Abstract
Glycosylphosphatidylinositol (GPI), covalently attached to many eukaryotic proteins, not only acts as a membrane anchor but is also thought to be a sorting signal for GPI-anchored proteins that are associated with sphingolipid and sterol-enriched domains. GPI anchors contain a core structure conserved among all species. The core structure is synthesized in two topologically distinct stages on the leaflets of the endoplasmic reticulum (ER). Early GPI intermediates are assembled on the cytoplasmic side of the ER and then are flipped into the ER lumen where a complete GPI precursor is synthesized and transferred to protein. The flipping process is predicted to be mediated by a protein referred as flippase; however, its existence has not been proven. Here we show that yeast Arv1p is an important protein required for the delivery of an early GPI intermediate, GlcN-acylPI, to the first mannosyltransferase of GPI synthesis in the ER lumen. We also provide evidence that ARV1 deletion and mutations in other proteins involved in GPI anchor synthesis affect inositol phosphorylceramide synthesis as well as the intracellular distribution and amounts of sterols, suggesting a role of GPI anchor synthesis in lipid flow from the ER.
INTRODUCTION
Glycosylphosphatidylinositol (GPI) is a complex glycolipid found in all eukaryotic cells. It is covalently attached to the C-terminal end of certain secretory proteins and acts as a membrane anchor (Kinoshita and Inoue, 2000; Ikezawa, 2002; Pittet and Conzelmann, 2007). Because many proteins anchored by GPI are poorly extracted with detergents at 4°C, they have been proposed to be localized in detergent-resistant membranes (DRMs), which may be related to microdomains called lipid rafts that have been proposed to play an important role in the trafficking of GPI-anchored proteins (Simons and Ikonen, 1997; Chatterjee and Mayor, 2001; Mayor and Riezman, 2004).
Biosynthesis of GPI takes place on the membranes of the endoplasmic reticulum (ER) (Kinoshita and Inoue, 2000; Pittet and Conzelmann, 2007), and this starts with the transfer of N-acetylglucosamine (GlcNAc) from UDP-GlcNAc to the inositol of phosphatidylinositol (PI) to generate N-acetylglucosaminyl-PI (GlcNAc-PI), followed by de-N-acetylation to form glucosaminyl-PI (GlcN-PI). The two reactions occur on the cytoplasmic side of the ER membrane (Vidugiriene and Menon, 1993; Orlean and Menon, 2007). The GlcN-PI is then acylated on the inositol ring to form glucosaminyl-acyl-PI (GlcN-acylPI) before or after transfer of the first mannose (Smith et al., 1997; Kinoshita and Inoue, 2000; Pittet and Conzelmann, 2007). Dolicholphosphomannose (Dol-P-Man) is the mannose donor for the core mannose residues (Menon et al., 1990). In Saccharomyces cerevisiae and Plasmodium falciparum, inositol acylation appears to be a prerequisite for mannosylation of GPIs (Doerrler et al., 1996; Gerold et al., 1999), whereas, in Trypansoma brucei and mammalian cells, it is not essential for addition of the first mannose (Guther and Ferguson, 1995; Smith et al., 1996; Murakami et al., 2003), although the mammalian mannosyltransferase (GPI-MT-I), PIG-M seems to prefer GlcN-acylPI to GlcN-PI. The PIG-M has a lumenally oriented, functionally important DXD motif that is found in many glycosyltransferases (Maeda et al., 2001). Based on the topological aspect of synthesis of GlcN-PI and subsequent mannosylation, it was suggested that the early GPI intermediates must flip from the cytoplasmic side to the ER lumen before the first mannosylation. In addition, genes coding for mammalian PIG-W and the yeast homologue GWT1, which are involved in inositol acylation of GPI were recently identified (Murakami et al., 2003; Umemura et al., 2003). They encode multi-spanning ER membrane proteins. Because the comparison of locations of predicted transmembrane domains and amino acid sequence of PIG-W homologues of various organisms showed that the location of conserved regions face the lumenal side of the ER, it was also proposed that inositol acylation occurs in the ER lumen; hence, inositol acylation is not required for flipping of GPI (Murakami et al., 2003). However, it remains unclear whether the catalytic site of PIG-W is located within the conserved regions. Mutations in the temperature-sensitive (ts) mutant alleles of GWT1, which were randomly generated by PCR mutagenesis, were not found in the conserved regions (Umemura et al., 2003). Thus, it is not yet known whether GlcN-PI or GlcN-acylPI, or both, are flipped across the ER membrane. After mannosylation and addition of phosphorylethanolamine (EtNP) residue(s), the entire GPI anchor precursor is attached to proteins by a GPI transamidase, which acts on the lumenal side of the ER (Kinoshita and Inoue, 2000; Ikezawa, 2002; Pittet and Conzelmann, 2007). Subsequently, the acyl group of the inositol is removed from the GPI-anchored proteins (Tanaka et al., 2004; Fujita et al., 2006a), and the GPI lipid moieties are remodeled to a more hydrophobic diacylglycerol or ceramide (Sipos et al., 1997; Reggiori et al., 1997). Most of the genes encoding enzymes involved in GPI biosynthetic pathway have been isolated and characterized (Kinoshita and Inoue, 2000; Pittet and Conzelmann, 2007). Recently, genes that are required for lipid remodeling of GPI-anchored proteins were also identified (Tashima et al., 2006; Bosson et al., 2006; Fujita et al., 2006b; Ghugtyal et al., 2007; Umemura et al., 2007). However, despite evidence that GPI precursor flipping is a protein-mediated process (Vishwakarma and Menon, 2005; Pomorski and Menon, 2006), the putative GPI flippase has not yet been identified.
Once the inositol-linked acyl chain is eliminated by a GPI inositol deacylase, the GPI-anchored proteins are transported from the ER to the Golgi apparatus via transport vesicles (Muniz and Riezman, 2000; Ikonen, 2001; Mayor and Riezman, 2004). In S. cerevisiae, GPI-anchored proteins are known to exit the ER in vesicles distinct from other secretory proteins (Muniz et al., 2001). Rab GTPase Ypt1p, the tethering factors Uso1p, COG complex Sec34p and Sec35p, and the ER v-SNAREs Bos1p, Bet1p, and Sec22p are necessary for the sorting of GPI-anchored proteins upon ER exit (Morsomme and Riezman, 2002; Morsomme et al., 2003). The lipid composition may have a function in sorting of GPI-anchored proteins into ER-derived vesicles because GPI-anchored proteins are associated with DRMs in the ER (Bagnat et al., 2000) and because ongoing ceramide synthesis is required for the efficient transport of GPI-anchored proteins (Horvath et al., 1994; Sutterlin et al., 1997a; David et al., 1998; Barz and Walter, 1999; Watanabe et al., 2002). Ceramide synthesis does not seem to be required for GPI-anchored protein transport in trypanosomes, but the anchors are not remodeled to ceramides in these cells (Sutterwala et al., 2007). Because the maturation of GPI-anchored proteins is delayed when yeast cells are incubated with myriocin, an inhibitor of serine palmitoyltransferase (SPT), or when a ts lcb1-100 mutant defective in SPT activity is incubated at nonpermissive temperature (Horvath et al., 1994; Sutterlin et al., 1997a), but not affected when yeast cells are treated with aureobasidin A (AbA), an inhibitor of yeast sphingolipid synthesis (Reggiori and Conzelmann, 1998), ceramide-rich domains rather than complex sphingolipid-rich domains could be important for the sorting step in the ER. This is also supported by the fact that ceramide is synthesized in the ER and then is delivered to the Golgi where it is converted to inositolphosphorylceramide (IPC), followed by mannosylation to form mannosyl IPC (MIPC) and mannosyl di(inositolphosphoryl)ceramide [M(IP)2C], which are the three major yeast sphingolipids (Funato et al., 2002; Dickson et al., 2006). Previously, we have shown that ER-to-Golgi transport of ceramide for IPC synthesis is mediated by both vesicular and nonvesicular trafficking (Funato and Riezman, 2001).
Although the roles of the different pathways are poorly understood, the vesicular transport of ceramide could be directly coupled to transport of proteins. In addition, GPI anchor attachment to proteins and the lipid remodeling are necessary for efficient transport of GPI-anchored proteins to the Golgi (Hamburger et al., 1995; Doering and Schekman, 1996; Bosson et al., 2006; Fujita et al., 2006b). Recent studies have shown that mutant cells defective in GPI anchor synthesis or attachment block export of some detergent-insoluble transmembrane proteins from the ER (Okamoto et al., 2006), suggesting a role of GPI-anchored proteins in raft-dependent protein sorting. However, evidence indicating that GPI-anchored proteins are required for transport of raft lipid(s) per se is missing.
In this study we show that yeast ARV1, which has been known to be somehow involved in the regulation of sphingolipid and sterol metabolism (Tinkelenberg et al., 2000; Swain et al., 2002; Fores et al., 2006), genetically interacts with genes involved in GPI anchor synthesis and that Arv1p is required for the efficient synthesis of GlcN-acylPI bearing one mannose, whereas arv1Δ cells retain both Dol-P-Man synthesis and GPI-MT activity. We propose that the primary function of Arv1p is either to deliver GlcN-acylPI from the cytoplasmic side to the luminal side of the ER or to present it to the yeast PIG-M homologue GPI14 (Maeda et al., 2001). In addition, we present evidence that GPI assembly is required not only for GPI-anchored protein transport but also for ceramide transport from the ER as well as to maintain the proper intracellular distribution and amounts of sterols.
MATERIALS AND METHODS
Strains, Media, Library Screen, Plasmids, and Reagents
The yeast strains and plasmids used in this study are listed in Supplementary Tables S1 and S2, respectively. Yeast cultures and genetic manipulations were carried out essentially as described by Sherman et al. (1983). To construct deletion strains, entire open reading frames were deleted and replaced with the designated genes. Deletions were confirmed by PCR. Yeast strains were grown in rich medium, YPUAD (Dulic et al., 1991), semisynthetic medium, SDYE (Horvath et al., 1994), or synthetic minimal medium, SD (Funato et al., 2003), supplemented with the appropriate nutrients to select for plasmids. pmi40Δ mutant cells were supplemented with 0.5% mannose (Pitkanen et al., 2004).
Temperature sensitivity was determined by spotting diluted yeast cultures on SD (−ura) plates at 25 and 37°C. To test drug sensitivity, cells were cultured on SD plates containing calcofluor white (CFW) or AbA. A genomic LEU2-marked library made in YEp13 (a gift from Dr. Y. Ohya, University of Tokyo, Japan) was used to isolate suppressors of the ts growth phenotype of arv1Δ mutant (FK137). A plasmid was isolated containing GPI15. GPI15, GPI1, GPI2, GPI3, and ERI1 genes were amplified from S. cerevisiae genomic DNA by PCR. GPI15 was cloned into BamHI/EcoRI sites of pRS426GPD, GPI1 into BamHI/XhoI, GPI2 into BamHI/SalI, GPI3 into SpeI/XhoI, and ERI1 into BamHI/XhoI of pRS426ADH vector. To construct a plasmid (pGAL-HA-GPI18) for expressing N-terminal two-hemagglutinin (HA)-tagged GPI18, the BamHI-SalI PCR fragment containing the open reading frame of GPI18 was ligated into pRS316-GAL1-2xHA-BS, a pRS316-based expression vector carrying the EcoRI site, the initiation codon, two HA-epitope–encoding regions, and multicloning sites (provided by Dr. K. Tanaka, Hokkaido University, Japan). The EcoRI-SalI fragment including the epitope-tagged GPI18 was subcloned into pRS416GAL1. A diploid heterozygous ARV1/arv1Δ::LEU2 GPI18/gpi18Δ::KANR double deletion strain was created by deleting ARV1 in the GPI18/gpi18::KANR diploid strain BY4743 MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 lys2Δ0 met15Δ0 gpi18Δ::KANR (generated by the Saccharomyces Gene Deletion Project and provided by Dr. T. Kinoshita, Osaka University, Japan) and transformed with pGAL-HA-GPI18. The transformants were sporulated and tetrads dissected to obtain haploid strains arv1Δ::LEU2 gpi18Δ::KANR (FK1015), gpi18Δ::KANR (FK1017), and wild-type (FK1019) harboring pGAL-HA-GPI18. A C-terminal triple HA-tagged ARV1 was amplified from genomic DNA (prepared from RH6076, which carried the epitope-tagged ARV1 gene) by PCR, and cloned into BamHI/XhoI sites of pRS426ADH. AbA and C2-ceramide were purchased from Takara (Tokyo, Japan) and Matreya (State College, PA), respectively. YW3548 was prepared as described previously (Sutterlin et al., 1997b).
Labeling of Lipids In Vivo
In vivo labeling of lipids with [3H]myo-inositol (Perkin Elmer-Cetus Life Sciences, Boston, MA) or [3H]dihydrosphingosine (DHS; American Radiolabeled Chemicals, St. Louis, MO) was performed as described previously (Zanolari et al., 2000). Cells were grown overnight in SDYE, harvested and resuspended in SD without inositol for labeling with [3H]myo-inositol, or SD medium for labeling with [3H]DHS. The cells were preincubated for 15 min at 25°C and then labeled by 25 μCi of [3H]myo-inositol or 10 μCi of [3H]DHS. If present, C2-ceramide (200 μM) was added at the start of the labeling. The incubation was stopped by the addition of 10 mM NaF and 10 mM NaN3. The cells were then washed with cold water, and lipids were extracted with chloroform-methanol-water (10:10:3, vol/vol/vol). Half of the dried sample was treated by mild alkaline hydrolysis with 0.6 M NaOH, neutralized, and desalted by n-butanol partitioning. The lipids were analyzed by thin-layer chromatography (TLC) using solvent system I, chloroform-methanol-0.25% KCl (55:45:10, vol/vol/vol) for complex sphingolipid labeled with [3H]myo-inositol, and solvent system II, chloroform-methanol-4.2 N ammonium hydroxide (9:7:2, vol/vol/vol) for sphingolipid labeled with [3H]DHS. Radiolabeled lipids were visualized and quantified on a Cyclone Storage Phosphor System (Packard Instrument, Meriden, CT).
In vivo [3H]mannose labeling using the pmi40Δ and arv1Δ pmi40Δ double mutant strains was performed as described (Sipos et al., 1994; Sutterlin et al., 1997b). In brief, the cells were grown overnight in SDCU medium (2% glucose, 1% peptone, 0.67% yeast nitrogen base, 0.5% mannose, and the required nutrients), harvested, and resuspended in SPCU medium (0.1% glucose, 2% pyruvate, 0.67% yeast nitrogen base, and the required nutrients). Wild-type and arv1Δ mutant strains were grown in SDCU medium without mannose. For [3H]mannose labeling of strains transformed with pGAL-HA-GPI18, cells were first grown in SGYE (2% galactose, 0.67% yeast nitrogen base, 0.2% yeast extract, and the required nutrients) medium, then shifted to SDYE medium for 16 h at 25°C, and resuspended in SPCU medium. The cells were preincubated for 20 min at 25°C with 20 μg/ml tunicamycin in the presence or absence of YW3548 (10 μM) and then labeled with 25 μCi of [3H]mannose (Perkin-Elmer Cetus Life Sciences) for 30 min at 25°C. The labeling was stopped by the addition of NaF and NaN3, and lipids were extracted with chloroform-methanol-water (10:10:3, vol/vol/vol), desalted by n-butanol partitioning, and analyzed by TLC using solvent system III, chloroform-methanol-water (10:10:3, vol/vol/vol).
Labeling of Lipids In Vitro
Membranes for in vitro labeling experiments with [14C]UDP-GlcNAc (Perkin-Elmer Cetus Life Sciences) or with [3H]GDP-mannose (GDP-Man; American Radiolabeled Chemicals) were prepared as described by Schonbachler et al. (1995), except that for cell breakage, TDP (50 mM Tris-HCl, pH 7.4, 5 mM DTT, and 1 mM PMSF) was used. The membranes were suspended in TDP containing 20% glycerol and stored at −80°C. Samples of membranes containing equivalent amounts of protein were assayed for the early steps of GPI anchor biosynthesis using [14C]UDP-GlcNAc as described (Schonbachler et al., 1995) with a few modifications. Two hundred micrograms of membranes were incubated with 50 nCi [14C]UDP-GlcNAc at 25°C in 50 μl of GPI buffer consisting of 100 mM Tris-HCl, pH 7.5, 1 mM EGTA, 3 mM Mg-acetate, 0.5 mM MnCl2, 1 mM CoA, 1 mM ATP, 20 μg/ml tunicamycin, and 250 nM GDP-Man. Dol-P-Man synthesis was assayed in GPI buffer containing 0.5 mM UDP-GlcNAc and 0.25 μCi [3H]GDP-Man (250 nM). Lipids were extracted by the addition 750 μl of chloroform-methanol (1:2, vol/vol), desalted, and analyzed by TLC using solvent system IV, chloroform-methanol-1N ammonium hydroxide (10:10:3, vol/vol/vol) for [14C]UDP-GlcNAc labeled lipids (Watanabe et al., 1998), and solvent system III for the synthesis of Dol-P-Man (Sutterlin et al., 1997b).
For mannosylated GPI lipid synthesis assay, ER-enriched membranes were prepared as described (Funato and Riezman, 2001), and the membranes were incubated with GPI buffer, 0.5 mM UDP-GlcNAc and 0.5 μCi [3H]GDP-Man in a final volume of 100 μl (Sutterlin et al., 1997b). Some reactions contained 10 mM n-octyl β-d-glucopyranoside and 0.4 μg/ml dioctanoyl-PI [GlcN-PI(C8); a gift from Dr. M. A. Lehrman, University of Texas Southwestern Medical Center, Dallas, TX] with 6 × 10−4 % Triton X-100, which was used to add GlcN-PI(C8). After 10-min incubation at 25°C, lipids were extracted by the addition 666 μl of chloroform-methanol (1:1, vol/vol), desalted, and analyzed by TLC using solvent system III.
Pulse-Chase Experiments for Protein Maturation and GPI Anchor Attachment
Radiolabeling and immunoprecipitation to measure maturation of GPI-anchored proteins (Gas1p, Yps1p) and carboxypeptidase Y (CPY) were performed as described previously (Sutterlin et al., 1997a). The samples were subjected to SDS-PAGE, analyzed, and quantified using the Cyclone Storage Phosphor System (Packard Instruments). The percentage of mature proteins was determined by taking the ratio of the mature form to the total signal (ER, Golgi, and mature forms). GPI anchor attachment was examined as described (Watanabe et al., 2002).
Fluorescence Microscopy
Labeling of cells with C6-NBD-ceramide was performed as described (Levine et al., 2000). Cells were incubated with or without AbA (20 μg/ml) at 25°C for 15 min and labeled with C6-NBD-ceramide (20 μM) at 25°C for 15 min in the presence of defatted BSA (5 mg/ml), followed by washing and back-extracting. The staining was visualized by fluorescence microscopy. For visualization of chitin (Sobering et al., 2004) and sterol distribution (Beh and Rine, 2004), cells were grown in SD medium at 25°C, washed in PBS, and stained with CFW (1 mg/ml) for 5 min and filipin complex (0.1 mg/ml) for 15 min, respectively. The cells were washed and observed by fluorescence microscopy with a UV filter.
Gas Liquid Chromatography–Mass Spectrometry Analysis of Sterols
Cells were grown in SD medium at 25°C. Total sterols including free sterols and sterol esters were extracted from whole cells and analyzed by gas liquid chromatography-mass spectrometry (GLC-MS) as described previously (Munn et al., 1999; Heese-Peck et al., 2002, Mullner et al., 2005). Cholesterol was used as an internal standard.
RESULTS
An arv1Δ Mutant Is Deficient in Ceramide Transport from the ER to the Golgi Site of IPC Synthesis
ARV1 was identified in a screen for genes that are essential in the absence of yeast sterol esterification (Tinkelenberg et al., 2000). Arv1p is a multispanning integral membrane protein with six predicted transmembrane domains, and homologues have been found in a wide variety of eukaryotes. Because disruption of ARV1 leads to not only altered levels and distribution of sterols but also reduced sphingolipid levels, it has been proposed that this protein is involved in maintenance of cellular sterol and sphingolipid homeostasis (Swain et al., 2002). To obtain further evidence for the role of Arv1p in the regulation of sphingolipid homoeostasis, we re-examined sphingolipid biosynthesis in arv1Δ mutant cells, which were generated in our strain background. Consistent with the previous report, the arv1Δ mutant showed a strong reduction of sphingolipid synthesis, ∼20% of IPC-C/-D/MIPC and 50% of M(IP)2C levels found in wild-type cells when cells were pulse-labeled with [3H]myo-inositol (Figure 1A). A similar reduction of IPC-C synthesis was also seen when the sphingolipid synthesis of arv1Δ mutant was assessed by [3H]DHS labeling (Figure 1B), confirming that this mutant exhibits a sphingolipid synthesis defect. As shown by a previous experiment (Swain et al., 2002), we also observed that the arv1Δ mutant cells made normal amounts of ceramides as determined by [3H]DHS pulse-labeling for 2 h (data not shown).
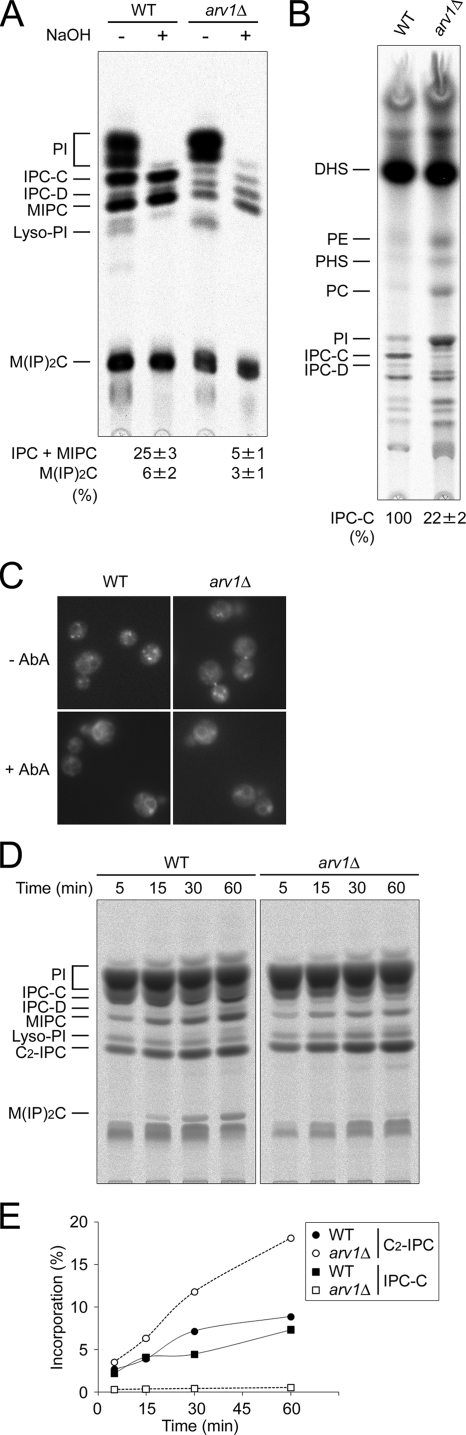
Figure 1.
The arv1Δ mutant is defective in ceramide transport from the ER to the Golgi. (A and B) Wild-type (RH6082) and arv1Δ (RH6078) strains were labeled with [3H]myo-inositol (A) or with [3H]DHS (B) for 2 h at 25°C. The [3H]myo-inositol–labeled lipids were subjected (+) or not (−) to mild alkaline hydrolysis with NaOH. (C) The same strains as in A were labeled with C6-NBD-ceramide for 15 min at 25°C, with or without AbA as described in Materials and Methods, and the staining was visualized by fluorescence microscopy. (D) The same strains were also labeled with [3H]myo-inositol for the indicated times at 25°C in the presence of C2-ceramide. The labeled lipid products were applied to TLC plates using solvent system I (A and D) or solvent system II (B). Lipids were identified by their mobility and resistance to mild-base treatment. The band, which was identified as C2-IPC, appeared only when the precursor C2-ceramide was added, and the appearance was resistant to mild-base treatment and sensitive to the addition of AbA (not shown). Incorporation (%) into sphingolipids was determined as the percentage of total radioactivity into all lipids containing inositol (A and D). Incorporation of [3H]DHS into IPC-C was quantified, and the relative amounts were calculated by setting the amounts in wild-type cells to 100% (B). Results of a typical experiment are shown. Data in A and B are means ± SE for six and four independent experiments, respectively, and in E are quantification of D. PI, phosphatidylinositol; IPC-C and -D, inositolphosphorylceramide subclasses C and D; MIPC, mannosyldi(inositolphosphoryl) ceramide; Lyso-PI, lyso-phosphatidylinositol; DHS, dihydrosphingosine; PE, phosphatidylethanolamine; PHS, phytosphingosine; PC, phosphatidylcholine.
Because a sphingolipid synthesis defect could result from a lack of IPC synthase activity, we next examined IPC synthase activity in arv1Δ mutant cells. In wild-type cells, exogenously added C6-NBD-ceramide is efficiently converted to C6-NBD-IPC, and the product is trapped in the Golgi compartment where Aur1p, the IPC synthase, is localized. When the fluorescent NBD probe is added to cells where IPC synthase is inhibited with AbA, an IPC synthase inhibitor, the probe is preferentially incorporated into the ER membrane, indicating that localization of the NBD probe is dependent on the activity of IPC synthase (Levine et al., 2000). Our data indicate that there is no apparent difference between wild-type and arv1Δ mutant cells in the localization of exogenous C6-NBD-ceramide during a short labeling period, 15 min (Figure 1C), suggesting that IPC synthase in the arv1Δ mutant is active. For a quantitative assessment of IPC synthase activity, incorporation of exogenous C2-ceramide into C2-IPC in these strains was measured with [3H]myo-inositol labeling (Figure 1D). Although the incorporation of endogenous ceramide into IPC-C in arv1Δ mutant cells was reduced, the incorporation of C2-ceramide into C2-IPC was 1.5–2-fold greater than wild-type cells after long labeling times, 30 and 60 min, respectively (Figure 1E). Increased incorporation is probably due to the low levels of competition between endogenous and exogenous ceramides. Consistently, wild-type cells treated with australifungin, an inhibitor of ceramide synthase, displayed a significant increase (1.8-fold) in C2-IPC synthesis (data not shown). These results demonstrate that arv1Δ mutant cells are not compromised for IPC synthase activity and therefore suggest that sphingolipid synthesis defect of arv1Δ mutant cells results from a deficiency in ceramide transport.
Considering that the arv1Δ mutant is deficient in ceramide transport, it is reasonable to assume that this block would entail the accumulation of intermediates in ceramide synthesis. The finding that arv1Δ mutant cells showed an increased incorporation of [3H]DHS into glycerophospholipids (Figure 1B) is consistent with this, because DHS incorporation is dependent upon its phosphorylation and subsequent cleavage by a long-chain base phosphate lyase (Funato et al., 2003; Saba, 2006). Higher amounts of DHS are likely to occur when ceramide synthesis is backed up, due to its lack of transport.
ARV1 Genetically Interacts with Genes Involved in GPI Anchor Synthesis
Although the arv1Δ mutant can grow at 25, 30, and 35°C, it fails to grow at 37°C (Swain et al., 2002, Figure 2, A and B). To gain further insight into the function of Arv1p in ceramide transport, we screened a 2 μ/LEU2-marked genomic yeast library for multicopy suppressors of the growth defect. A plasmid was isolated from this screen, which contained GPI15, and rescued the arv1Δ ts growth (Figure 2A). GPI15 encodes a protein involved in the synthesis of GlcNAc-PI, the first intermediate in GPI anchor biosynthesis (Yan et al., 2001). GlcNAc-PI is synthesized by the GPI-GlcNAc transferase, which is thought to act as a complex composed of six known components in yeast: Gpi1p, Gpi2p, Gpi3p, Gpi15p, Gpi19p, and Eri1p (Pittet and Conzelmann, 2007). To test whether an excess of GPI-GlcNAc transferase activity was required for the suppression, other genes were cloned into a 2 μ vector and transformed into the arv1Δ mutant cells. Overexpression of Gpi2p suppressed the arv1Δ growth defect at 37°C almost as well as Arv1p overexpression (Figure 2A). Partial rescue of the arv1Δ mutant growth defect was observed by overexpressing Gpi1p, Gpi3p, and Eri1p.
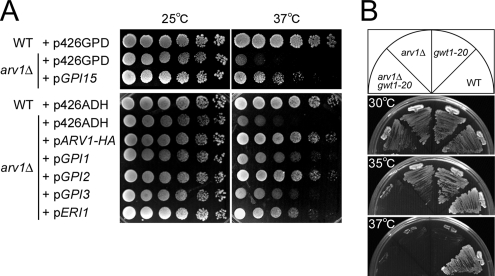
Figure 2.
Overexpresssion of genes involved in GPI anchor synthesis suppresses the temperature-sensitive growth defect of arv1Δ, and combining arv1Δ and gwt1-20 causes a synthetic growth defect. (A) Wild-type (RH6082) and arv1Δ (FK137) strains transformed with pRS426GPD, pRS426ADH, pGPI15, pARV1-HA, pGPI1, pGPI2, pGPI3, or pERI1 were spotted onto SD-ura plates and incubated for 4 d at 25 or 37°C. (B) Wild-type (W303-1B), gwt1-20, arv1Δ (FK245), and arv1Δ gwt1-20 (FK246) were streaked onto YPUAD plates and incubated for 3 d at 30, 35, or 37°C.
Furthermore, we found that arv1Δ mutant causes a synthetic growth defect with the gwt1-20 mutant allele, which shows a ts growth phenotype (Umemura et al., 2003). gwt1-20 mutant cells have a defect in inositol acylation of the GPI precursor lipid even at 24°C. Analysis of an arv1Δ gwt1-20 double mutant at several temperatures revealed that although the arv1Δ and gwt1-20 single mutants grow until 35°C, the arv1Δ gwt1-20 double mutant is inviable at this temperature (Figure 2B), supporting a functional link between ARV1 and GPI anchor biosynthesis.
Arv1p Is Involved in GPI Anchoring of Proteins
These genetic interactions suggest that Arv1p may have a role in GPI anchor synthesis or anchoring. To test this hypothesis, we examined the kinetics of maturation of GPI-anchored proteins in an arv1Δ mutant and wild-type cells, because GPI anchor synthesis and anchoring are required for efficient transport of GPI-anchored proteins from the ER to the Golgi (Schonbachler et al., 1995; Hamburger et al., 1995; Doering and Schekman, 1996; Sobering et al., 2004). Gas1p, a GPI-anchored protein that undergoes O-linked and N-linked glycosylation, is synthesized as a 105-kDa glycoprotein in the ER, and after arrival at the Golgi compartment, fully glycosylated Gas1p has an apparent size of 125 kDa (Nuoffer et al., 1993). A pulse-chase experiment revealed that the maturation of Gas1p is retarded in arv1Δ mutant cells (Figure 3A). A similar result was observed for the maturation of another GPI-anchored protein, Yps1p, whose ER form is found at 85 kDa, and then is modified in the Golgi and migrates as a smear of 120–180 kDa (Sievi et al., 2001). By contrast, transport from the ER (P1) to the Golgi (P2) and onto the vacuole (mature form) of CPY (Stevens et al., 1982) in arv1Δ mutant cells occurs with wild-type kinetics. Because CPY is an N-glycosylated but not a GPI-anchored protein, these results indicate that protein N-glycosylation is normal and ER-to-Golgi transport of GPI-anchored proteins is specifically delayed in arv1Δ mutant.
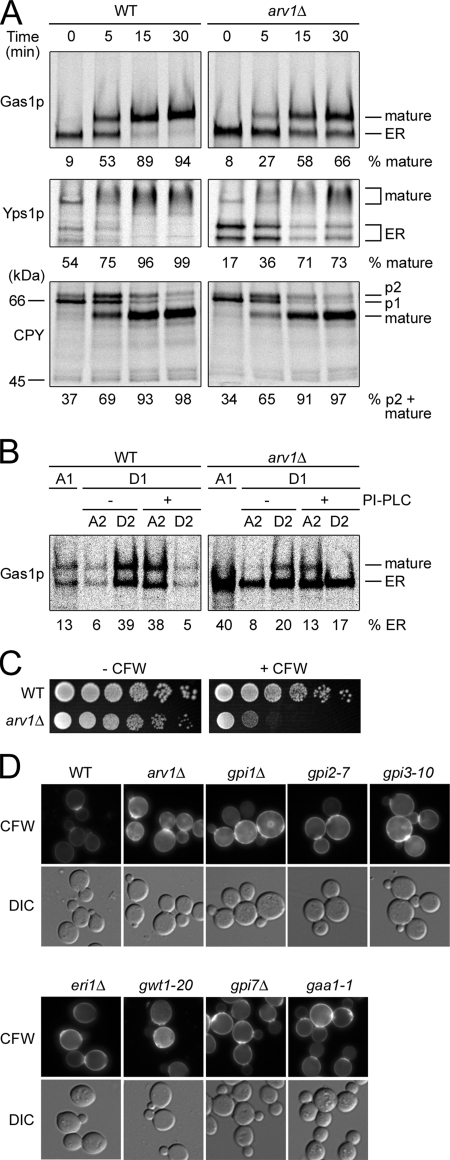
Figure 3.
arv1Δ mutant cells show GPI-anchored protein transport and attachment defects and display the same phenotypes as gpi mutants. (A) ER-to-Golgi transport of protein was examined by pulse-chase experiments as described in Materials and Methods. Wild-type (RH6082) and arv1Δ (RH6078) strains were labeled with [35S]methionine for 6 min at 25°C and chased for the indicated period of time. The percentage of mature Gas1p, Yps1p, or CPY is shown. (B) GPI anchor attachment was studied in wild-type (RH6082) and arv1Δ (RH6078) cells. The cells were labeled with [35S]methionine for 6 min and chased for 5 min at 25°C. Cell extracts were prepared and solubilized with Triton X-114. After partitioning into detergent (D1) and aqueous phases (A1), the detergent phase was divided and incubated in the presence or absence of PI-PLC to remove anchors. Phases were re-extracted to yield A2 and D2 fractions and processed for Gas1p immunoprecipitation followed by SDS-PAGE. The total amount of ER form of Gas1p quantified in each partition was set to 100%. (C) Wild-type (RH6082) and arv1Δ (FK137) strains were spotted onto YPUAD, YPUAD supplemented with 10 μg/ml CFW, and incubated for 4 d at 25°C. (D) Wild-type (RH6082), arv1Δ (FK137), gpi1Δ (DL2831), gpi2-7 (DL2828), gpi3-10 (DL2829), eri1Δ/eri1Δ (DL2680), gwt1-20, gpi7Δ (FBY182) and gaa1-1 (RH401-7C) were stained for chitin with CFW (1 mg/ml). CFW staining was visualized by fluorescence microscopy. The images were taken with equal exposures. The morphology of the cells was observed by differential interference contrast (DIC).
To further examine whether arv1Δ mutant cells have a cargo-specific defect in protein transport, we analyzed Golgi-to-ER retrograde transport, endocytosis, and endosome-to-vacuole transport. Because it has been shown that several mutants that block retrograde transport exhibit a defect in the ER-to-Golgi anterograde traffic of CPY (Duden et al., 1994; Gaynor and Emr, 1997; Duden et al., 1998), it is unlikely that the arv1Δ mutant has a retrograde defect. This is supported by our finding that, although GFP-Rer1p is mislocalized to the vacuole in a coatomer mutant, ret1-1, which has a defect in the Golgi-to-ER retrograde transport (Sato et al., 2001), it shows normal Golgi localization in arv1Δ mutant cells (Figure S1A). Moreover, α-factor internalization (Figure S1B), delivery of lucifer yellow (Figure S1C) and FM4–64 (Figure S1D), fluid-phase and endocytic membrane markers were not affected in the arv1Δ mutant cells.
The delay in maturation of GPI-anchored proteins could result from a defect in GPI anchor synthesis (Schonbachler et al., 1995; Sobering et al., 2004), a defect in GPI anchoring (Hamburger et al., 1995; Doering and Schekman, 1996) or a lack of stable membrane association of GPI-anchored proteins (Watanabe et al., 2002). Therefore, we first examined the membrane association of Gas1p. We found that in the arv1Δ mutant, a significantly larger fraction of Gas1p was released from the membranes under three different conditions, buffer alone, high pH, and detergent, compared with wild-type membranes, whereas the behavior of Gap1p, an integral membrane protein, and Ypt1p, a prenylated protein, were similar in arv1Δ and wild-type membranes (Figure S2A), indicating that association of GPI-anchored proteins to the membranes is weakened in arv1Δ mutant cells.
To test whether the weakened membrane association of Gas1p was due to inefficient GPI anchoring, we examined GPI anchoring of Gas1p by pulse-chase protein labeling and Triton X-114 phase separation (Watanabe et al., 2002). Anchored Gas1p partitioning into the detergent phase can only be shifted into the aqueous phase by treatment with PI-specific phospholipase C (PI-PLC), which removes the diacylglycerol moiety of GPI lipids. Unanchored Gas1p partitions into the aqueous phase and the portion remaining in the detergent phase is not affected by PI-PLC treatment (Nuoffer et al., 1991). After the first partition, in arv1Δ mutant cells, 40% of the ER form of Gas1p was partitioned into the aqueous phase (A1) in contrast to wild-type cells (13%; Figure 3B). Furthermore, among the fraction that was partitioned into the first detergent phase (D1), most of the ER form of Gas1p in wild-type cells was shifted from the detergent phase (D2, −PI-PLC) to the aqueous phase (A2, +PI-PLC) with PI-PLC treatment, confirming that they were indeed GPI-anchored. However in arv1Δ mutant cells, only small fraction of the ER form of Gas1p found in D1 fraction was sensitive to PI-PLC treatment, suggesting that they were not GPI-anchored Gas1p. These results demonstrate that GPI anchoring is significantly defective in the arv1Δ mutant. The anchoring deficiency was confirmed by examining the incorporation of [3H]myo-inositol or [3H]DHS into proteins in arv1Δ mutant (Figure S2B), both of which label the glycolipid portion of GPI-anchored proteins (Reggiori et al., 1997).
Because strains defective in GPI anchoring (Benghezal et al., 1995) as well as GPI anchor synthesis mutants (Taron et al., 2000; Umemura et al., 2003; Fujita et al., 2004) are hypersensitive to CFW, we reasoned that arv1Δ mutant cells might display the same phenotype. Indeed, the arv1Δ mutant cells showed hypersensitivity to CFW (Figure 3C). Hypersensitivity to CFW seen in gpi mutants is most likely due to elevated cell wall chitin levels that are thought to be brought about by either an up-regulated chitin synthase 3 activity in response to cell wall stress (Osmond et al., 1999; Valdivieso et al., 2000; Lesage et al., 2005) or an incidental increase of UDP-GlcNAc concentration (Sobering et al., 2004). Therefore, we looked for evidence of the elevated chitin levels by staining with CFW. For this, we tested six GPI anchor synthesis mutants (e.g., gpi1Δ, gpi2-7, gpi3-10, eri1Δ, gwt1-20, and gpi7Δ) and one GPI-anchoring mutant (e.g., gaa1-1) that are hypersensitive to CFW (data not shown). All gpi mutants that we tested show hyperaccumulation of chitin in the cell wall (Figure 3D), suggesting that the hyperaccumulation of chitin is a common phenotype of gpi mutants. Similarly, arv1Δ mutant cells hyperaccumulated chitin. This is consistent with our working hypothesis that Arv1p is required for GPI anchoring.
The arv1Δ Mutant Accumulates GlcN-acylPI and Has a Defect in Synthesis of Man-GlcN-acylPI
It is possible that GPI-anchoring defects result from a block in GPI anchor synthesis. Therefore, we examined whether the arv1Δ mutant cells have a specific defect in GPI anchor synthesis. We assayed production of GlcNAc-PI, GlcN-PI, and GlcN-acylPI by using an in vitro assay with membranes and a radiolabeled donor, [14C]UDP-GlcNAc (Schonbachler et al., 1995; Sobering et al., 2004). All three GPI anchor intermediates were detected in membranes of arv1Δ mutant cells (Figure 4A), indicating that the arv1Δ mutant has enzyme activities for the first three steps of GPI anchor synthesis. As a control, membranes of the gwt1-20 mutant (Umemura et al., 2003) inefficiently generated GlcN-acylPI (data not shown). Remarkably, arv1Δ mutant membranes accumulated GlcN-acylPI when compared with wild-type membranes (Figure 4, A and B). The accumulation of GlcN-acylPI could not be detected in arv1Δ mutant cells by labeling with [3H]myo-inositol (Figure 1, A and D). The reason for this discrepancy is unclear but may be due to differences between in vitro and in vivo assays (e.g., different enzymatic reaction rates) or radioactive substrates used (e.g., different labeling efficiencies). Time courses for Dol-P-Man synthase activity in the same membranes revealed that the higher accumulation of GlcN-acylPI in the arv1Δ membranes is not due to the lack of Dol-P-Man (Figure 4C), suggesting that later steps of GPI anchor synthesis are affected in the arv1Δ mutant cells.
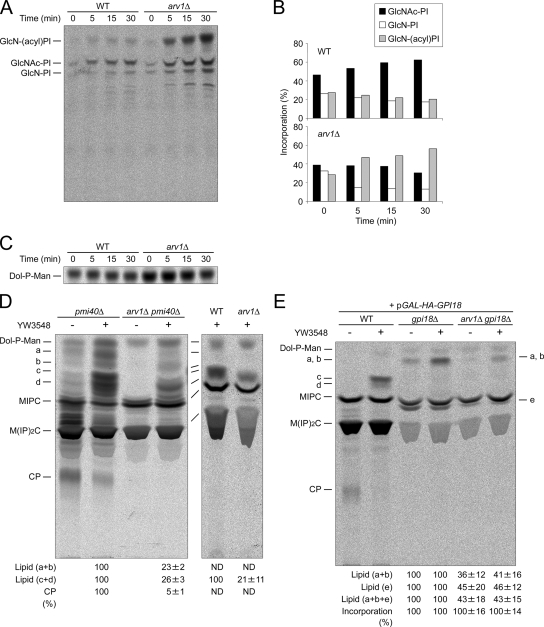
Figure 4.
arv1Δ mutant cells accumulate GlcN-acylPI and have a strong reduction in mannosylated GlcN-acylPI synthesis. (A and B) In vitro assay for the early steps of GPI anchor biosynthesis was performed as described in Materials and Methods. Membranes from strains wild-type (RH6082) or arv1Δ (FK137) were incubated with [14C]UDP-GlcNAc for the indicated times at 25°C. The lipids were extracted and analyzed by TLC plates using solvent system IV. The positions of GlcNAc-PI, GlcN-PI, and GlcN-acylPI were marked in A. Radioactivity in each GPI lipid was quantified, and incorporation (%) was determined as percentage of the total radioactivity. Data in B are quantification of A. (C) The same membranes as in A were labeled with [3H]GDP-Man for the indicated times at 25°C. The lipids were extracted and analyzed by TLC plates using solvent system III. (D and E) Strains pmi40Δ (FK395), arv1Δ pmi40Δ (FK397), wild-type (RH6082), and arv1Δ (FK137) were grown as described in Materials and Methods (D). Wild-type [+ pGAL-HA-GPI18] (FK1019), gpi18Δ [+ pGAL-HA-GPI18] (FK1017), and arv1Δ gpi18Δ [+ pGAL-HA-GPI18] (FK1015) cells were grown in galactose-containing medium, and shifted to glucose-containing medium for 16 h at 25°C to repress GPI18 expression (E). The cells were labeled with [3H]mannose for 30 min at 25°C in the presence of YW3548 (10 μM) or methanol. The lipids were extracted and analyzed by TLC using solvent system III. To give better separation of Dol-P-Man and lipids a–d, some experiments were applied to a double development using the same solvent. Radioactivity in each lipid was quantified, and the relative amounts were calculated by setting the amounts in pmi40Δ with YW3548, wild-type with YW3548 cells (D) or in gpi18Δ-pGAL-HA-GPI18 without or with YW3548 (E) to 100% and were expressed as percentages of the relative controls. Data in D and E are means ± range for two independent experiments. The relative amounts of total incorporation into lipids were also shown (E). ND, not detectable; a–d, mannolipids accumulated upon YW3548 treatment; e, lipid accumulated in Gpi18-depleted cells, preferentially in the absence of YW3548 [the mobility is consistent with that of (EtNP)Man-GlcN-acylPI (M1), which has been reported to accumulate in Gpi18p-depleted cells (Fabre et al., 2005)]); CP, complete GPI precursor.
We next examined whether deletion of ARV1 blocks the formation of late stage GPI intermediates. Two double mutants were created by introducing the arv1Δ mutation into the gaa1-1 and gpi7Δ mutants, which accumulate a complete GPI precursor (CP) (Hamburger et al., 1995) and an abnormal GPI intermediate, Man-(EtNP)Man-Man-(EtNP)Man-GlcN-acylPI (M4; Benachour et al., 1999), respectively, and tested for the accumulation of CP and M4 by pulse-labeling with [3H]myo-inositol. The results revealed that arv1Δ mutation suppresses the accumulation of CP and M4 (Figure S3), indicating that Arv1p functions upstream of Gaa1p and Gpi7p.
To further investigate which step of GPI anchor biosynthesis is defective in the arv1Δ mutant cells, we carried out an analogous experiment with a specific inhibitor of GPI anchor synthesis, YW3548, that inhibits the addition of EtNP onto the first mannose of the GPI core structure and leads to the accumulation of a GPI lipid, Man-Man-GlcN-acylPI (Man2; Sutterlin et al., 1997b). Because a mutation in phosphomannose isomerase, pmi40, which causes mannose auxotrophy enhances [3H]mannose labeling of mannosylated GPI intermediates (Sipos et al., 1994), pmi40Δ and arv1Δ pmi40Δ, mutant strains were created and analyzed for synthesis of mannosylated GPIs. When YW3548 was added to the pmi40Δ mutant (Figure 4D) or a pmi40 ts mutant (Figure S4A) cells, four major mannose-labeled lipids termed a, b, c, and d were accumulated, whereas synthesis of CP was significantly decreased. All a–d mannosylated lipids are acylated inositol ring-bearing GPI species because they were sensitive to GPI-phospholipase D (PLD; Figure S4, A and B), NaOH (Figure S4A), nitrous acid (Figure S4B) but resistant to PI-PLC (Figure S4, A and B). PI-PLC cleavage requires the two-position of inositol to be unacylated (Doerrler et al., 1996). Because c and d migrated just above MIPC at a position expected for Man2 (Sutterlin et al., 1997b) and because a and b were more hydrophobic than c and d (Figure 4D and Figure S4A), it appears that mannolipids c and d are Man2 species and that a and b species are GlcN-acylPI bearing one mannose (Man1). This was consistent with the fact that lipids (a and b) comigrated with the spot accumulated by shutting off GPI18 expression in the presence of YW3548 (Figure 4E), which would be predicted to be a Man1 species, as Gpi18p is required for addition of the second mannose during GPI assembly (Fabre et al., 2005; Pittet and Conzelmann, 2007).
GPI-PLD cleaves the linkage between the phosphate and inositol in GPI structures and generates phosphatidic acid and mannosylated GlcN-acyl-inositol. If lipid a and b species and lipid c and d species, respectively, have different acyl groups on the inositol ring, then treatment of these lipid mixtures with GPI-PLD should yield products of Man- and Man2-GlcN-acyl-inositol consisting of at least two species. By treating the mixtures with GPI-PLD, we observed distinct subsets that migrated differently in our TLC system (Figure S4B). The GPI-PLD products derived from lipids (a and b) contained two bands that have slightly different mobilities (Rf = 0.46 and 0.44). The products from lipids c and d had multiple bands migrating at lower mobilities (Rf = 0.34–0.24). The subsets were also seen when [3H]mannose-radiolabeled total lipids were tested (Figure S4A). These results suggest the existence of multiple forms of mannosylated GlcN-acylPI carrying different acyl groups on the inositol.
Importantly, deletion of ARV1 in the pmi40Δ mutant strains prevented the accumulation of mannolipids (a and b) upon treatment with YW3548 (Figure 4D, left). In the absence of YW3548 a minor amount of these lipids was still observed in the pmi40Δ mutant, but was not detectable in the arv1Δ pmi40Δ mutant. Also, the arv1Δ mutation suppressed the accumulation of mannolipids (c and d) seen in the pmi40Δ mutant (Figure 4D, left) or wild-type (Figure 4D, right) cells in the presence of YW3548. The formation of lipids a and b, which accumulate in Gpi18p-depleted cells was reduced by disrupting ARV1 (Figure 4E). These results show indirectly that Arv1p is required for the efficient synthesis of Man1.
Arv1p Is Required for the Delivery of GlcN-acylPI to the GPI-MT
The Man1 synthesis defect could be explained by 1) a block in production and/or translocation of Dol-P-Man across the ER membrane; 2) a defect in the activity of GPI-MT transferring mannose from Dol-P-Man to GlcN-acylPI; and 3) a defect in the delivery of GlcN-acylPI to the GPI-MT. The first possibility is unlikely because underglycosylation of CPY, a phenotype seen in the dpm1 mutant defective in Dol-P-Man synthase (Helenius et al., 2002) was not observed in the arv1Δ mutant cells (Figure 3A). Indeed, no defect in Dol-P-Man production was observed when arv1Δ mutant cells were labeled with [3H]mannose (Figure 4D, right) or when arv1Δ mutant membranes were labeled with [3H]GDP-Man (Figure 4C).
To assess GPI-MT activity, we used ER-enriched membranes from wild-type and arv1Δ mutant cells labeled with [3H]GDP-Man. The membranes from wild-type cells generated two mannolipids comigrating with lipids b and d, respectively, whereas arv1Δ membranes inefficiently synthesized them (Figure 5A, lanes 1 and 3). The lipids synthesized in vitro were also sensitive to GPI-PLD but not to PI-PLC and were accumulated upon treatment with YW3548 (data not shown). These results indicate that arv1Δ mutant membranes have a reduced mannosylGlcN-acylPI synthesis activity, which is consistent with our in vivo results (Figure 4, D and E). Because the incorporation of [3H]GDP-Man into these lipids (b and d) was linear over 15 min and that of [3H]GDP-Man into Dol-P-Man was constant (Figure S5), all subsequent assays were performed at the incubation time of 10 min.
Figure 5.
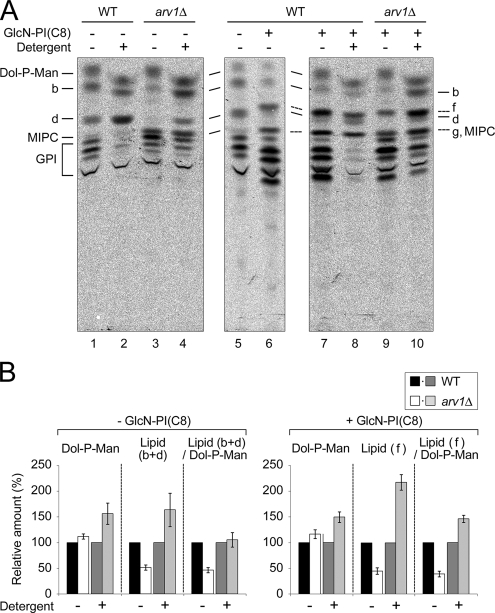
arv1Δ mutant cells have full Dol-P-Man synthesis and GPI-MT activity. (A) To measure GPI-MT activity, the ER-enriched membranes from wild-type (RH6082) and arv1Δ (FK137) were incubated with [3H]GDP-Man for 10 min at 25°C in the presence (lanes 2, 4, 8, and 10) or absence (lanes 1, 3, 5–7, and 9) of 10 mM n-octyl β-d-glucopyranoside. The experiments were also done in the presence (lanes 6–10) or absence (lanes 1–5) of 0.4 μg/ml GlcN-PI(C8). The lipids were extracted and analyzed by TLC using solvent system III. The detergent causes a slightly altered migration of Dol-P-Man and lipid b. (B) Radioactivity in each lipid was quantified, and the relative amounts were calculated by setting the amounts in wild-type membranes in the presence or absence of detergent to 100%. For the estimation of GPI-MT activity, the amounts of lipids (b and d) or lipid (f) were normalized by dividing by the amounts of Dol-P-Man. Because lipid g comigrated with MIPC, we could not obtain the proper amounts of lipid g for quantification. The results represent the means ± SD from four independent experiments. Lipids f and g represent Man-GlcN-acylPI(C8) and Man2-GlcN-acylPI(C8), respectively, and GPI designates more hydrophilic GPI intermediates than lipid d. Addition of 10 mM n-octyl β-d-glucopyranoside inhibited the incorporation into GPI to 24 ± 9% (wild-type) and 35 ± 19% (arv1Δ) of the amounts without detergent (lanes 1–4; see also Figure S5).
To definitively estimate GPI-MT activity, we measured the incorporation of [3H]GDP-Man into the lipids with the membrane topology being destroyed by detergent. n-octyl β-d-glucopyranoside was chosen for this assay because it has been shown to be the most suitable detergent for preserving GPI-MT activity in T. brucei (Smith et al., 1996). In the presence of 10 mM detergent, wild-type membranes could still generate the lipids b and d, but lost the activities to generate more hydrophilic GPI intermediates (Figure S5). As expected from the previous results with T. brucei, addition of higher concentrations of the detergent (e.g., 25 and 50 mM) resulted in an almost complete block of synthesis of all GPI intermediates including lipids b and d. Also, 25 mM n-octyl β-d-glucopyranoside inhibited the synthesis of all mannolipids generated with the arv1Δ mutant membranes (Figure S5). Therefore, we assayed the enzymatic activity of mannolipid synthase at the concentration of 10 mM n-octyl β-d-glucopyranoside. In comparison with wild-type membranes (Figure 5A, lanes 1 and 2), the arv1Δ mutant membranes showed an increased amount of labeled mannolipids (b and d) in the presence of the detergent (lanes 3 and 4). Because an increase in Dol-P-Man production was also observed with the arv1Δ mutant membranes, we normalized the amounts of mannolipid synthesis with those of Dol-P-Man synthesis. Normalized mannolipid synthesis was identical in wild-type and arv1Δ membranes (Figure 5B), suggesting that arv1Δ mutant membranes have full GPI-MT activity. Under the conditions where the membranes were not intact, a sufficient level of GPI-MT activity in arv1Δ membranes was also detected using a synthetic substrate, GlcN-PI with dioctanoyl-PI [GlcN-PI(C8); Figure 5B], which is an efficient substrate for GPI-MT-I (Doerrler et al., 1996) and is converted into Man-GlcN-acylPI(C8) (Figure 5A, lipid f). Therefore, Man-GlcN-acylPI synthesis defect of arv1Δ mutant is neither due to the defect in Dol-P-Man synthesis nor GPI-MT activity. Thus, we conclude that Arv1p is required for the delivery of GlcN-acylPI to the GPI-MT.
gpi Mutants Affect Sphingolipid Synthesis and Sterol Amounts and Distribution
If the primary function of Arv1p protein was to transport ceramides out of the ER, the defect of Man1 synthesis seen in the arv1Δ mutant would result from the accumulation of ceramides in the ER. To investigate this possibility, we assayed mannosylGlcN-acylPI synthesis in cells treated with AbA and in sec12 mutant cells, which are defective in ER-to-Golgi protein transport at nonpermissive temperature of 37°C. When wild-type cells were labeled with [3H]mannose, lipids a and b were not detectable at 25°C (Figure 4D, right), but they were detected at 37°C (Figure S6). Neither AbA treatment nor the sec12 allele affected lipid a and b synthesis nor lipid c and d synthesis at 37°C. This result suggests that the accumulation of ceramides in the ER cannot be the reason for the defect of mannosylated GlcN-acylPI synthesis, thereby suggesting that Arv1p functions primarily in the delivery of GlcN-acylPI to the GPI-MT.
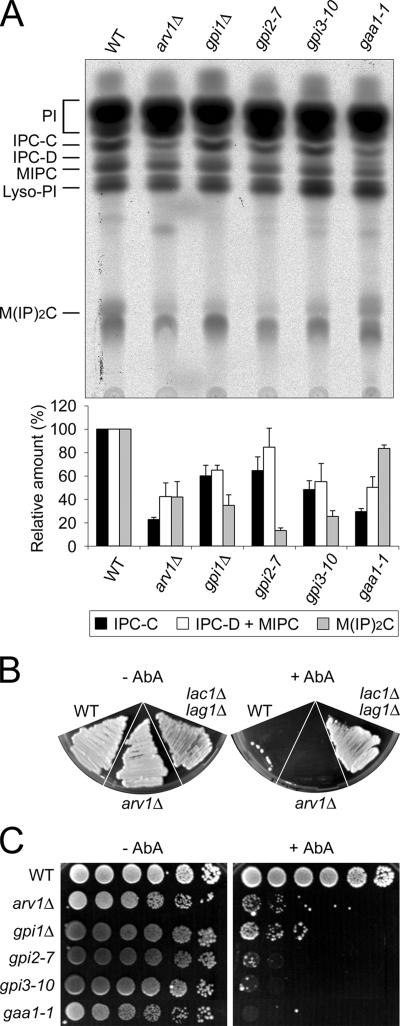
On the other hand, we consistently, found that other gpi mutants affect sphingolipid synthesis. When wild-type and gaa1 mutant cells were labeled with [3H]myo-inositol, the gaa1-1 mutant cells showed ∼30% of the level of IPC-C synthesis as found in wild-type cells (Figure 6A). The extent of reduction was similar to arv1Δ mutant cells. Moreover, gpi1Δ, gpi2-7, and gpi3-10 mutant cells made ∼50–60% of the amount of IPC-C synthesized in the wild-type cells. These results underscore that a GPI anchor synthesis or anchoring defect causes a defect in sphingolipid biosynthesis.
Figure 6.
gpi mutants have a reduced level of sphingolipids and are hypersensitive to aureobasidin A. (A) Wild-type (RH6082), arv1Δ (FK137), gpi1Δ (DL2831), gpi2-7 (DL2828), gpi3-10 (DL2829), and gaa1-1 (RH401-7C) were labeled with [3H]myo-inositol for 1 h at 25°C. The lipids were extracted and analyzed by TLC plates using solvent system I. Incorporation (%) into sphingolipids was determined as the percentage of total radioactivity into all lipids containing inositol, and the relative amounts of each species were calculated by setting the amount in wild-type cells to 100%. Data in A are means ± SE for three independent experiments. (B) AbA (final concentration ≈1 μg/ml, right) or ethanol (left) was spread onto YPUAD plates. Wild-type (RH6082), lac1Δ lag1Δ (RH5308), and arv1Δ (RH6078) were streaked on the plates and incubated for 3 d at 25°C. (C) The same strains as in A were spotted onto YPUAD, YPUAD supplemented with 0.1 μg/ml AbA or ethanol and incubated for 4 d at 25°C.
Changes in ceramide levels could be monitored using AbA. The fact that wild-type strains are sensitive to AbA, whereas lac1Δ lag1Δ double and lip1Δ single mutant strains defective in ceramide synthesis are resistant to AbA (Schorling et al., 2001; Vallee and Riezman, 2005) implies that AbA sensitivity is associated with an increased level of ceramides rather than a decreased level of sphingolipids. Thus one would predict that arv1Δ mutant cells should be hypersensitive to AbA, because the arv1Δ mutant strain accumulates ceramides (Swain et al., 2002), and we found that the ceramide synthesis pathway appears to be overloaded in the arv1Δ mutant (Figure 1B). Wild-type and arv1Δ mutant strains were sensitive to 1 μg/ml AbA, whereas the lac1Δ lag1Δ mutant was resistant (Figure 6B). This is consistent with our finding that arv1Δ mutant cells had normal ceramide synthase activity (data not shown). At a lower concentration (0.1 μg/ml) of AbA, wild-type cells grew, but arv1Δ mutant cells could not grow (Figure 6C), indicating that the arv1Δ mutant is hypersensitive to AbA. Other gpi mutants (e.g., gpi1Δ, gpi2-7, gpi3-10, and gaa1-1) were also hypersensitive to the inhibitor. These results imply that a GPI anchor synthesis or anchoring defect causes an accumulation of ceramides, most likely as a consequence of the ceramide transport defect.
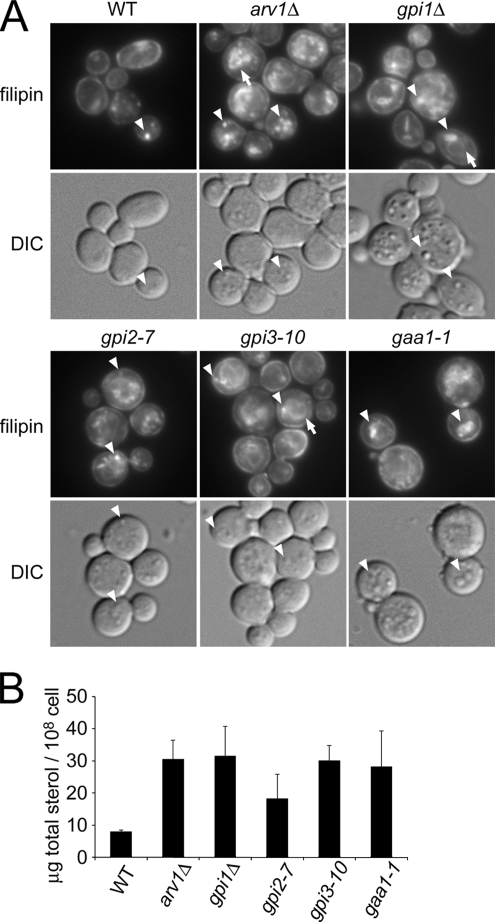
Because the ARV1 deletion has been shown to accumulate sterols in internal membrane fractions (Tinkelenberg et al., 2000; Beh and Rine, 2004), we asked whether gpi mutants affect intracellular sterol distribution. As detected by filipin staining, the majority of sterols in arv1Δ and gpi mutants were found in internal membranes of cells. Wild-type cells displayed much less internal filipin levels than the mutants (Figure 7A). In the mutant cells, internal filipin fluorescence was observed in ring-like perinuclear structures of the ER (Figure 7A, arrows). Fluorescence was also seen in the lipid particles, which can be easily visualized with differential interference contrast (DIC; Figure 7A, arrowheads). Although the number and size of lipid particles per cell varies among cells and experiments, gpi mutants seem to have an increased number of lipid particles.
Figure 7.
gpi mutants affect intracellular distribution and level of total sterols. (A) Wild-type (RH6082), arv1Δ (FK137), gpi1Δ (DL2831), gpi2-7 (DL2828), gpi3-10 (DL2829), and gaa1-1 (RH401–7C) were grown to similar densities (7–10 × 106 cells/ml) and then stained for sterols with filipin complex (0.1 mg/ml). The staining was visualized by fluorescence microscopy, and the images were taken with equal exposures. The morphology of the cells and lipid particles were observed by DIC. Arrows and arrowheads indicate ring-like perinuclear ER and lipid particles, respectively. (B) The same strains as in A were grown likewise, and total sterol levels were analyzed by GLC/MS using cholesterol as an internal standard. The amounts of total sterols were determined in two independent experiments analyzed in duplicate, and data are shown as means; error bars, ± range.
Moreover, we measured total sterols in these mutants by GLC/MS analysis. Compared with the wild-type cells, gpi mutants showed a 2–4-fold increase in total cellular sterols (Figure 7B). TLC analysis further indicated that the free sterol level was 50–200% increased in gpi mutants and arv1Δ cells compared with wild-type (data not shown). Thus, GPI assembly not only affects sphingolipid metabolism and transport but also the intracellular distribution and level of sterols.
DISCUSSION
In this study, we have shown that yeast deleted for ARV1 accumulates GlcN-acylPI and is impaired for GlcN-acylPI mannosylation without having defects in Dol-P-Man synthesis or GPI-MT activity. Because N-glycosylation of Gas1p, Yps1p and CPY was not affected in arv1Δ mutant, the delivery of Dol-P-Man required for synthesis of N-linked oligosaccharides to the mannosyltransferases appears to be normal. Therefore, our results suggest a specific role of Arv1p in the delivery of GlcN-acylPI to the GPI-MT. Furthermore, we provide evidence that GPI anchor synthesis and attachment to proteins are not only required for GPI-anchored protein transport but also regulate ceramide transport from the ER and intracellular sterol distribution.
When we labeled pmi40Δ or pmi40 ts mutant cells with [3H]mannose, we detected four mannolipids, a–d, that were accumulated upon treatment with YW3548. On the basis of labeling experiments (Figure 4, D and E; Figure S4, A and B), we suggest that the lipids a and b are Man-GlcN-acylPI and c and d are Man2-GlcN-acylPI species. The mannosylGlcN-acylPI species might contain different types of acyl groups on the inositol ring, which to our knowledge has not been documented in yeast GPI anchor synthesis (Sipos et al., 1997; Reggiori et al., 1997; Bosson et al., 2006; Fujita et al., 2006b). Several studies demonstrated that acyl chains linked to inositol of GPI are heterogeneous in trypanosomes (Guther et al., 1996) and mammalian cells (Houjou et al., 2007). At present, we cannot exclude that the appearance of multiple forms of mannosylGlcN-acylPI might be a secondary effect, because it was neither found in wild-type cells without YW3548 (data not shown) nor with membranes in our in vitro system (Figure 5A and Figure S5). However, it has been shown that yeast membranes can utilize acyl-CoAs containing different lengths of fatty acid (C14–C20) as exogenous substrates for inositol acylation (Costello and Orlean, 1992; Doerrler et al., 1996; Umemura et al., 2003), suggesting that the various species of mannosylated GlcN-acylPI bearing different acyl chain lengths can be generated in yeast.
Murakami et al. (2003) demonstrated that in mammalian cells, the first mannosylation occurs without inositol acylation, similar to the T. brucei GPI biosynthetic pathway (Guther and Ferguson, 1995; Smith et al., 1996). This indicates that mammalian GPI-MT-I can accept GlcN-PI as a substrate. On the other hand, yeast GPI-MT does not seem to act on GlcN-PI. Previous studies have shown that all mannosylated GPI lipids accumulating in yeast mutants, gaa1 (Hamburger et al., 1995), gpi11 (Taron et al., 2000), gpi7 (Benachour et al., 1999), smp3 (Grimme et al., 2001), gpi10 (Sutterlin et al., 1998), and gpi18 (Fabre et al., 2005) were acylated GPI species. Also, we never detected any mannosylated GPI species that was not acylated. Only two acylated species, Man-GlcN-acylPI and (EtNP)Man-GlcN-acylPI, were detected in Gpi18p-depleted cells (Figure 4E). These observations strongly support the idea that the substrate specificity of yeast GPI-MT is restricted to GlcN-acylPI.
Three possibilities could explain how the GPI precursor lipid flips across the ER membrane. First, only GlcN-acylPI synthesized on the cytoplasmic side of the ER may flip into the ER lumen. Second, both GlcN-PI and GlcN-acylPI can flip but only GlcN-acylPI can be mannosylated. Finally, GlcN-PI flipped into the ER lumen can be acylated to yield GlcN-acylPI. In the last model, inositol acylation would occur on the luminal side of the ER, and fatty acyl-CoAs must be present in the ER lumen. Consistent with this notion, it is known that secretory proteins such as Hedgehog and Wnt are acylated in the ER lumen by the related acyl-CoA–dependent acyltransferases (Hofmann, 2000; Orlean and Menon, 2007). However, fatty acyl-CoAs are synthesized in the cytosol or the cytoplasmic side of organelle membranes (Black and Dirusso, 2007), and it has been reported that they do not normally penetrate into microsomal membranes (Gooding et al., 2004). In addition, there is evidence that the yeast ER does not contain enzymes involved in carnitine-dependent acyl-CoA transport across membranes (Tehlivets et al., 2007). These findings lead to the proposal that fatty acyl-CoAs may not reside in the ER lumen. Thus, it remains to be determined if inositol acylation can occur on the luminal side of the ER at all.
What might be the biochemical function of Arv1p? If GlcN-acylPI is synthesized on the cytoplasmic side of the ER, it is possible that Arv1p functions as a GPI flippase or as its regulator. Based on the biochemical analysis using proteoliposomes from a detergent extract of ER with fluorescent GPI analogues (Vishwakarma and Menon, 2005), it was recently proposed that flipping of early GPI intermediates occurs via an ATP-independent process mediated by specific proteins. Such energy-independent flipping processes are often observed with lipids across the ER. For example, flip-flop of phospholipids across the ER membrane does not require ATP (Pomorski et al., 2004; Gummadi and Kumar, 2005; Pohl et al., 2005). In addition, the translocation of lipid-linked oligosaccharides from the cytosolic leaflet to the luminal leaflet of the ER, which is mediated by a putative flippase Rft1p, also seems to be independent of metabolic energy. RFT1 encodes a multimembrane-spanning protein but does not have ATP-binding domains (Helenius et al., 2002; Helenius and Aebi, 2002). Similarly, Arv1p contains six predicted transmembrane domains and lacks any ATP-binding site motifs. Instead, this protein contains an N-terminal domain highly conserved among Arv1 proteins from different organisms, defined as the Arv1 homology domain (AHD) that consists of the N-terminal subdomain with a putative zinc-binding motif and the C-terminal subdomain with a transmembrane domain. Although the role of the N-terminal subdomain remains unclear, it was recently suggested that the N-terminal subdomain of the AHD is not required for Arv1p function, whereas the C-terminal subdomain plays an essential role (Fores et al., 2006). This may imply an important role of the membrane-spanning region within the C-terminal subdomain in Arv1p function. Furthermore, our analysis of hydrophobicity profiles and ClustalW algorithms suggested that well-conserved regions among Arv1 homologues were mainly located close to or in the membrane and that they were found in both leaflets of the membranes (data not shown). Interestingly, it was also predicted by intragenic complementation analysis that Arv1p functions as a multimer, presumably a dimer, resulting in an arrangement of 6 + 6 transmembrane domains in the membrane (Tinkelenberg et al., 2000). This appears to be a common characteristic of flippases and transporters such as ATP-binding cassette transporters (Pohl et al., 2005; Tusnady et al., 2006). Although GPI flipping activity of Arv1p still has to be proven directly, these observations make it likely that Arv1p is either a GPI flippase or an accessory protein that facilitates the flipping of GPI. In this case, the remaining activity for mannosylGlcN-acylPI synthesis in the absence of Arv1p may be due to the spontaneous flipping of GlcN-acylPI or the flipping mediated by other proteins that are not yet identified. As an attractive possibility, it has been proposed that GPI flippase may be the same as the putative glycerophospholipid flippase (Vishwakarma and Menon, 2005). It would be interesting to see if Arv1p is involved in flip-flop of glycerophospholipids. However, because no direct evidence is provided yet for the site of GlcN-acylPI synthesis, we cannot rule out that Arv1p plays a role in the lateral movement of GlcN-acylPI in the ER lumen, as in the case of SL15/Lec35 for the utilization of Dol-P-Man (Anand et al., 2001). Alternatively, Arv1p might be involved in specific delivery of Dol-P-Man to the GPI-MT, although we consider this unlikely.
We have previously reported that ceramides synthesized in the ER are transported to the Golgi compartment by both vesicular and nonvesicular pathways in yeast (Funato and Riezman, 2001). The vesicular transport of ceramide occurs via COPII-coated vesicles and requires ATP, whereas nonvesicular transport does not require ATP. In this study, we observed that ARV1 deletion and GAA1 mutation result in strong reductions of IPC synthesis by ∼75–80 and 70%, respectively, which are similar to the effect on IPC synthesis (−80%) seen in sec18 mutant cells (Funato and Riezman, 2001). In addition, our analysis showed that ATP depletion using NaN3 and NaF inhibited IPC synthesis of wild type by ∼50%, but had no effect on IPC synthesis of arv1Δ mutant cells (data not shown). This suggests that IPC synthesis via an ATP-requiring vesicular pathway is almost completely abolished in the absence of Arv1p. Collectively, these results imply that GPI anchor synthesis regulates sphingolipid synthesis mainly by vesicular ceramide transport, which is consistent with the model previously proposed that GPI-anchored proteins and ceramides are cotransported from the ER in yeast.
Filipin staining revealed that sterols probably hyperaccumulate in gpi mutant cells in lipid particles as well as in the ER. Ergosterol, the major sterol of yeast is not required for ER-to-Golgi transport of GPI-anchored proteins (Heese-Peck et al., 2002), suggesting that the sterol structural requirements for ER-to-Golgi transport may be not strict or that sterols might not be cotransported with GPI-anchored proteins and ceramides. Because sterol transport to the plasma membrane is not blocked in mutants affecting the secretory pathway, such as sec18, sec12 (Baumann et al., 2005) and sec23 (Schnabl et al., 2005), we propose that the sterol distribution defect in the arv1Δ mutant is an indirect effect stemming from its sphingolipid synthesis and transport defect. Without the proper amount of sphingolipids in the plasma membrane, sterols would no longer properly equilibrate between the ER and the plasma membrane, as proposed previously (Baumann et al., 2005). Lipid particles are thought to be storage compartments for steryl esters and triacyglycerols, but they also may play a role in sterol synthesis and/or transport in yeast, because some enzymes involved in sterol synthesis are localized to the lipid particles (Daum et al., 1998; Sorger et al., 2004). It is plausible that if sterol transport from the ER to the plasma membrane is defective, the excess of sterols found in the ER could be packaged into lipid particles. Lipid particles are thought to be formed from the ER membrane. Consistent with this idea, we observed that the total level of sterols as well as the free sterol pool was increased in gpi mutants. A similar observation has been reported in a GPI-deficient lymphocyte lacking caveolins, which has more cholesterol than control cells (Abrami et al., 2001). We also found that ARE1 and ARE2 genes, which encode yeast acyl-CoA: sterol acyltransferases, interact genetically with GAA1 (Figure S7) as they do with ARV1 (Tinkelenberg et al., 2000). These observations, together with the fact that sphingolipid-deficient lcb1-100 mutant cells cause a weak membrane association of GPI-anchored proteins with the ER membrane (Watanabe et al., 2002), accumulate sterols in lipid particle structures similar to those seen in gpi mutant, and increase the level of sterols (Baumann et al., 2005), suggests the existence of a coordinated regulation for the trafficking of GPI-anchored proteins and raft lipids.
Finally, the results presented here support the model that the primary function of Arv1p is to deliver GlcN-acylPI to GPI-MT and that GPI anchor synthesis is required for efficient ceramide transport from the ER as well as intracellular distribution of sterols. How these events are connected in detail remains to be elaborated. It is possible that GPI anchor synthesis might be required for the formation and/or maintenance of functional lipid domains in the yeast ER.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Orlean (University of Illinois) for the gpi1Δ, gpi2-7, and gpi3-10; D. E. Levin (The Johns Hopkins University) for the eri1Δ/eri1Δ; Y. Jigami (National Institute of Advanced Industrial Science and Technology) for the gwt1-20; and A. Conzelmann (University of Fribourg) for the gpi7Δ strains, Y. Ohya (University of Tokyo) for the YEp13 (a genomic library), A. Nakano (Riken Discovery Research Institute) for the pSKY5-RER1, K. Tanaka for the pRS316-GAL1-2xHA-BS, M. Funk (Institut Für Molekularbiologie and Tumorforschung) for pRS series plasmids, M. A. Lehrman for GlcN-PI(C8), R. Schekman (University of California, Berkeley) for strains and antibodies, and T. Kinoshita for recombinant GPI-PLD and GPI18/gpi18::KANR diploid strains. This work was supported by the long-term fellowship from Human Frontier Science Program (HFSP) to R.W.; by a Federation of European Biochemical Societies long-term fellowship to H.P.; by grants from the Swiss National Science Foundation and the Human Frontiers Science Program Organization to H.R.; and by grants from the Naito Foundation and the ONO Medical Research Foundation to K.F.
Abbreviations used:
- AbA
aureobasidin A
- CFW
calcofluor white
- CP
complete GPI precursor
- CPY
carboxypeptidase Y
- DHS
dihydrosphingosine
- Dol-P-Man
dolicholphosphomannose
- EtNP
phosphorylethanolamine
- GDP-Man
GDP-mannose
- GlcNAc-PI
N-acetylglucosaminyl-phosphatidylinositol
- GlcN-PI
glucosaminyl-phosphatidylinositol
- GlcN-acylPI
glucosaminyl-acyl-phosphatidylinositol
- GPI
glycosylphosphatidylinositol
- GPI-MT
GPI mannosyltransferase
- HA
hemagglutinin
- IPC
inositolphosphorylceramide
- LY
lucifer yellow
- MIPC
mannosyl inositolphosphorylceramide
- M(IP)2C
mannosyl di(inositolphosphoryl)ceramide
- PI
phosphatidylinositol
- PLC
phospholipase C
- PLD
phospholipase D
- UDP-GlcNAc
UDP-N-acetylglucosamine.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-08-0740) on February 20, 2008.
REFERENCES
- Abrami L., Fivaz M., Kobayashi T., Kinoshita T., Parton R. G., van der Goot F. G. Cross-talk between caveolae and glycosylphosphatidylinositol-rich domains. J. Biol. Chem. 2001;276:30729–30736. doi: 10.1074/jbc.M102039200. [DOI] [PubMed] [Google Scholar]
- Anand M., Rush J. S., Ray S., Doucey M. A., Weik J., Ware F. E., Hofsteenge J., Waechter C. J., Lehrman M. A. Requirement of the Lec35 gene for all known classes of monosaccharide-P-dolichol-dependent glycosyltransferase reactions in mammals. Mol. Biol. Cell. 2001;12:487–501. doi: 10.1091/mbc.12.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M., Keranen S., Shevchenko A., Shevchenko A., Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast, K. Proc. Natl. Acad. Sci. USA. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barz W. P., Walter P. Two endoplasmic reticulum (ER) membrane proteins that facilitate ER-to-Golgi transport of glycosylphosphatidylinositol-anchored proteins. Mol. Biol. Cell. 1999;10:1043–1059. doi: 10.1091/mbc.10.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann N. A., Sullivan D. P., Ohvo-Rekila H., Simonot C., Pottekat A., Klaassen Z., Beh C. T., Menon A. K. Transport of newly synthesized sterol to the sterol-enriched plasma membrane occurs via nonvesicular equilibration. Biochemistry. 2005;44:5816–5826. doi: 10.1021/bi048296z. [DOI] [PubMed] [Google Scholar]
- Beh C. T., Rine J. A role for yeast oxysterol-binding protein homologs in endocytosis and in the maintenance of intracellular sterol-lipid distribution. J. Cell Sci. 2004;117:2983–2996. doi: 10.1242/jcs.01157. [DOI] [PubMed] [Google Scholar]
- Benachour A., Sipos G., Flury I., Reggiori F., Canivenc-Gansel E., Vionnet C., Conzelmann A., Benghezal M. Deletion of GPI7, a yeast gene required for addition of a side chain to the glycosylphosphatidylinositol (GPI) core structure, affects GPI protein transport, remodeling, and cell wall integrity. J. Biol. Chem. 1999;274:15251–15261. doi: 10.1074/jbc.274.21.15251. [DOI] [PubMed] [Google Scholar]
- Benghezal M., Lipke P. N., Conzelmann A. Identification of six complementation classes involved in the biosynthesis of glycosylphosphatidylinositol anchors in Saccharomyces cerevisiae. J. Cell Biol. 1995;130:1333–1344. doi: 10.1083/jcb.130.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black P. N., Dirusso C. C. Yeast acyl-CoA synthetases at the crossroads of fatty acid metabolism and regulation. Biochim. Biophys. Acta. 2007;1771:286–298. doi: 10.1016/j.bbalip.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Bosson R., Jaquenoud M., Conzelmann A. GUP1 of Saccharomyces cerevisiae encodes an O-acyltransferase involved in remodeling of the GPI anchor. Mol. Biol. Cell. 2006;17:2636–2645. doi: 10.1091/mbc.E06-02-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S., Mayor S. The GPI-anchor and protein sorting. Cell. Mol. Life Sci. 2001;58:1969–1987. doi: 10.1007/PL00000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello L. C., Orlean P. Inositol acylation of a potential glycosyl phosphoinositol anchor precursor from yeast requires acyl coenzyme A. J. Biol. Chem. 1992;267:8599–8603. [PubMed] [Google Scholar]
- Daum G., Lees N. D., Bard M., Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- David D., Sundarababu S., Gerst J. E. Involvement of long chain fatty acid elongation in the trafficking of secretory vesicles in yeast. J. Cell Biol. 1998;143:1167–1182. doi: 10.1083/jcb.143.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Sumanasekera C., Lester R. L. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog. Lipid Res. 2006;45:447–465. doi: 10.1016/j.plipres.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Doering T. L., Schekman R. GPI anchor attachment is required for Gas1p transport from the endoplasmic reticulum in COP II vesicles. EMBO J. 1996;15:182–191. [PMC free article] [PubMed] [Google Scholar]
- Doerrler W. T., Ye J., Falck J. R., Lehrman M. A. Acylation of glucosaminyl phosphatidylinositol revisited. Palmitoyl-CoA dependent palmitoylation of the inositol residue of a synthetic dioctanoyl glucosaminyl phosphatidylinositol by hamster membranes permits efficient mannosylation of the glucosamine residue. J. Biol. Chem. 1996;271:27031–27038. doi: 10.1074/jbc.271.43.27031. [DOI] [PubMed] [Google Scholar]
- Duden R., Hosobuchi M., Hamamoto S., Winey M., Byers B., Schekman R. Yeast beta- and beta′-coat proteins (COP). Two coatomer subunits essential for endoplasmic reticulum-to-Golgi protein traffic. J. Biol. Chem. 1994;269:24486–24495. [PubMed] [Google Scholar]
- Duden R., Kajikawa L., Wuestehube L., Schekman R. epsilon-COP is a structural component of coatomer that functions to stabilize alpha-COP. EMBO J. 1998;17:985–995. doi: 10.1093/emboj/17.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulic V., Egerton M., Elguindi I., Raths S., Singer B., Riezman H. Yeast endocytosis assays. Methods Enzymol. 1991;194:697–710. doi: 10.1016/0076-6879(91)94051-d. [DOI] [PubMed] [Google Scholar]
- Fabre A. L., Orlean P., Taron C. H. Saccharomyces cerevisiae Ybr004c and its human homologue are required for addition of the second mannose during glycosylphosphatidylinositol precursor assembly. FEBS J. 2005;272:1160–1168. doi: 10.1111/j.1742-4658.2005.04551.x. [DOI] [PubMed] [Google Scholar]
- Fores O., Arro M., Pahissa A., Ferrero S., Germann M., Stukey J., McDonough V., Nickels J. T., Jr., Campos N., Ferrer A. Arabidopsis thaliana expresses two functional isoforms of Arvp, a protein involved in the regulation of cellular lipid homeostasis. Biochim. Biophys. Acta. 2006;1761:725–735. doi: 10.1016/j.bbalip.2006.03.025. [DOI] [PubMed] [Google Scholar]
- Fujita M., Yoko-o T., Okamoto M., Jigami Y. GPI7 involved in glycosylphosphatidylinositol biosynthesis is essential for yeast cell separation. J. Biol. Chem. 2004;279:51869–51879. doi: 10.1074/jbc.M405232200. [DOI] [PubMed] [Google Scholar]
- Fujita M., Yoko-O T., Jigami Y. Inositol deacylation by Bst1p is required for the quality control of glycosylphosphatidylinositol-anchored proteins. Mol. Biol. Cell. 2006a;17:835–850. doi: 10.1091/mbc.E05-05-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Umemura M., Yoko-o T., Jigami Y. PER1 is required for GPI-phospholipase A2 activity and involved in lipid remodeling of GPI-anchored proteins. Mol. Biol. Cell. 2006b;17:5253–5264. doi: 10.1091/mbc.E06-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato K., Riezman H. Vesicular and nonvesicular transport of ceramide from ER to the Golgi apparatus in yeast. J. Cell Biol. 2001;155:949–959. doi: 10.1083/jcb.200105033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato K., Vallee B., Riezman H. Biosynthesis and trafficking of sphingolipids in the yeast Saccharomyces cerevisiae. Biochemistry. 2002;41:15105–15114. doi: 10.1021/bi026616d. [DOI] [PubMed] [Google Scholar]
- Funato K., Lombardi R., Vallee B., Riezman H. Lcb4p is a key regulator of ceramide synthesis from exogenous long chain sphingoid base in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:7325–7334. doi: 10.1074/jbc.M209925200. [DOI] [PubMed] [Google Scholar]
- Gaynor E. C., Emr S. D. COPI-independent anterograde transport: cargo-selective ER to Golgi protein transport in yeast COPI mutants. J. Cell Biol. 1997;136:789–802. doi: 10.1083/jcb.136.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerold P., Jung N., Azzouz N., Freiberg N., Kobe S., Schwarz R. T. Biosynthesis of glycosylphosphatidylinositols of Plasmodium falciparum in a cell-free incubation system: inositol acylation is needed for mannosylation of glycosylphosphatidylinositols. Biochem. J. 1999;344:731–738. [PMC free article] [PubMed] [Google Scholar]
- Ghugtyal V., Vionnet C., Roubaty C., Conzelmann A. CWH43 is required for the introduction of ceramides into GPI anchors in Saccharomyces cerevisiae. Mol. Microbiol. 2007;65:1493–1502. doi: 10.1111/j.1365-2958.2007.05883.x. [DOI] [PubMed] [Google Scholar]
- Gooding J. M., Shayeghi M., Saggerson E. D. Membrane transport of fatty acylcarnitine and free L-carnitine by rat liver microsomes. Eur. J. Biochem. 2004;271:954–961. doi: 10.1111/j.1432-1033.2004.03997.x. [DOI] [PubMed] [Google Scholar]
- Grimme S. J., Westfall B. A., Wiedman J. M., Taron C. H., Orlean P. The essential Smp3 protein is required for addition of the side-branching fourth mannose during assembly of yeast glycosylphosphatidylinositols. J. Biol. Chem. 2001;276:27731–27739. doi: 10.1074/jbc.M101986200. [DOI] [PubMed] [Google Scholar]
- Gummadi S. N., Kumar K. S. The mystery of phospholipid flip-flop in biogenic membranes. Cell. Mol. Biol. Lett. 2005;10:101–121. [PubMed] [Google Scholar]
- Guther M. L., Ferguson M. A. The role of inositol acylation and inositol deacylation in GPI biosynthesis in Trypanosoma brucei. EMBO J. 1995;14:3080–3093. doi: 10.1002/j.1460-2075.1995.tb07311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guther M. L., Treumann A., Ferguson M. A. Molecular species analysis and quantification of the glycosylphosphatidylinositol intermediate glycolipid C from Trypanosoma brucei. Mol. Biochem. Parasitol. 1996;77:137–145. doi: 10.1016/0166-6851(96)02585-6. [DOI] [PubMed] [Google Scholar]
- Hamburger D., Egerton M., Riezman H. Yeast Gaa1p is required for attachment of a completed GPI anchor onto proteins. J. Cell Biol. 1995;129:629–639. doi: 10.1083/jcb.129.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese-Peck A., Pichler H., Zanolari B., Watanabe R., Daum G., Riezman H. Multiple functions of sterols in yeast endocytosis. Mol. Biol. Cell. 2002;13:2664–2680. doi: 10.1091/mbc.E02-04-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius J., Aebi M. Transmembrane movement of dolichol linked carbohydrates during N-glycoprotein biosynthesis in the endoplasmic reticulum. Semin. Cell Dev. Biol. 2002;13:171–178. doi: 10.1016/s1084-9521(02)00045-9. [DOI] [PubMed] [Google Scholar]
- Helenius J., Ng D. T., Marolda C. L., Walter P., Valvano M. A., Aebi M. Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 protein. Nature. 2002;415:447–450. doi: 10.1038/415447a. [DOI] [PubMed] [Google Scholar]
- Hofmann K. A superfamily of membrane-bound O-acyltransferases with implications for Wnt signaling. Trends Biochem. Sci. 2000;25:111–112. doi: 10.1016/s0968-0004(99)01539-x. [DOI] [PubMed] [Google Scholar]
- Horvath A., Sutterlin C., Manning-Krieg U., Movva N. R., Riezman H. Ceramide synthesis enhances transport of GPI-anchored proteins to the Golgi apparatus in yeast. EMBO J. 1994;13:3687–3695. doi: 10.1002/j.1460-2075.1994.tb06678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houjou T., Hayakawa J., Watanabe R., Tashima Y., Maeda Y., Kinoshita T., Taguchi R. Changes in molecular species profiles of glycosylphosphatidylinositol anchor precursors in early stages of biosynthesis. J. Lipid Res. 2007;48:1599–1606. doi: 10.1194/jlr.M700095-JLR200. [DOI] [PubMed] [Google Scholar]
- Ikezawa H. Glycosylphosphatidylinositol (GPI)-anchored proteins. Biol. Pharm. Bull. 2002;25:409–417. doi: 10.1248/bpb.25.409. [DOI] [PubMed] [Google Scholar]
- Ikonen E. Roles of lipid rafts in membrane transport. Curr. Opin. Cell Biol. 2001;13:470–477. doi: 10.1016/s0955-0674(00)00238-6. [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Inoue N. Dissecting and manipulating the pathway for glycosylphosphatidylinositol-anchor biosynthesis. Curr. Opin. Chem. Biol. 2000;4:632–638. doi: 10.1016/s1367-5931(00)00151-4. [DOI] [PubMed] [Google Scholar]
- Lesage G., Shapiro J., Specht C. A., Sdicu A. M., Menard P., Hussein S., Tong A. H., Boone C., Bussey H. An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet. 2005;6:8. doi: 10.1186/1471-2156-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine T. P., Wiggins C. A., Munro S. Inositol phosphorylceramide synthase is located in the Golgi apparatus of Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:2267–2281. doi: 10.1091/mbc.11.7.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y., Watanabe R., Harris C. L., Hong Y., Ohishi K., Kinoshita K., Kinoshita T. PIG-M transfers the first mannose to glycosylphosphatidylinositol on the lumenal side of the ER. EMBO J. 2001;20:250–261. doi: 10.1093/emboj/20.1.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S., Riezman H. Sorting GPI-anchored proteins. Nat. Rev. Mol. Cell Biol. 2004;5:110–120. doi: 10.1038/nrm1309. [DOI] [PubMed] [Google Scholar]
- Menon A. K., Mayor S., Schwarz R. T. Biosynthesis of glycosyl-phosphatidylinositol lipids in Trypanosoma brucei: involvement of mannosylphosphoryldolichol as the mannosyl donor. EMBO J. 1990;9:4249–4258. doi: 10.1002/j.1460-2075.1990.tb07873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsomme P., Riezman H. The Rab GTPase Ypt1p and tethering factors couple protein sorting at the ER to vesicle targeting to the Golgi apparatus. Dev. Cell. 2002;2:307–317. doi: 10.1016/s1534-5807(02)00133-8. [DOI] [PubMed] [Google Scholar]
- Morsomme P., Prescianotto-Baschong C., Riezman H. The ER v-SNAREs are required for GPI-anchored protein sorting from other secretory proteins upon exit from the ER. J. Cell Biol. 2003;162:403–412. doi: 10.1083/jcb.200212101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullner H., Deutsch G., Leitner E., Ingolic E., Daum G. YEH2/YLR020c encodes a novel steryl ester hydrolase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:13321–13328. doi: 10.1074/jbc.M409914200. [DOI] [PubMed] [Google Scholar]
- Muniz M., Riezman H. Intracellular transport of GPI-anchored proteins. EMBO J. 2000;19:10–15. doi: 10.1093/emboj/19.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz M., Morsomme P., Riezman H. Protein sorting upon exit from the endoplasmic reticulum. Cell. 2001;104:313–320. doi: 10.1016/s0092-8674(01)00215-x. [DOI] [PubMed] [Google Scholar]
- Munn A. L., Heese-Peck A., Stevenson B. J., Pichler H., Riezman H. Specific sterols required for the internalization step of endocytosis in yeast. Mol. Biol. Cell. 1999;10:3943–3957. doi: 10.1091/mbc.10.11.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y., Siripanyapinyo U., Hong Y., Kang J. Y., Ishihara S., Nakakuma H., Maeda Y., Kinoshita T. PIG-W is critical for inositol acylation but not for flipping of glycosylphosphatidylinositol-anchor. Mol. Biol. Cell. 2003;14:4285–4295. doi: 10.1091/mbc.E03-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuoffer C., Jeno P., Conzelmann A., Riezman H. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol. Cell. Biol. 1991;11:27–37. doi: 10.1128/mcb.11.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuoffer C., Horvath A., Riezman H. Analysis of the sequence requirements for glycosylphosphatidylinositol anchoring of Saccharomyces cerevisiae Gas1 protein. J. Biol. Chem. 1993;268:10558–10563. [PubMed] [Google Scholar]
- Okamoto M., Yoko-o T., Umemura M., Nakayama K., Jigami Y. Glycosylphosphatidylinositol-anchored proteins are required for the transport of detergent-resistant microdomain-associated membrane proteins Tat2p and Fur4p. J. Biol. Chem. 2006;281:4013–4023. doi: 10.1074/jbc.M504684200. [DOI] [PubMed] [Google Scholar]
- Orlean P., Menon A. K. Thematic review series: lipid posttranslational modifications GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J. Lipid Res. 2007;48:993–1011. doi: 10.1194/jlr.R700002-JLR200. [DOI] [PubMed] [Google Scholar]
- Osmond B. C., Specht C. A., Robbins P. W. Chitin synthase III: synthetic lethal mutants and “stress related” chitin synthesis that bypasses the CSD3/CHS6 localization pathway. Proc. Natl. Acad. Sci. USA. 1999;96:11206–11210. doi: 10.1073/pnas.96.20.11206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitkanen J. P., Torma A., Alff S., Huopaniemi L., Mattila P., Renkonen R. Excess mannose limits the growth of phosphomannose isomerase PMI40 deletion strain of Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:55737–55743. doi: 10.1074/jbc.M410619200. [DOI] [PubMed] [Google Scholar]
- Pittet M., Conzelmann A. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2007;1771:405–420. doi: 10.1016/j.bbalip.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Pohl A., Devaux P. F., Herrmann A. Function of prokaryotic and eukaryotic ABC proteins in lipid transport. Biochim. Biophys. Acta. 2005;1733:29–52. doi: 10.1016/j.bbalip.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Pomorski T., Holthuis J. C., Herrmann A., van Meer G. Tracking down lipid flippases and their biological functions. J. Cell Sci. 2004;117:805–813. doi: 10.1242/jcs.01055. [DOI] [PubMed] [Google Scholar]
- Pomorski T., Menon A. K. Lipid flippases and their biological functions. Cell Mol. Life Sci. 2006;63:2908–2921. doi: 10.1007/s00018-006-6167-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Canivenc-Gansel E., Conzelmann A. Lipid remodeling leads to the introduction and exchange of defined ceramides on GPI proteins in the ER and Golgi of Saccharomyces cerevisiae. EMBO J. 1997;16:3506–3518. doi: 10.1093/emboj/16.12.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Conzelmann A. Biosynthesis of inositol phosphoceramides and remodeling of glycosylphosphatidylinositol anchors in Saccharomyces cerevisiae are mediated by different enzymes. J. Biol. Chem. 1998;273:30550–30559. doi: 10.1074/jbc.273.46.30550. [DOI] [PubMed] [Google Scholar]
- Saba J. D. Hirabayashi Y., Igarashi Y., Merrill A. H., Jr. Sphingolipid Biology. Tokyo: Springer-Verlag; 2006. Sphingosine-1-phosphate lyase; pp. 219–230. [Google Scholar]
- Sato K., Sato M., Nakano A. Rer1p, a retrieval receptor for endoplasmic reticulum membrane proteins, is dynamically localized to the Golgi apparatus by coatomer. J. Cell Biol. 2001;152:935–944. doi: 10.1083/jcb.152.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabl M., Daum G., Pichler H. Multiple lipid transport pathways to the plasma membrane in yeast. Biochim. Biophys. Acta. 2005;1687:130–140. doi: 10.1016/j.bbalip.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Schonbachler M., Horvath A., Fassler J., Riezman H. The yeast spt14 gene is homologous to the human PIG–A gene and is required for GPI anchor synthesis. EMBO J. 1995;14:1637–1645. doi: 10.1002/j.1460-2075.1995.tb07152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorling S., Vallee B., Barz W. P., Riezman H., Oesterhelt D. Lag1p and Lac1p are essential for the Acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisiae. Mol. Biol. Cell. 2001;12:3417–3427. doi: 10.1091/mbc.12.11.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G., Hicks J. B. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1983. [Google Scholar]
- Sievi E., Suntio T., Makarow M. Proteolytic function of GPI-anchored plasma membrane protease Yps1p in the yeast vacuole and Golgi. Traffic. 2001;2:896–907. doi: 10.1034/j.1600-0854.2001.21205.x. [DOI] [PubMed] [Google Scholar]
- Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Sipos G., Puoti A., Conzelmann A. Glycosylphosphatidylinositol membrane anchors in Saccharomyces cerevisiae: absence of ceramides from complete precursor glycolipids. EMBO J. 1994;13:2789–2796. doi: 10.1002/j.1460-2075.1994.tb06572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos G., Reggiori F., Vionnet C., Conzelmann A. Alternative lipid remodelling pathways for glycosylphosphatidylinositol membrane anchors in Saccharomyces cerevisiae. EMBO J. 1997;16:3494–3505. doi: 10.1093/emboj/16.12.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. K., Cottaz S., Brimacombe J. S., Ferguson M. A. Substrate specificity of the dolichol phosphate mannose: glucosaminyl phosphatidylinositol alpha1–4-mannosyltransferase of the glycosylphosphatidylinositol biosynthetic pathway of African trypanosomes. J. Biol. Chem. 1996;271:6476–6482. doi: 10.1074/jbc.271.11.6476. [DOI] [PubMed] [Google Scholar]
- Smith T. K., Sharma D. K., Crossman A., Dix A., Brimacombe J. S., Ferguson M. A. Parasite and mammalian GPI biosynthetic pathways can be distinguished using synthetic substrate analogues. EMBO J. 1997;16:6667–6675. doi: 10.1093/emboj/16.22.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobering A. K., Watanabe R., Romeo M. J., Yan B. C., Specht C. A., Orlean P., Riezman H., Levin D. E. Yeast Ras regulates the complex that catalyzes the first step in GPI-anchor biosynthesis at the ER. Cell. 2004;117:637–648. doi: 10.1016/j.cell.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Sorger D., Athenstaedt K., Hrastnik C., Daum G. A yeast strain lacking lipid particles bears a defect in ergosterol formation. J. Biol. Chem. 2004;279:31190–31196. doi: 10.1074/jbc.M403251200. [DOI] [PubMed] [Google Scholar]
- Stevens T., Esmon B., Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Sutterlin C., Doering T. L., Schimmoller F., Schroder S., Riezman H. Specific requirements for the ER to Golgi transport of GPI-anchored proteins in yeast. J. Cell Sci. 1997a;110:2703–2714. doi: 10.1242/jcs.110.21.2703. [DOI] [PubMed] [Google Scholar]
- Sutterlin C., Horvath A., Gerold P., Schwarz R. T., Wang Y., Dreyfuss M., Riezman H. Identification of a species-specific inhibitor of glycosylphosphatidylinositol synthesis. EMBO J. 1997b;16:6374–6383. doi: 10.1093/emboj/16.21.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin C., Escribano M. V., Gerold P., Maeda Y., Mazon M. J., Kinoshita T., Schwarz R. T., Riezman H. Saccharomyces cerevisiae GPI10, the functional homologue of human PIG-B, is required for glycosylphosphatidylinositol-anchor synthesis. Biochem. J. 1998;332:153–159. doi: 10.1042/bj3320153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterwala S. S., Creswell C. H., Sanyal S., Menon A. K., Bangs J. D. De novo sphingolipid synthesis is essential for viability, but not for transport of glycosylphosphatidylinositol-anchored proteins, in African trypanosomes. Eukaryot. Cell. 2007;6:454–464. doi: 10.1128/EC.00283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain E., Stukey J., McDonough V., Germann M., Liu Y., Sturley S. L., Nickels J. T., Jr. Yeast cells lacking the ARV1 gene harbor defects in sphingolipid metabolism. Complementation by human ARV1. J. Biol. Chem. 2002;277:36152–36160. doi: 10.1074/jbc.M206624200. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Maeda Y., Tashima Y., Kinoshita T. Inositol deacylation of glycosylphosphatidylinositol-anchored proteins is mediated by mammalian PGAP1 and yeast Bst1p. J. Biol. Chem. 2004;279:14256–14263. doi: 10.1074/jbc.M313755200. [DOI] [PubMed] [Google Scholar]
- Taron C. H., Wiedman J. M., Grimme S. J., Orlean P. Glycosylphosphatidylinositol biosynthesis defects in Gpi11p- and Gpi13p-deficient yeast suggest a branched pathway and implicate Gpi13p in phosphoethanolamine transfer to the third mannose. Mol. Biol. Cell. 2000;11:1611–1630. doi: 10.1091/mbc.11.5.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashima Y., Taguchi R., Murata C., Ashida H., Kinoshita T., Maeda Y. PGAP2 is essential for correct processing and stable expression of GPI-anchored proteins. Mol. Biol. Cell. 2006;17:1410–1420. doi: 10.1091/mbc.E05-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehlivets O., Scheuringer K., Kohlwein S. D. Fatty acid synthesis and elongation in yeast. Biochim. Biophys. Acta. 2007;1771:255–270. doi: 10.1016/j.bbalip.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Tinkelenberg A. H., Liu Y., Alcantara F., Khan S., Guo Z., Bard M., Sturley S. L. Mutations in yeast ARV1 alter intracellular sterol distribution and are complemented by human ARV1. J. Biol. Chem. 2000;275:40667–40670. doi: 10.1074/jbc.C000710200. [DOI] [PubMed] [Google Scholar]
- Tusnady G. E., Sarkadi B., Simon I., Varadi A. Membrane topology of human ABC proteins. FEBS Lett. 2006;580:1017–1022. doi: 10.1016/j.febslet.2005.11.040. [DOI] [PubMed] [Google Scholar]
- Umemura M., Okamoto M., Nakayama K., Sagane K., Tsukahara K., Hata K., Jigami Y. GWT1 gene is required for inositol acylation of glycosylphosphatidylinositol anchors in yeast. J. Biol. Chem. 2003;278:23639–23647. doi: 10.1074/jbc.M301044200. [DOI] [PubMed] [Google Scholar]
- Umemura M., Fujita M., Yoko-O T., Fukamizu A., Jigami Y. Saccharomyces cerevisiae CWH43 is involved in the remodeling of the lipid moiety of GPI anchors to ceramides. Mol. Biol. Cell. 2007;18:4304–4316. doi: 10.1091/mbc.E07-05-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivieso M. H., Ferrario L., Vai M., Duran A., Popolo L. Chitin synthesis in a gas1 mutant of Saccharomyces cerevisiae. J. Bacteriol. 2000;182:4752–4757. doi: 10.1128/jb.182.17.4752-4757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee B., Riezman H. Lip1p: a novel subunit of acyl-CoA ceramide synthase. EMBO J. 2005;24:730–741. doi: 10.1038/sj.emboj.7600562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidugiriene J., Menon A. K. Early lipid intermediates in glycosyl-phosphatidylinositol anchor assembly are synthesized in the ER and located in the cytoplasmic leaflet of the ER membrane bilayer. J. Cell Biol. 1993;121:987–996. doi: 10.1083/jcb.121.5.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwakarma R. A., Menon A. K. Flip-flop of glycosylphosphatidylinositols (GPI's) across the ER. Chem. Commun. 2005;4:453–455. doi: 10.1039/b413196g. [DOI] [PubMed] [Google Scholar]
- Watanabe R., Inoue N., Westfall B., Taron C. H., Orlean P., Takeda J., Kinoshita T. The first step of glycosylphosphatidylinositol biosynthesis is mediated by a complex of PIG-A, PIG-H, PIG-C and GPI1. EMBO J. 1998;17:877–885. doi: 10.1093/emboj/17.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R., Funato K., Venkataraman K., Futerman A. H., Riezman H. Sphingolipids are required for the stable membrane association of glycosylphosphatidylinositol-anchored proteins in yeast. J. Biol. Chem. 2002;277:49538–49544. doi: 10.1074/jbc.M206209200. [DOI] [PubMed] [Google Scholar]
- Yan B. C., Westfall B. A., Orlean P. Ynl038wp (Gpi15p) is the Saccharomyces cerevisiae homologue of human Pig-Hp and participates in the first step in glycosylphosphatidylinositol assembly. Yeast. 2001;18:1383–1389. doi: 10.1002/yea.783. [DOI] [PubMed] [Google Scholar]
- Zanolari B., Friant S., Funato K., Sutterlin C., Stevenson B. J., Riezman H. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 2000;19:2824–2833. doi: 10.1093/emboj/19.12.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.