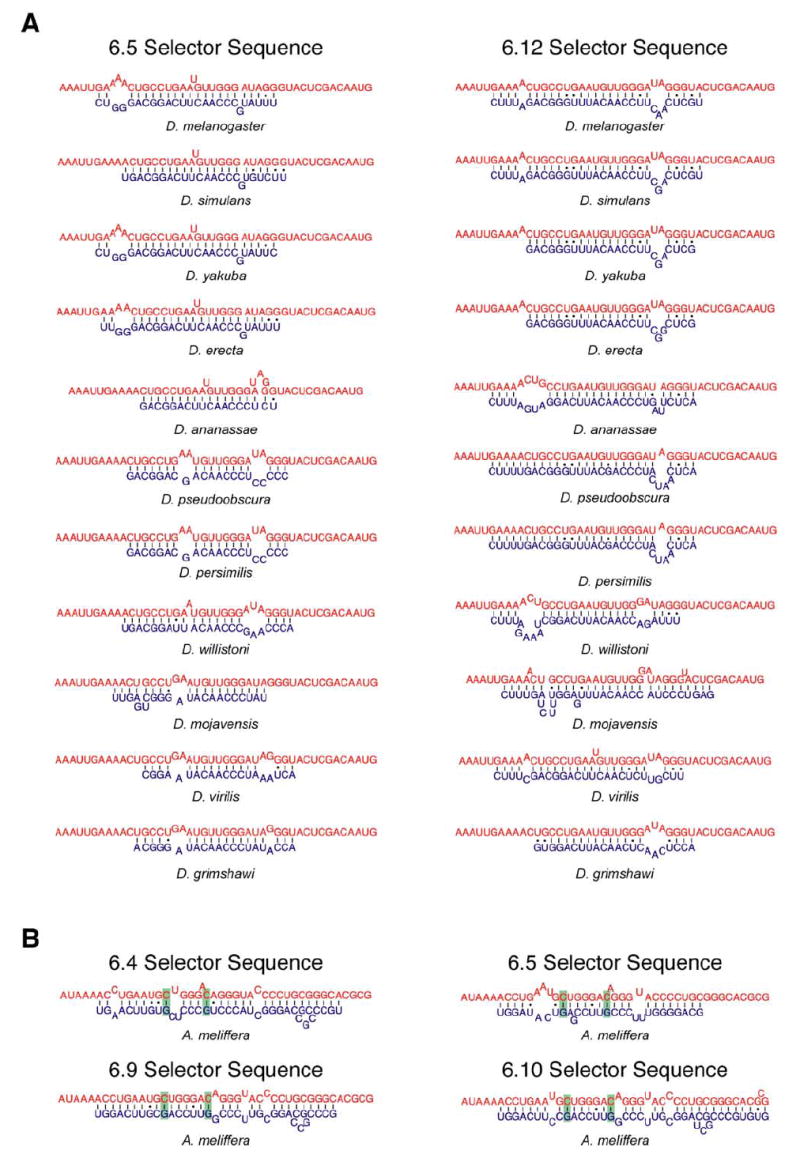
(A) The RNA secondary structures of the proposed interactions between the docking site and the exon 6.5 and 6.12 selector sequences are shown for each Drosophila species. Although the precise structure is not absolutely conserved in each species, similar structures have the potential to form.
(B) The RNA secondary structures of the docking site with the exon 6.4, 6.5, 6.9, and 6.10 selector sequences from A. meliffera are shown. These examples demonstrate that the two nucleotides in the docking site that are invariant in all other species (shaded in green) engage in base-pairing interactions with the selector sequences. These compensatory mutations provide additional evidence supporting the proposed docking site:selector sequence interactions.
The interactions between the selector sequences and the docking site are supported by a number of observations. The docking site is nearly invariant, consistent with the notion that it engages in multiple mutually exclusive interactions. Any mutation in the docking site would affect the interaction with most, if not all, of the selector sequences and therefore interfere with the splicing of the entire exon 6 cluster. In contrast, mutations within a selector sequence would only affect the splicing of the downstream exon 6 variant. Consistent with this, the selector sequences are much less conserved than the docking site. Nonetheless, orthologous selector sequences that contain nucleotide differences can still form similar interactions with the docking site. As an example of this, the predicted secondary structures that the docking site forms with the D. melanogaster exon 6.5 and 6.12 selector sequences and the orthologous selector sequences from ten other Drosophila species are shown in Figure 6A.
Although the docking sites of the non-Drosophila species have diverged from the Drosophila docking site to some extent, potential selector sequences exist upstream of each exon 6 variant in these other species. This is particularly striking given the apparent high rate of recombination within the exon 6 region (Graveley et al., 2004). The docking site in the honey bee A. mellifera is the most divergent from the docking site in D. melanogaster and contains two U to C changes in the most highly conserved portion. However, putative selector sequences exist upstream of each A. mellifera exon 6 variant and are predicted to interact with the A. mellifera docking sequence with a thermodynamic stability similar to those in D. melanogaster (Figure 6B). Most importantly, the two nucleotides that are different in the A. mellifera docking site engage in base-pairing interactions in the majority of the predicted docking site:selector sequence secondary structures. Many of the docking site:selector sequence structures in all species other than the honeybee contain U-A base pairs at these positions, while C-G base pairs exist at these positions in the honeybee structures (Figure 6B). This provides several independent examples of compensatory double mutations that maintain the structural integrity of the docking site:selector sequence interactions. Together, these observations strongly support a model in which the selector sequences interact with the docking site in a mutually exclusive manner.
Discussion
Model for the Mutually Exclusive Splicing of the Dscam Exon 6 Cluster
Several mechanisms have been identified that serve to guarantee that pairs of alternative exons are spliced in a mutually exclusive manner. However, none of the known mechanisms can explain how the alternative splicing of genes containing more than two mutually exclusive exons occurs such that only one exon is included. The Dscam gene is an extreme example of this since the exon 4, 6, and 9 clusters contain 12, 48, and 33 exons, respectively. Here, I have described the docking site and the selector sequences—two classes of conserved sequence elements within the Dscam exon 6 cluster that have the potential to engage in base-pairing interactions. The mutually exclusive nature of the interactions of the selector sequences with the docking site suggests that the formation of these structures is a central component of the mechanism ensuring that only one of the exon 6 variants is included.
It is quite intriguing that each of the Dscam mRNAs isolated from the fly contains only one of the 48 exon 6 variants despite the fact that each exon is flanked by what appear to be functional splice sites (Neves et al., 2004; Schmucker et al., 2000; Zhan et al., 2004). Thus, the mechanism that exists to prevent multiple exon 6 variants from being included must operate with a high degree of fidelity. We have recently identified a protein in an RNAi screen that appears to function to prevent all of the exon 6 variants from being spliced together—when depleted by RNAi, multiple, even adjacent, exon 6 variants are included in the mRNA and they are accurately spliced together (Y. Savva, J. Park, and B.R.G., unpublished data). This finding demonstrates that the exon 6 variants are in fact capable of being spliced together but that protein factors exist that function to repress this reaction.
Based on these two sets of observations, a model can be proposed to explain how the alternative splicing of the exon 6 cluster is mutually exclusive (Figure 7). A key component of this model is that a protein(s) acts to both repress the splicing of each exon 6 variant and to prevent the exon 6 variants from being spliced together. I propose that the selector sequence upstream of the exon 6 variant that is to be included interacts with the docking site and that this interaction somehow relieves the repression on the downstream exon 6 variant, and as a result, it can be spliced to exon 5. Finally, the exon 6 variant that is then spliced to exon 5 could only be spliced to exon 7 because the exon 6 variants downstream of the included exon would still be repressed. As a result, only one exon 6 variant would be included in the mRNA.
Figure 7. Model for the Mechanism of Dscam Exon 6 Mutually Exclusive Splicing.
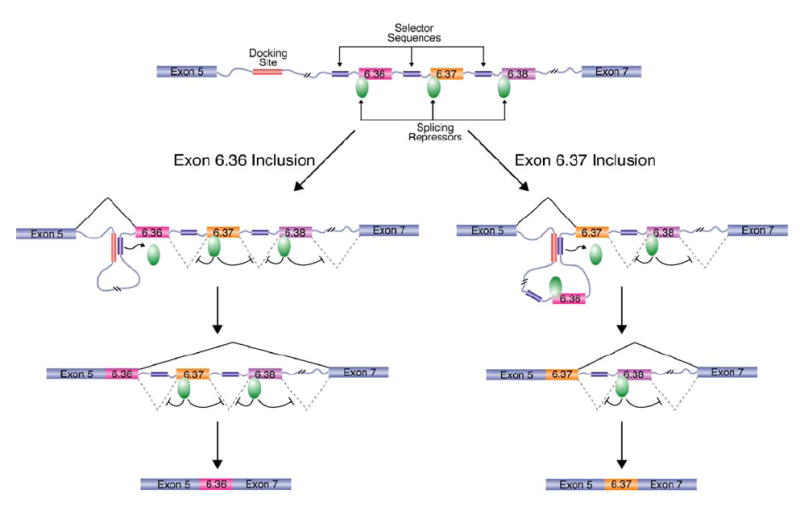
A model of the Dscam exon 6 cluster is depicted in which only variable exons 6.36, 6.37, and 6.38 are shown. A key component of this model is that a splicing repressor functions to prevent the exon 6 variants from being spliced together (green oval). In order for an exon 6 variant to be included in the Dscam mRNA, the selector sequence upstream of the exon must interact with the docking site. For example, if exon 6.36 is to be included (left), the selector sequence upstream of exon 6.36 will interact with the docking site. Likewise, if exon 6.37 is to be included, the selector sequence upstream of exon 6.37 will interact with the docking site. By some unknown mechanism, the docking site:selector sequence interaction inactivates the splicing repressor on the downstream exon and, consequently, activates the splicing of the downstream exon 6 variant to exon 5. Subsequently, the exon that is joined to exon 5 can only be spliced to constitutive exon 7 because the remaining exon 6 variants are actively repressed by the splicing repressor. As a result, only one exon 6 variant is included in the mRNA.
Although the docking site:selector sequence interactions are strongly supported by their evolutionary conservation and some compensatory mutations in the honeybee A. mellifera, this model will obviously need to be experimentally tested with mutations and compensatory mutations that disrupt and restore the docking site:selector sequence interactions. Due to the size (14,000 bp in D. melanogaster) and complexity (48 exons) of the exon 6 cluster, we have made several attempts to generate minigene constructs that lack several of the alternative exons. However, none of the constructs we have made to date are accurately spliced in tissue culture cells. Thus, these experiments may need to be conducted in the fly using the entire exon 6 cluster or perhaps even the entire Dscam gene. Nonetheless, once a system is in place, it will be interesting to test whether the strength of the docking site:selector sequence interactions contribute to the frequency at which each exon 6 variant is used. At first glance, however, it does not appear that the predicted thermodynamic stability of each docking site:selector sequence interaction correlates with the frequency with which each exon 6 variant is used in flies (Neves et al., 2004). Moreover, contrary to what one would expect if splicing occurs cotranscriptionally and the docking site:selector sequence interactions are the driving force of exon 6 selection, the exon 6 variants closest to exon 5 are not chosen more frequently than other exons (Neves et al., 2004). Thus, the mechanism involved in selecting a specific exon 6 variant may be distinct from the interaction between the selector sequence and the docking site.
It will also be interesting to determine precisely what the docking site:selector sequence interaction does. The structures of the docking site:selector sequence interactions are somewhat reminiscent of those that direct site-specific RNA editing (Reenan, 2001). Though it is formally possible that some components of the RNA editing machinery could play a role in Dscam alternative splicing, RNAi depletion of ADAR does not affect alternative splicing of Dscam (Park et al., 2004). An alternate possibility is that the docking site:selector sequence structures serve as binding sites for a protein that somehow inactivates the repression of the downstream exon 6 variant. An intriguing possibility is that the interaction juxtaposes the exon 6 variant to a splicing regulatory element upstream of the docking site. Interestingly, an additionally highly conserved sequence element is located immediately adjacent to the docking site that is predicted to form a 20 bp stem-loop structure that is supported by multiple compensatory mutations (B.R.G., unpublished data). However, the function and relevance of this stem-loop structure is not immediately obvious.
Are competing base-pairing interactions a common mechanism that evolved to negotiate the splicing of genes containing multiple mutually exclusive exons? At first glance, it does not appear so. Conserved elements similar to the docking site and selector sequences are not readily apparent in either the exon 4 or exon 9 clusters of Dscam, nor in other genes containing multiple mutually exclusive exons (C. elegans unc-32 and D. melanogaster Myosin heavy chain, ATPα, GluClα, slowpoke, hephaestus, and Thiolester containing protein II). Thus, the use of competing base-pairing interactions may be unique to the Dscam exon 6 cluster. Moreover, additional experimental and comparative genomic work from our laboratory suggests that the mechanisms of mutually exclusive splicing of each cluster in Dscam are quite possibly different (J. Kreahling and B.R.G, unpublished data). This suggests that multiple distinct and independent mechanisms to ensure the mutually exclusive splicing of clusters of three or more exons may have evolved multiple times. This is not entirely surprising, however, since multiple, distinct mechanisms are known to exist to guarantee that only one exon is included when only two alternative exons need to be chosen from.
Curiously, vertebrate genes that contain a region with more than two mutually exclusive exons have not been identified. This suggests that the vertebrate spliceo-some may have lost the ability to negotiate pre-mRNAs containing more than two mutually exclusive exons. Alternatively, insects and worms (and perhaps other metazoans) may have evolved the ability to cope with the challenge of including only one alternative exon among a multitude of possible choices after they last shared a common ancestor with higher eukaryotes. Due to the fact that multiple mutually exclusive exons can be successfully used to generate such a tremendous diversity of proteins from a single gene, it is striking that genes with this organization are not more common in general and appear to be all together absent from vertebrates.
Experimental Procedures
Gene Assemblies
The sequences of the Dscam genes from D. melanogaster, D. pseudoobscura, D. virilis, A. gambiae, and A. mellifera have been previously described (Graveley et al., 2004; Schmucker et al., 2000). The sequences of the Dscam genes for the other Drosophila species (D. erecta, D. yakuba, D. ananassae, D. simulans, D. persimilis, D. willistoni, D. grimshawi, and D. mojavensis [http://flybase.org/blast/]), the mosquito A. aegypti (http://mosquito.colostate.edu/tikiwiki/tiki-index.php), the silk worm B. mori (Xia et al., 2004), and the red flour beetle T. castaneum (http://www.hgsc.bcm.tmc.edu/projects/tribolium/) were assembled from thousands of individual raw sequence reads available from the NCBI trace archives. Seed sequencing reads were identified by BLAST searches using the sequence of the most closely related organisms. These seed sequences were then used to search the trace archives to identify overlapping sequence reads. These overlapping sequences were manually assembled into larger contigs using the Pustel sequence alignment program in MacVector until the entire gene was eventually assembled.
Sequence Alignments
Initial alignments of the 16 Dscam genes were performed using Multi-PipMaker (http://pipmaker.bx.psu.edu/cgi-bin/multipipmaker; Schwartz et al., 2000). The alignments of specific regions between species were further refined using the ClustalW program in Mac-Vector, as were the alignments of the selector sequences to one another. The consensus sequences of the docking site and selector sequences were derived using WebLogos (http://weblogo.berkeley.edu/; Crooks et al., 2004). The observation that the selector sequences are complementary to the docking site occurred through a combination of staring at the sequences for months and sheer luck.
Secondary Structure Predictions
The structural models of the interactions between the docking site and the selector sequences were generated using Mfold (Zuker, 2003).
Supplementary Material
Supplemental Data include two figures and can be found with this article online at http://www.cell.com/cgi/content/full/123/1/65/DC1/.
Acknowledgments
I would like to thank the members of my laboratory, Rob Reenan, and Asis Das for encouragement, discussions, and comments on the manuscript. This work was supported by a grant from the NIH (GM67842) to B.R.G.
Footnotes
References
- Black DL. Protein diversity from alternative splicing: A challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Kaur A, Gunning D, Zipursky SL, Rowen L, Clemens JC. The organization and evolution of the dipteran and hymenopteran Down syndrome cell adhesion molecule (Dscam) genes. RNA. 2004;10:1499–1506. doi: 10.1261/rna.7105504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Jones RB, Wang F, Luo Y, Yu C, Jin C, Suzuki T, Kan M, McKeehan WL. The nonsense-mediated decay pathway and mutually exclusive expression of alterantively spliced FGFR2 IIIb and IIIc mRNAs. J Biol Chem. 2001;276:4158–4167. doi: 10.1074/jbc.M006151200. [DOI] [PubMed] [Google Scholar]
- Kennedy CF, Berget SM. Pyrimidine tracts between the 5′ splice site and branch point facilitate splicing and recognition of a small Drosophila intron. Mol Cell Biol. 1997;17:2774–2780. doi: 10.1128/mcb.17.5.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Bork P. Common exon duplication in animals and its role in alternative splicing. Hum Mol Genet. 2002;11:1561–1567. doi: 10.1093/hmg/11.13.1561. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- Neves G, Zucker J, Daly M, Chess A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nat Genet. 2004;36:240–246. doi: 10.1038/ng1299. [DOI] [PubMed] [Google Scholar]
- Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci USA. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- Powell JR. Progress and Prospects in Evolutionary Biology: The Drosophila Model. Oxford: Oxford University Press; 1997. [Google Scholar]
- Reenan RA. The RNA world meets behaviour: A to I pre-mRNA editing in animals. Trends Genet. 2001;17:53–56. doi: 10.1016/s0168-9525(00)02169-7. [DOI] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker—a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA, Burge CB. Classification of introns: U2-type or U12-type. Cell. 1997;91:875–879. doi: 10.1016/s0092-8674(00)80479-1. [DOI] [PubMed] [Google Scholar]
- Smith CW, Nadal-Ginard B. Mutually exclusive splicing of α-tropomyosin exons enforced by an unusual lariat branch point location: Implications for constitutive splicing. Cell. 1989;56:749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- Southby J, Gooding C, Smith CW. Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of α-actinin mutually exclusive exons. Mol Cell Biol. 1999;19:2699–2711. doi: 10.1128/mcb.19.4.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc V, Gauhar Z, Mason C, Halasz G, van Batenburg MF, Rifkin SA, Hua S, Herreman T, Tongprasit W, Barbano PE, et al. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- Thanaraj TA, Stamm S, Clark F, Riethoven JJ, Le Texier V, Muilu J. ASD: the alternative splicing database. Nucleic Acids Res. 2004;32:D64–D69. doi: 10.1093/nar/gkh030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B, Zhao P, Zha X, Cheng T, Chai C, et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- Zhan XL, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, Vasconcelos ML, Chess A, Zipursky SL. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 2004;43:673–686. doi: 10.1016/j.neuron.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data include two figures and can be found with this article online at http://www.cell.com/cgi/content/full/123/1/65/DC1/.


