Abstract
Meckel-Gruber syndrome (MKS), nephronophthisis (NPHP), and Joubert syndrome (JBTS) are a group of heterogeneous cystic kidney disorders with partially overlapping loci. Many of the proteins associated with these diseases interact and localize to cilia and/or basal bodies. One of these proteins is MKS1, which is disrupted in some MKS patients and contains a B9 motif of unknown function that is found in two other mammalian proteins, B9D2 and B9D1. Caenorhabditis elegans also has three B9 proteins: XBX-7 (MKS1), TZA-1 (B9D2), and TZA-2 (B9D1). Herein, we report that the C. elegans B9 proteins form a complex that localizes to the base of cilia. Mutations in the B9 genes do not overtly affect cilia formation unless they are in combination with a mutation in nph-1 or nph-4, the homologues of human genes (NPHP1 and NPHP4, respectively) that are mutated in some NPHP patients. Our data indicate that the B9 proteins function redundantly with the nephrocystins to regulate the formation and/or maintenance of cilia and dendrites in the amphid and phasmid ciliated sensory neurons. Together, these data suggest that the human homologues of the novel B9 genes B9D2 and B9D1 will be strong candidate loci for pathologies in human MKS, NPHP, and JBTS.
INTRODUCTION
Meckel-Gruber syndrome (MKS), nephronophthisis (NPHP), and Joubert syndrome (JBTS) are three genetically heterogeneous autosomal recessive disorders that exhibit renal cystic dysplasia in addition to several other phenotypes. MKS is a perinatal lethal disorder defined by not only renal cysts but also by severe developmental defects including polydactyly and occipital encephalocele (Alexiev et al., 2006). In contrast, NPHP patients develop end-stage renal disease as children and young adults, but this disease is not associated with significant perinatal lethality. Phenotypes associated with NPHP include the formation of corticomedullary cysts, disruption of tubular basement membrane, tubulointerstitial nephropathy, and sometimes retinal defects (reviewed in Hildebrandt and Zhou, 2007). JBTS type B, also known as cerebello-oculo-renal syndrome (CORS), is characterized by retinal dystrophy and renal anomalies as well as underdeveloped cerebellar vermis, brain stem abnormalities, and hypotonia (reviewed in Chen, 2007). Genetic analysis has revealed that in some cases, MKS, NPHP, and JBTS share loci that are allelic in nature. These include MKS3/JBTS6 (Baala et al., 2007b), NPHP1/JBTS4 (Parisi et al., 2004), MKS4/NPHP6/JBTS5 (Sayer et al., 2006; Baala et al., 2007a), and MKS5/NPHP8/JBTS7 (Arts et al., 2007; Delous et al., 2007). Notably, the current known genes connected to MKS, NPHP, and JBTS have been linked to only a small percentage of afflicted individuals, suggesting that a large number of loci responsible for these disorders are yet to be identified (Hildebrandt and Otto, 2005; Khaddour et al., 2007).
A common theme connecting most cystic kidney disorders is that the genes associated with the observed pathology encode cilia or basal body proteins. This is seen for NPHP1, 2, and 4–8 (Mollet et al., 2002; Morgan et al., 2002; Otto et al., 2002, 2003, 2005; Fliegauf et al., 2006; Sayer et al., 2006; Attanasio et al., 2007; Delous et al., 2007) as well as MKS1 and MKS3 (meckelin) (Smith et al., 2006; Dawe et al., 2007b). Furthermore, many of the NPHP proteins appear to function as part of a complex (Mollet et al., 2005; Hildebrandt and Zhou, 2007) that may also include RPGRIP1L (NPHP8), which was originally identified as a binding partner for NPHP4 (Roepman et al., 2005). These genetic, cellular, and biochemical data together indicate that MKS, NPHP, and JBTS may represent a continuum of a common underlying ciliary/basal body disorder where the phenotypic outcome is determined by the nature of the mutation or genetic background of the affected patient.
Cilia are microtubule-based organelles that have multiple functions ranging from fluid and cell movement to sensory reception and signaling regulation. All cilia are associated with a basal body, which is a modified centriole that forms in nondividing cells. Before ciliogenesis, the centrioles migrate to the cell membrane where they form attachments and nucleate ciliary microtubule assembly (reviewed in Rosenbaum and Witman, 2002). The processes of centriole migration and subsequent tethering to the membrane are not fully understood (Dawe et al., 2007a; Marshall, 2007). In addition to the role of basal bodies in initializing ciliogenesis, they also serve as the docking and assembly site for proteins involved in intraflagellar transport (IFT) as well as cargo targeted to the cilium (Deane et al., 2001). IFT is essential for cilia formation and involves the bidirectional movement of large protein complexes (IFT particles) and associated cargo along the cilium axoneme (Kozminski et al., 1993; Cole et al., 1998; Piperno et al., 1998; Orozco et al., 1999). This occurs in the anterograde direction via kinesin motors and in the retrograde direction by a cytoplasmic dynein motor (Cole et al., 1998; Porter et al., 1999). Mutations that disrupt IFT components compromise ciliogenesis and result in severe developmental and disease pathologies in mice and humans collectively referred to as ciliopathies (Moyer et al., 1994; Murcia et al., 2000; Pazour et al., 2000; Taulman et al., 2001; Haycraft et al., 2005; Badano et al., 2006; Beales et al., 2007; Davenport et al., 2007).
Much of our understanding about IFT, ciliogenesis, and the function of proteins associated with ciliopathies has come from analyses conducted in invertebrates such as Chlamydomonas and C. elegans. Unlike the ubiquitous nature of cilia in mammals, cilia are present in C. elegans only at the distal tips of neuron dendrites where they function as sensors of conditions in the external environment. These ciliated cells include the amphid and labial (inner and outer) neurons in the head of the worm in addition to phasmid neurons in the tail. All cilia in C. elegans are immotile and are separated from the distal tips of the dendrites by a region called the transition zone, which is loosely analogous to the proximal end of the mammalian cilium where the basal bodies are located (Ward et al., 1975; Ware et al., 1975). Although most animals possess basal bodies composed of nine triplet microtubules, C. elegans centrioles have a nine-singlet microtubule arrangement yet are thought to have conserved function in cell division. There is no direct ultrastructural evidence that these centrioles are incorporated into the base of C. elegans cilia; however, in daf-19 mutant worms lacking all ciliary structures, the centrioles are found at the distal tips of neuron dendrites where the cilia are normally formed (Perkins et al., 1986). Thus, C. elegans centrioles are likely to have similar function in ciliogenesis as in mammalian systems.
Characterization of C. elegans and Chlamydomonas mutants where cilia are improperly formed or have defects in cilia-mediated signaling activity has uncovered many components of IFT that have evolutionarily conserved functions in mammals and has thus provided important insights into the molecular basis of the human ciliopathies (Scholey et al., 2004; Wang et al., 2006; Ou et al., 2007). This also includes analysis of the genes involved in Bardet-Biedl syndrome (BBS), another heterogeneous cystic kidney disorder involving proteins that localize to cilia or basal bodies (reviewed in Blacque and Leroux, 2006), as well as two genes linked to human polycystic kidney disease (PKD) (Barr et al., 2001). Additionally, the human NPHP proteins neprocystin-1 (NPHP1) and nephrocystin-4 (NPHP4) have highly conserved homologues in C. elegans (NPH-1 and NPH-4, respectively) that colocalize specifically to transition zones where they appear to function in a common protein complex. This is supported by the requirement of NPH-4 for localization of NPH-1 to the transition zones (Winkelbauer et al., 2005). nph-1 and nph-4 mutant worms display several of the sensory behavior abnormalities typically associated with C. elegans mutants that have cilia morphology defects. These include altered chemotaxis responses toward attractants and increased lifespan in the single nph mutants as well as male mating defects in nph-1;nph-4 double mutants (Jauregui and Barr, 2005; Winkelbauer et al., 2005). Although these mutants do not have overt abnormalities in cilia morphology based both on the ability of worms to absorb fluorescent dye and to properly localize an IFT protein along the length of cilia axonemes, recent transmission electron microscopy data indicate that a subpopulation of nph mutant worms occasionally exhibit ultrastructural and morphological defects in their cilia. These abnormalities include the loss or truncation of cilia from a subset of amphid neurons and open or lost B-tubules (Jauregui et al., 2008).
In contrast to the extensive work conducted in C. elegans on the genes involved in human BBS, PKD, and NPHP, the homologues of the MKS-associated genes have largely been unexplored. Previous studies have shown that the MKS1 homolog xbx-7(R148.1) is expressed in ciliated sensory neurons of C. elegans; however the function or localization of XBX-7 protein is unknown (Efimenko et al., 2005). This protein contains a B9 domain that has not been investigated and is found in only two other proteins in C. elegans or humans. Herein we describe XBX-7 and the two other B9 proteins, TZA-1(Y38F2AL.2) and TZA-2(K03E6.4), and demonstrate that all three are part of a complex formed specifically at the base of cilia. TZA-1 is necessary for anchoring TZA-2 in this complex, whereas XBX-7 requires both TZA-1 and TZA-2 for proper localization. Importantly, we have determined that the B9 proteins function redundantly with the transition zone proteins NPH-1 and NPH-4 to regulate the formation and/or maintenance of cilia and dendrites of the amphid and phasmid neurons. Mutations that disrupt the B9 proteins do not cause overt cilia morphology defects; however, simultaneous disruption of a single B9 protein along with disruption of a NPH protein results in malformed ciliated sensory neuron dendrites expressing shortened and laterally oriented cilia. Together, these data suggest the B9 proteins in combination with the NPH proteins have roles in mediating cilia assembly and provide insights into why disruption of basal body proteins leads to the broad range of overlapping ciliopathies associated with MKS, NPHP, and JBTS in humans. Moreover, our analyses indicate that the two B9 domain-encoding genes in addition to MKS1 are important candidate loci for MKS, NPHP, and JBTS patients in which the underlying genetic defect has not yet been identified.
MATERIALS AND METHODS
General Molecular Biology Methods
Molecular biology procedures were performed according to standard protocols (Sambrook et al., 1989). C. elegans genomic DNA, C. elegans cDNA, or cloned C. elegans DNA was used for PCR amplifications, for direct sequencing, or for subcloning (Sambrook et al., 1989). All PCR for cloning was performed with AccuTaq LA Polymerase (Sigma-Aldrich, St. Louis, MO). Clones, primer sequences, and PCR conditions are available upon request. DNA sequencing was performed by the University of Alabama at Birmingham Genomics Core Facility of the Heflin Center for Human Genetics.
DNA and Protein Sequence Analyses
Genome sequence information was obtained from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). Gene and protein sequences were identified using the C. elegans database Wormbase and references therein (http://www.wormbase.org). BLAST searches to identify homologues in human, mouse, and Strongylocentrotus purpuratus were performed using the National Center for Biotechnology Information (NCBI) BLAST service (http://www.ncbi.nlm.nih.gov/BLAST/). Protein sequence alignments were performed and conserved motifs were identified using ClustalW (http://www.ebi.ac.uk/clustalw/).
Strains
Worm strains were obtained from the Caenorhabditis Genetics Center, C. elegans Knock-out Consortium, and National BioResource Project in Japan. Strains were grown using standard C. elegans growth methods (Brenner, 1974) at 22°C unless otherwise stated. The wild-type strain was N2 Bristol. The following mutations were used: RB743 nph-1(ok500)II, JT6924 daf-19(m86)II;daf-12(sa204)X, FX2705 xbx-7(tm2705)III, FX2452 tza-1(tm2452)IV, FX925 nph-4(tm925)V, JT204 daf-12(sa204)X, RB1682 tza-2(ok2092)X, YH560 nph-1(ok500)II;xbx-7(tm2705)III, YH559 nph-1(ok500)II;tza-1(tm2452)IV, YH561 nph-1(ok500)II;tza-2(ok2092),YH513 xbx-7(tm2705)III;nph-4(tm925)V, YH482 tza-1(tm2452);nph-4(tm925)V, and YH496 nph-4(tm925)V;tza-2(ok2092)X. RB743, FX2705, FX2452, FX925, and RB1682 were outcrossed three times and genotyped by PCR before analysis. The following extrachromosomal arrays were used: yhEx279 and yhEx281 were used for xbx-7::YFP, tza-1::CFP, and tza-2::CFP expression experiments; yhEx240 and yhEx288 were used for DAF-19 regulation of xbx-7::YFP, tza-1::DsRed2, and tza-2::CFP expression experiments; yhEx232, yhEx285, yhEx290, and yhEx293 were used for TZA-2::CFP, XBX-7::YFP, and TZA-1::YFP localization analyses; yhEx149 and yhEx232 were used for localization of CHE-13::YFP in cilia of single mutants; yhEx142, yhEx149, yhEx232, yhEx285, and yhEx290 were used for analysis of localization of NPH-1::CFP, NPH-4::YFP, TZA-2::CFP, XBX-7::YFP, and TZA-1::YFP in single mutants; yhEx298, yhEx300, yhEx301, yhEx307, yhEx308, and yhEx309 were used for localization of CHE-13::YFP in double mutants; yhEx281 and yhEx297 were used for xbx-7::YFP expression experiments in double mutants; yhEx149, yhEx232, yhEx298, yhEx300, yhEx301, yhEx307, yhEx308, and yhEx309 were used for CHE-13::YFP/CFP localization in cilia length measurements; yhEx311 and yhEx313 were used for tza-1::DsRed2 and f16f9.3::GFP expression experiments.
Generation of Constructs and Strains
Vectors used for generating transcriptional and translational fusion constructs were modified from pPD95.81 (a gift from A. Fire, Stanford University). pCJF6 (CFP), pCJF7 (YFP), pCJ102 (DsRed2), pCJF36 (CHE-13::CFP), pCJF37 (CHE-13::YFP), pCJ146 (NPH-4::YFP), and pCJ148 (NPH-1::CFP) were generated as previously described (Winkelbauer et al., 2005). pCJ309 (xbx-7::YFP), pCJ261.2 (tza-1::DsRed2), and pCJ257 (tza-2::CFP) were generated by inserting pCJF7, pCJ102, and pCJF6, respectively, with 1500-, 1200-, and 1000-base pair promoter fragments amplified from N2 genomic DNA corresponding to the regions immediately upstream and slightly downstream of xbx-7 ATG, tza-1 ATG, and tza-2 ATG, respectively. pCJ312 (tza-1::CFP) was generated by transferring the 1200-base pair insert of pCJ261.2 into pCJF6. pCJ318 (XBX-7::YFP) and pCJ308 (TZA-1::YFP) were generated by inserting pCJF7 with a fragment consisting of the 1500-base pair promoter and genomic region (covering the first 10 exons) of xbx-7 or a fragment consisting of the 1200-base pair promoter and genomic region of tza-1, respectively. pCJ271 (TZA-2::CFP) was generated by inserting pCJF6 with a fragment consisting of the 1000-base pair promoter and genomic region of tza-2. pCP41 (f16f9.3::GFP) was generously provided by S. Shaham (The Rockefeller University). All PCR was performed using AccuTaq-LA DNA Polymerase (Sigma) according to the manufacturer's instructions. Transgenic worms were generated as previously described (Mello et al., 1991).
Imaging
Worms were anesthetized using 10 mM levamisole and immobilized on 2% agar pads for imaging. Worms were examined using a Nikon Eclipse TE200 inverted microscope (Melville, NY), and images were captured with a CoolSnap HQ camera (Photometrics, Tucson, AZ). Shutters and filters were computer driven. Images were processed using Metamorph software (Universal Imaging, Downingtown, PA). Confocal analysis was performed on a Leica DMIRBE inverted epifluorescence/Nomarski microscope outfitted with Leica TCS NT SP1 Laser Confocal optics (Leica, Exton, PA). Optical sections through the Z axis were generated using a stage galvanometer. Confocal images were processed with ImageJ (National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij/) and Voxx 2.09d (Indiana University School of Medicine, Indianapolis, IN). Further processing of images was performed using Photoshop 7.0 (Adobe Systems, San Jose, CA).
Yeast Two-Hybrid Assay
Full-length xbx-7, tza-1, and tza-2 cDNA was amplified from cDNA library using AccuTaq LA Polymerase (Sigma-Aldrich) and sequence-specific primers flanked by unique restriction enzyme sites. The resulting PCR fragments were cloned into the vectors pGBKT7 and pGADT7 (BD Biosciences, Clontech, San Jose, CA). Interaction experiments were performed according to the MATCHMAKER 3 Yeast two-hybrid manual (BD Biosciences Clontech) as described previously (Haycraft et al., 2003).
DAF-19 Regulation
To asses DAF-19 regulation in vivo, the transgenic line YH493 was generated by injection of xbx-7::YFP into JT6924, and YH380 was generated by injection of tza-1::DsRed2 and tza-2::CFP into JT6924. These strains were then crossed to JT204 to achieve daf-19(+) background. The strains used contain a mutation in daf-12(sa204)X to suppress the Daf-c phenotype of daf-19(m86)II.
RT-PCR
RNA was isolated as described previously (Haycraft et al., 2003) from xbx-7(tm2705), tza-1(tm2452), and tza-2(ok2092) mutants. Reverse-transcribed RNA was generated using Superscript II Reverse Transcriptase (Invitrogen, Carlsbad, CA) using the manufacturer's instructions. Fragments spanning the deleted region of each respective gene were then amplified by PCR and sequenced.
Assays
Dye-filling using DiI (Molecular Probes, Carlsbad, CA), and osmotic avoidance assays were performed as described previously (Starich et al., 1995). The ability of C. elegans strains to form dauer stages was tested as described previously (Starich et al., 1995). Chemotaxis assays to volatile attractants were performed as described previously (Winkelbauer et al., 2005). Foraging behavior analysis was performed with slight modification of that described previously (Fujiwara et al., 2002). Briefly, a single young adult worm (rather than L4 stage worms) was placed in the center of a uniformly sized lawn of bacteria on a 6-cm plate and was allowed to move freely for 18 h at 20°C before being removed from the plate by aspiration. Tracking was then quantified by counting the number of 3 × 3-mm squares on a grid the worm tracks touched.
RESULTS
xbx-7, tza-1, and tza-2 Encode the B9 Family of Proteins in C. elegans
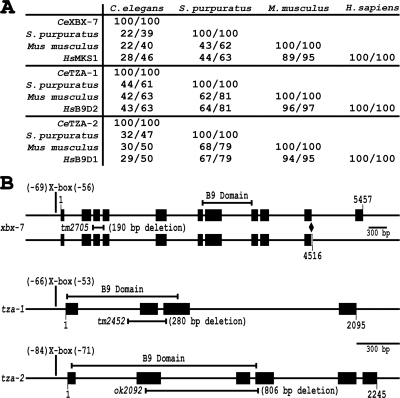
The B9 protein domain is present within only three human proteins, MKS1, B9D2 (B9 protein Domain 2), and B9D1 (B9 protein Domain 1). The function of this 104-amino acid motif is unknown. Although the molecular role of B9D1 has not been explored, MKS1 as well as the B9D2 homolog in Paramecium tetraurelia (ICIS-1) were recently described as proteins functioning in the process of ciliogenesis (Dawe et al., 2007b; Ponsard et al., 2007). To explore the possibility that the B9 domain proteins are functionally related in regards to cilia signaling activities and ciliopathies in MKS, we characterized the homologues of these proteins in C. elegans. As in mammals, C. elegans has three proteins with the B9 motif. These include the previously described XBX-7 (X-box promoter element regulated 7), which corresponds to mammalian MKS1, TZA-1 (transition zone-associated 1), which corresponds to mammalian B9D2, and TZA-2 (transition zone-associated 2), which corresponds to mammalian B9D1 (Figure 1A).
Figure 1.
B9 protein family conservation and schematic gene representations. (A) Homology with other organisms presented as % amino acid identity/% amino acid similarity. (B) Schematic representation of xbx-7, tza-1, and tza-2 genomic regions. Promoter X-box location, gene region encoding each B9 domain, gene region deleted in each mutant allele, and in the case of xbx-7, splice isoforms are shown. xbx-7(tm2705) and tza-1(tm2452) both contain internal in-frame deletions, whereas tza-2(ok2092) contains an internal frameshift deletion that results in a truncated protein. Exons are represented as black boxes. Diamond signifies site of xbx-7 alternative splicing.
To assess the conserved nature of the family of B9 proteins, we compared the putative homologues across several vertebrate and invertebrate species. Mammalian MKS1 and C. elegans XBX-7 are the most diverged of the B9 homologues and show the strongest similarities in the C-terminal regions of the proteins and within the B9 domains. In contrast, both TZA-1 and TZA-2 share homology with their mammalian counterparts, B9D2 and B9D1, respectively, throughout nearly their entire lengths.
xbx-7, tza-1, and tza-2 Are Coexpressed in Ciliated Sensory Neurons and Are Dependent on DAF-19
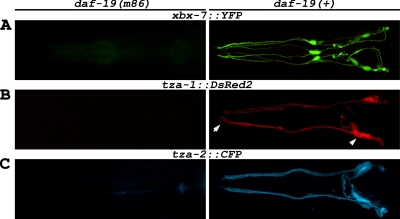
Previous studies have shown that expression of several C. elegans genes associated with IFT and ciliogenesis, as well as homologues of genes involved in human cystic kidney disorders (e.g., BBS- and NPHP-associated genes), are regulated by the DAF-19 transcription factor (Collet et al., 1998; Swoboda et al., 2000; Blacque et al., 2005; Efimenko et al., 2005; Winkelbauer et al., 2005). DAF-19 is required for cilia formation and regulates expression of genes in ciliated sensory neurons through an X-box promoter element typically found within the first 200 nucleotides upstream of the translational start site (Swoboda et al., 2000; Efimenko et al., 2005). Genome sequence-based searches have identified xbx-7, tza-1, and tza-2 as potential DAF-19 targets because they contain a conserved X-box motif in each of their respective promoters (Efimenko et al., 2005). The X-boxes found in the promoters of these genes are located at position −69 of xbx-7, position −66 of tza-1, and position −84 of tza-2, relative to the start of translation (Figure 1B). Previous data indicates that xbx-7, tza-1, and tza-2 are all expressed in the ciliated amphid and labial neurons in the head and in the phasmid neurons at the tail of C. elegans (Blacque et al., 2005; Efimenko et al., 2005). Utilizing transgenic lines coexpressing transcriptional fusions xbx-7::YFP and tza-1::CFP or xbx-7::YFP and tza-2::CFP, we also observed expression of these genes specifically in ciliated sensory neurons and additionally found that their expression was overlapping (Supplementary Figure 1). Furthermore, our analyses revealed that expression of tza-1::DsRed2 and tza-2::CFP in ciliated sensory neurons was dependant on DAF-19 as was previously shown for xbx-7 (Efimenko et al., 2005; Figure 2). This was demonstrated by generating transgenic lines with the transcriptional fusion constructs in the daf-19(m86) mutant background. Expression from these transgenes was abolished in the daf-19(m86) mutants but was recovered after a single outcross to daf-19 wild-type worms (Figure 2).
Figure 2.
DAF-19 regulation of xbx-7, tza-1, and tza-2 in C. elegans. Expression analysis of (A) xbx-7::YFP, (B) tza-1::DsRed2, and (C) tza-2::CFP in the head ciliated sensory neurons in (left panels) daf-19(m86) mutant and (right panels) daf-19(+) backgrounds. The expression of each transgene was absent in daf-19(m86) mutants while outcrossing to daf-19(+) background restored expression to wild-type levels. Arrowhead, position of the amphid neuron cell bodies from which the dendrites extend to the anterior of the worm. Cilia project from the (arrow) distal tips of the dendrites. For this and subsequent figures, anterior is toward the left.
XBX-7, TZA-1, and TZA-2 Localize to the Base of Cilia
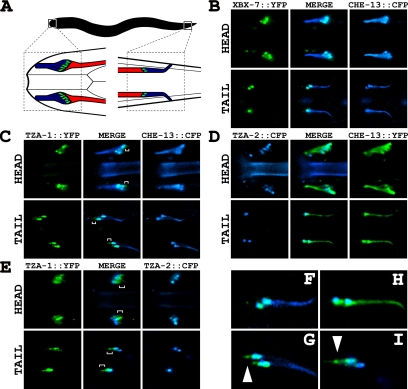
To determine the localization of the XBX-7, TZA-1, and TZA-2 proteins, we generated transgenic lines coexpressing B9 protein translational fusions with cyan fluorescent protein (CFP) or yellow fluorescent protein (YFP) under the control of their endogenous promoters (XBX-7::YFP, TZA-1::YFP, and TZA-2::CFP) and a translational fusion of the IFT protein CHE-13 with CFP or YFP, which we utilized as a cilia and transition zone marker. TZA-1::YFP, TZA-2::CFP, and XBX-7::YFP (corresponding to the 10-exon splice variant), were all detected specifically at the transition zones between cilia and the distal end of the dendrites of ciliated sensory neurons and were not detected in cilia axonemes as marked by CHE-13::CFP (Figure 3). This localization to the base of cilia is similar to that reported for MKS1 (Dawe et al., 2007b). On closer inspection of TZA-1::YFP, we noted that it did not entirely colocalize with CHE-13::CFP in the head and tail neurons; TZA-1::YFP localized to a domain that extended beyond the transition zones into the dendrites away from the cilia and toward the cell bodies (Figure 3, C and G). The difference in the localization of TZA-1 was further examined in worms coexpressing both TZA-1::YFP and TZA-2::CFP (Figure 3, E and I). In addition to overlapping TZA-2::CFP at the transition zones, TZA-1::YFP was observed immediately posterior to TZA-2::CFP in the amphid dendrites of the head and anterior to TZA-2::CFP in the phasmid dendrites of the tail.
Figure 3.
Localization of XBX-7, TZA-1, and TZA-2 proteins to transition zones at the base of cilia in C. elegans. (A) An illustration depicting the anatomical positions of (left) amphid cilia bundles in the head and (right) phasmid cilia bundles in the tail. Cilia axonemes are represented by blue, transition zones by green, and the dendritic processes by red. (B–D) Transgenic lines were generated that expressed XBX-7::YFP, TZA-1::CFP, or TZA-2::CFP along with cilia/transition zone markers CHE-13::CFP or CHE-13::YFP, each under the control the endogenous promoter of the corresponding gene. (B) XBX-7::YFP localized to the transition zones at the base of cilia in the (top) amphid neurons of the head and (bottom) phasmid neurons of the tail but not along cilia axonemes. (C) TZA-1::YFP localized to the transition zones at the base of cilia in the (top) amphid neurons of the head and (bottom) phasmid neurons of the tail but not along cilia axonemes. In contrast to CHE-13::CFP localization, which was restricted to the cilia axonemes and transition zones, TZA-1::YFP was found at the transition zones and (top, brackets) extended slightly into the dendrites of the amphid neurons. In the (bottom) phasmid neurons, the (brackets) extension of TZA-1::YFP into the dendrites versus the restriction of CHE-13::CFP to the transition zones and cilia axonemes was more distinctive. (D) TZA-2::CFP localized to the transition zones at the base of cilia in the (top) amphid neurons of the head and (bottom) phasmid neurons of the tail but not along cilia axonemes. (E) Analysis of TZA-1::YFP and TZA-2::CFP colocalization at the base of cilia. In contrast to TZA-2::CFP localization, which was restricted to the transition zones, TZA-1::YFP was found at the transition zones and (brackets) extended into the dendrites of both the (top) amphid and (bottom) phasmid neurons. (F–I) Enlargement of phasmid cilia regions from B–E, respectively. (G) TZA-1::YFP and CHE-13::CFP and (I) TZA-1::YFP and TZA-2::CFP colocalized largely to the transition zones but not to (arrowheads) a small domain at the distal ends of the dendrites which contained TZA-1::YFP only.
Analysis of xbx-7, tza-1, and tza-2 Mutant Alleles
Previous data in mammalian cells have indicated that disrupting MKS1 by small interfering RNA (siRNA) results in defects in cilia formation. This appears to be due to abnormalities in cell polarization and in apical positioning of the centrosomes, which are required for basal body and cilia formation (Dawe et al., 2007b). Similarly, disruption of the B9D2 homolog by RNAi in P. tetraurelia was found to interfere with cilia stability or formation (Ponsard et al., 2007), but the mechanism by which this occurred was not determined.
To better assess the function of the B9 proteins and their potential role in ciliogenesis, we obtained genetic mutants FX2705 xbx-7(tm2705), FX2452 tza-1(tm2452), and RB1682 tza-2(ok2092) from the National Bioresource Project and the C. elegans Knockout Consortium. FX2705 xbx-7(tm2705) contains an internal genomic deletion of nucleotides 591-780 that begins in intron 2 and extends into exon 4, upstream of the region encoding the XBX-7 B9 domain (Figure 1B). RT-PCR analysis of the tm2705 transcript revealed that this deletion does not cause a shift in the reading frame and would encode a protein lacking amino acids 69-142 (Supplementary Figure 2A). FX2452 tza-1(tm2452) is an internal deletion that removes nucleotides 446-724 of tza-1. This deletion, which spans through part of intron 1 into exon 3, disrupts much of the TZA-1 B9 domain coding region (Figure 1B). RT-PCR analysis of the tm2452 transcript revealed that this deletion does not cause a frameshift, and the predicted protein would lack 47 (30-77) of 175 amino acids (Supplementary Figure 2B). The RB1682 tza-2(ok2092) allele contains a deletion of nucleotides 574-1380. This deletion spans part of exon 2 through a portion of exon 3, directly disrupting the region encoding the TZA-2 B9 motif (Figure 1B). RT-PCR of the ok2092 transcript revealed that the allele contains a frameshift producing a putative protein consisting of the first 42 correct amino acids followed by 49 altered amino acids (Supplementary Figure 2C).
Disruption of the B9 Genes Does Not Overtly Affect Cilia Morphology in C. elegans
To begin analyzing the B9 gene mutants and their potential effects on ciliogenesis, we first evaluated the ability of xbx-7(tm2705), tza-1(tm2452), and tza-2(ok2092) mutant worms to absorb fluorescent hydrophobic dye (DiI) into the dendrites and cell bodies of the amphid and phasmid neurons via cilia that are exposed to the external environment of the worm (Perkins et al., 1986; Starich et al., 1995). IFT mutants such as osm-5, che-13, and xbx-1 cannot dye-fill because of gross cilia morphology defects, and likewise, mutants of BBS homologues such as bbs-7 and bbs-8 also have cilia structure and dye-filling deficiencies (Haycraft et al., 2001, 2003; Schafer et al., 2003; Blacque et al., 2004). In contrast, mutations in the NPHP homologues nph-1 and nph-4 reportedly do not compromise dye-filling (Jauregui and Barr, 2005; Winkelbauer et al., 2005), although morphological defects in the cilia on a subset of the sensory neurons of these worms have recently been observed (Jauregui et al., 2008).
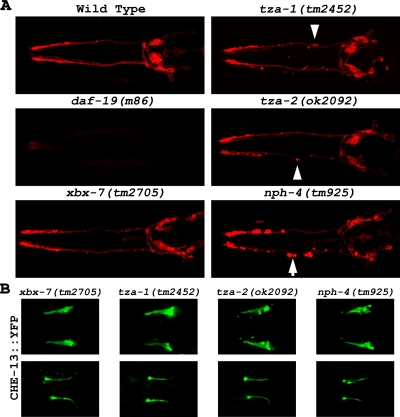
In contrast to the cilia null mutant daf-19(m86) worms, the xbx-7(tm2705), tza-1(tm2452), and tza-2(ok2092) mutant strains efficiently absorbed DiI into the amphid and phasmid neurons as was shown for the wild-type control (Figure 4A; phasmid data are not shown). However, as seen in the nph-4(tm925) mutants, discrete regions of dye accumulation were evident along the dendrites of the B9 gene mutants. These pockets of dye accumulation were not reported in previous studies and their cause is unknown. We additionally generated worm strains with all possible combinations of B9 gene mutations including the B9 gene triple homozygous mutant. None of these compound mutants exhibited any discernable defects in DiI uptake apart from occasional accumulations of dye along the dendrites as seen in the single mutants (data not shown). To further analyze the possible effects that disruption of the B9 proteins may have on cilia morphology, we generated xbx-7(tm2705), tza-1(tm2452), and tza-2(ok2092) mutant strains each expressing CHE-13::YFP as a cilia marker. Consistent with their normal dye-filling phenotype, we could detect no alterations in CHE-13::YFP localization or signal in either xbx-7(tm2705), tza-1(tm2452), or tza-2(ok2092) mutant backgrounds (Figure 4B).
Figure 4.
Cilia morphology analysis of transition zone protein mutants. (A) Potential cilia defects were analyzed in xbx-7(tm2705), tza-1(tm2452), tza-2(ok2092), and nph-4(tm925) mutants by evaluating their ability of to absorb DiI into the ciliated sensory neurons. The ability of xbx-7(tm2705), tza-1(tm2452), tza-2(ok2092), and nph-4(tm925) mutants to absorb DiI was not affected compared with wild-type controls. However, in these mutant worms, the dye appeared to (arrowheads) concentrate in aggregates along the dendrites. This was (arrows) particularly evident in the nph-4(tm925) mutants and was not seen in the wild-type controls. N2 wild-type worms and daf-19(m86) mutants, which lack cilia, were used as positive and negative controls, respectively. (B) Cilia morphology was more directly analyzed in xbx-7(tm2705), tza-1(tm2452), tza-2(ok2092), and nph-4(tm925) mutants expressing the cilia marker CHE-13::YFP. (Top) head and (bottom) tail cilia morphology appeared overtly normal in these strains.
In agreement with the absence of cilia morphological defects, none of the B9 gene mutants exhibited any defects in other processes linked to proper cilia structure such as chemotaxis toward a variety of attractants, osmotic avoidance, and dauer formation (data not shown; Dusenbery et al., 1975; Culotti and Russell, 1978; Riddle et al., 1981; Perkins et al., 1986; Starich et al., 1995). Lifespan length and male mating behavior, additional processes dependent on cilia function in C. elegans, were not assayed in the B9 gene mutants (Liu and Sternberg, 1995; Apfeld and Kenyon, 1999).
At the level of analysis conducted here using the dye-filling assay and observing the expression and localization of an IFT protein in the mutant backgrounds of the tm2705, tm2452, and ok2092 alleles, cilia appear to not be significantly affected by disruption of the B9 proteins. These data are in contrast to the siRNA results generated for MKS1 in mammalian cells and the RNAi data generated for ICIS-1 in P. tetraurelia (Dawe et al., 2007b; Ponsard et al., 2007).
TZA-1 Is Required for XBX-7 and TZA-2 Localization at the Base of Cilia
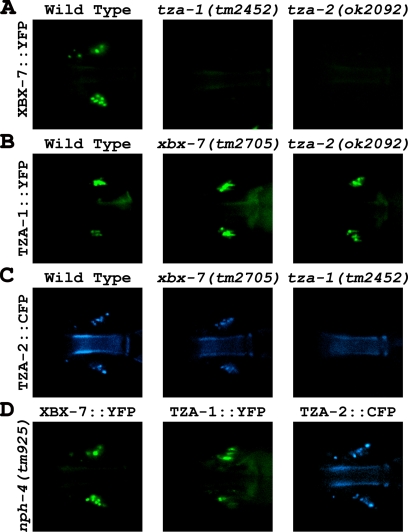
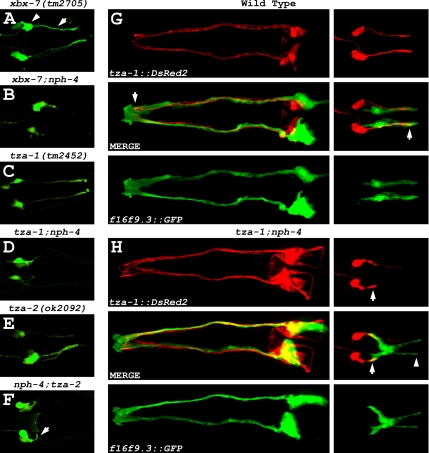
Previously, TZA-1 and TZA-2 were identified as putative binding partners using yeast two-hybrid analysis as part of a large-scale interactome study (Li et al., 2004), and together with our finding that all the B9 proteins colocalize to the transition zones (Figure 3), we hypothesized they function at the base of cilia as part of a protein complex. This possibility was explored using a genetic approach to assess whether a mutation in one B9 protein has an effect on the localization of another B9 protein at the transition zones. We crossed wild-type worms expressing either XBX-7::YFP, TZA-1::YFP, or TZA-2::YFP into the other B9 gene mutants (i.e., XBX-7::YFP into tza-1(tm2452) or tza-2(ok2092)). In both tza-1(tm2452) and tza-2(ok2092) mutants, XBX-7::YFP failed to localize to the transition zones (Figure 5A). In contrast, TZA-1::YFP localization at the base of cilia was not affected in the background of either xbx-7(tm2705) or tza-2(ok2092) (Figure 5B). Additionally, TZA-2::CFP was not present at the transition zone in tza-1(tm2452) mutants (Figure 5C).
Figure 5.
TZA-1 protein is required for proper localization of XBX-7 and TZA-2 to the transition zone. To analyze the effect of tza-1(tm2452), tza-2(ok2092), or nph-4(tm925) mutations on XBX-7::YFP localization, strains were generated by passing an extrachromosomal array encoding XBX-7::YFP from wild-type worms into the mutant backgrounds. Similarly, the TZA-1::YFP array was passed into xbx-7(tm2705), tza-2(ok2092), or nph-4(tm925) mutant background from wild-type, and the TZA-2::CFP array was passed into xbx-7(tm2705), tza-1(tm2452), or nph-4(tm925) mutant background from wild-type. (A) In contrast to (left) wild type, XBX-7::YFP was not detected at the transition zone in the (middle) tza-1(tm2452), or (right) tza-2(ok2092) mutants. (B) Compared with (left) wild type, TZA-1::YFP localization was unaltered in the background of (middle) xbx-7(tm2705) and (right) tza-2(ok2092) mutants. (C) TZA-2::CFP localization was not affected in (middle) xbx-7(tm2705) mutant background; however, it failed to localize correctly to the transition zone in (right) tza-1(tm2452) mutant background. (D) In the nph-4(tm925) mutant background, (left) XBX-7::YFP, (middle) TZA-1::YFP, and (right) TZA-2::CFP localization was not affected.
To examine the possibility XBX-7 is anchored at the transition zones by either TZA-1 or TZA-2, we used a yeast two-hybrid assay to test for direct protein-protein interactions. Although we were able to duplicate the interaction between TZA-1 and TZA-2 reported previously (Li et al., 2004), we observed no growth on selective media of yeast strains expressing both XBX-7 and either TZA-1 or TZA-2 (Supplementary Figure 3). This data raises the possibility that XBX-7 associates with the B9 protein complex by an additional unidentified factor.
Because the NPHP1 and NPHP4 homologues in C. elegans (NPH-1 and NPH-4, respectively) associate with each other at the base of cilia (Winkelbauer et al., 2005), we were interested in exploring the possibility that the B9 and NPH proteins function as part of the same complex. However, in nph-1(ok500) and nph-4(tm925) mutant strains expressing either XBX-7::YFP, TZA-1::YFP, or TZA-2::CFP, localization of the fluorescent reporters to the base of cilia was unaffected (Figure 5D; nph-1(ok500) data not shown). Likewise, xbx-7(tm2705), tza-1(tm2452), or tza-2(ok2092) mutant strains expressing NPH-1::CFP or NPH-4::YFP exhibited no delocalization of either reporter protein (Supplementary Figure 4). Together, these data indicate that the B9 proteins form part of a complex at the base of cilia independent of the function of the NPH proteins.
tza-1, tza-2, and nph-4 Mutants Exhibit Defects in Foraging Behavior
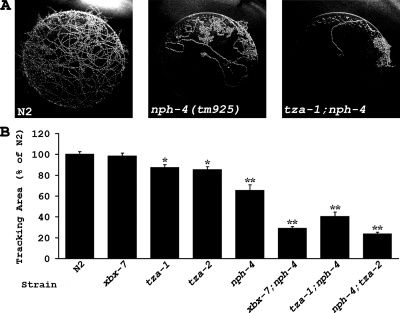
Given our finding that the B9 gene mutants did not have overt cilia morphology defects or exhibit abnormalities in chemotaxis or osmotic avoidance, we further assayed these worms for behavioral phenotypes associated with cilia dysfunction. Previous reports indicate that mutants lacking cilia display abnormal foraging behavior and exhibit a dwelling phenotype in the presence of food (Fujiwara et al., 2002; Murayama et al., 2005; Kobayashi et al., 2007). We tested the B9 gene mutants and nph-4(tm925) mutants in an 18-h tracking assay and found that tza-1(tm2452) and tza-2(ok2092) worms exhibited slight but significant defects in foraging behavior (Figure 6; Table 1). xbx-7(tm2705) worms were no different from the N2 wild-type controls. Interestingly, the nph-4(tm925) worms also displayed a foraging defect that was significantly more severe than that observed in the B9 gene mutants.
Figure 6.
tza-1, tza-2, and nph-4 mutants exhibit defects in foraging behavior. Single worms were allowed to roam freely for 18 h on a bacteria lawn of 3.0-cm diameter, and area covered by the worm was quantified by overlaying the lawn with a grid and counting the number of squares of the grid that contained tracks. (A) Representative tracking patterns of (left) N2 wild-type, (middle) nph-4(tm925), and (right) tza-1;nph-4 worms. Wild-type worms roamed across the entire area of the bacteria lawns, whereas nph-4(tm925) mutants restricted movement to a smaller portion of the lawn. Exploratory behavior in xbx-7;nph-4, tza-1;nph-4, and nph-4;tza-2 double mutants was even more limited. (B) Graph of quantified tracking data for N2 wild type, xbx-7(tm2705), tza-1(tm2452), tza-2(ok2092), nph-4(tm925), xbx-7;nph-4, tza-1;nph-4, and nph-4;tza-2. Data are given as percent area units covered compared with N2 (set to 100%). Error bars, SEM. *p < 0.005; **p < 0.0001, compared with N2.
Table 1.
Tracking assay statistics
| Genotype | na | N2a | SEMa | p values compared with |
||||
|---|---|---|---|---|---|---|---|---|
| N2 | xbx-7 | tza-1 | nph-4 | tza-2 | ||||
| Wild type | 28 | 100 | 2.82 | |||||
| xbx-7(tm2705) | 28 | 98.5 | 2.59 | 0.3547 | ||||
| tza-1(tm2452) | 29 | 87.2 | 3.01 | 0.0016* | ||||
| tza-2(ok2092) | 29 | 85.3 | 2.88 | 0.0003* | ||||
| nph-4(tm925) | 26 | 65.3 | 5.57 | <0.0001* | ||||
| xbx-7;nph-4 | 29 | 28.9 | 1.74 | <0.0001* | <0.0001* | <0.0001* | ||
| tza-1;nph-4 | 27 | 40.4 | 4.37 | <0.0001* | <0.0001* | 0.0005* | ||
| nph-4;tza-2 | 29 | 23.7 | 1.7 | <0.0001* | <0.0001* | 0.0006* | ||
a Values are mean %.
* p < 0.05 was deemed significant.
The fact that NPH and B9 proteins colocalize at the base of cilia and have homologues associated with human cystic kidney disorders (NPHP1, NPHP4, and MKS1) and that mutants show similar defects regarding foraging behavior together suggest there may be a functional connection between the NPH and B9 proteins. We addressed this possibility by generating xbx-7;nph-4, tza-1;nph-4, and nph-4;tza-2 double mutant worms and evaluated them for foraging activity. The results show that the three double mutant strains all had significantly more severe dwelling phenotypes than any of the single mutants alone (Figure 6B; Table 1). Furthermore, the severity of the double mutant phenotypes was similar to that seen in cilia mutants such as che-2, che-13, and osm-5 (Fujiwara et al., 2002; Kobayashi et al., 2007). Particularly surprising was the pronounced dwelling phenotype observed in xbx-7;nph-4 double mutant worms because the xbx-7(tm2705) single mutants alone were no different from wild-type controls. Together, these data suggest that the B9 proteins and NPH-4 may function in parallel at the transition zone to regulate foraging behavior.
Disruption of Both a B9 Protein and NPH-4 Causes Defects in Dye-Filling
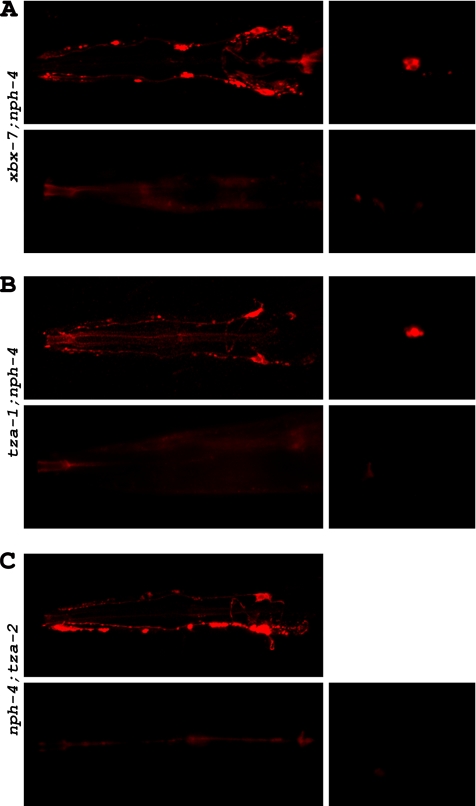
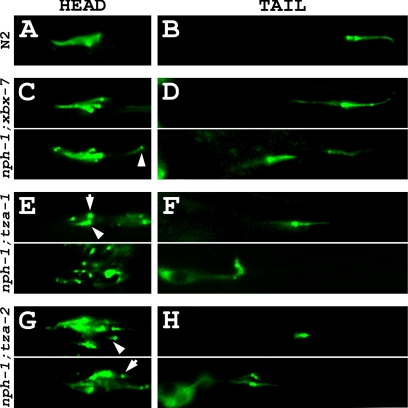
Our finding that worms with compound B9 gene;nph-4 mutations displayed foraging defects that were significantly more severe than the single mutants alone and were similar to that exhibited by cilia mutants raised the possibility that these double mutants had morphologically abnormal cilia. To begin addressing this question, we utilized the dye-filling assay. In contrast to the single mutants, all three B9 gene;nph-4 double mutants had defects in their ability to absorb DiI (Figure 7). Although some fluorescence was typically detected in the heads of xbx-7;nph-4 double mutants, there were far fewer amphid neurons able to uptake dye than expected (Figure 7A; Table 2). Additionally, only 52.7% of xbx-7;nph-4 double mutants examined were able to dye-fill the phasmid neurons. tza-1;nph-4 double mutants were almost completely defective in dye-filling in both the amphid and phasmid neurons; only 5.4 and 1.8% of the worms analyzed exhibited any dye-filling in the head and tail, respectively (Figure 7B; Table 2). Of those 5.4% that dye-filled in the head, none exhibited more than a few amphid neurons uptaking DiI. The nph-4;tza-2 double mutants exhibited dye-filling defects similar to those observed in tza-1;nph-4 double mutants (Figure 7C; Table 2). However, unlike the tza-1;nph-4 double mutants, none of the nph-4;tza-2 double mutants analyzed displayed dye-filling in the phasmid neurons. Together, these dye-filling data suggest that B9 gene;nph-4 double mutant worms either have gross cilia morphology defects, have defects in access of cilia to the external environment, or have defects in the process by which the dye is transported into the ciliated sensory endings.
Figure 7.
B9 gene;nph-4 double mutants exhibit dye-filling defects. Potential cilia abnormalities were analyzed in xbx-7;nph-4, tza-1;nph-4, and nph-4;tza-2 double mutants by evaluating their ability to absorb DiI into the ciliated sensory neurons. (A) xbx-7;nph-4 mutants displayed either (top) partial or (bottom) complete inability to dye-fill (left) amphid neurons and (right) phasmid neurons. (B) tza-1;nph-4 mutants displayed either (top) partial or (bottom) complete inability to dye-fill (left) amphid neurons and (right) phasmid neurons. (C) nph-4;tza-2 displayed either (top) partial or (bottom) complete inability to dye-fill (left) amphid neurons but were completely defective in dye-filling (right) phasmid neurons.
Table 2.
% dye-filling in mutant strains
| Genotype | Head | Tail | n |
|---|---|---|---|
| Wild type | 100 | 97.2 | 72 |
| xbx-7(tm2705) | 100 | 97.2 | 108 |
| tza-1(tm2452) | 100 | 98.6 | 71 |
| tza-2(ok2092) | 100 | 98.9 | 96 |
| nph-4(tm925) | 100 | 100 | 91 |
| xbx-7;nph-4 | 94.3 | 52.7 | 74 |
| tza-1;nph-4 | 5.4 | 1.8 | 112 |
| nph-4;tza-2 | 10.8 | 0 | 93 |
Cilia Morphology Is Altered in B9 Gene;nph Gene Double Mutants
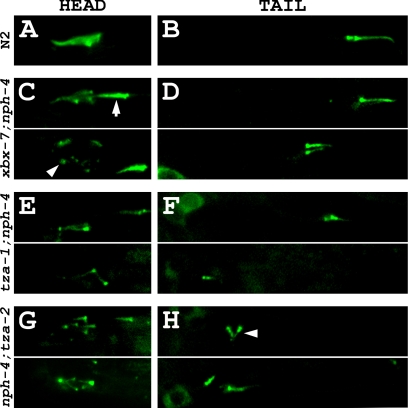
To explore the mechanism by which the B9 gene;nph-4 double mutants exhibit a synthetic dye-filling deficiency, we generated transgenic strains expressing CHE-13::YFP as a cilia marker in each of the three double mutant backgrounds. We observed that xbx-7;nph-4 double mutant worms, which exhibited a relatively mild dye-filling defect in comparison to tza-1;nph-4 and nph-4;tza-2 double mutants, possessed cilia but lacked properly organized amphid cilia bundles (Figure 8C). Interestingly, the disorganized amphid bundles consisted of a variable combination of normally positioned cilia, cilia localized posterior in relation to the rest of the bundle, and transition zones that sometimes altogether lacked cilia axonemes (Figure 8C). These worms also expressed significantly shortened phasmid cilia that were often positioned away from their expected location near the cuticle where the cilia normally are housed in the sheath/socket channel (Figure 8C; Table 3).
Figure 8.
B9 gene;nph-4 double mutants exhibit cilia morphology and positioning defects. Cilia were analyzed in xbx-7;nph-4, tza-1;nph-4, and nph-4;tza-2 double mutant worms expressing CHE-13:: YFP as a cilia/transition zone marker. (A) A representative amphid cilia bundle in the head of an N2 wild-type worm. (B) A representative pair of phasmid cilia in the tail of an N2 wild-type worm. These cilia are positioned near the cuticle of the worm where they are exposed to the external environment. See Figure 3A for schematic representation of the N2 wild-type amphid and phasmid cilia bundles. (C) Amphid cilia bundle morphology was altered in xbx-7;nph-4 double mutant background. In many of the xbx-7;nph-4 double mutants, some of the cilia were (arrow) positioned posterior to the rest of the bundle. In other xbx-7;nph-4 double mutants, cilia failed to extend off the (arrowhead) transition zones at the distal ends of the dendrites. (D) Phasmid cilia in xbx-7;nph-4 double mutants appeared (top) almost normal in some mutant worms but in others they were (bottom) shortened and mispositioned relative to wild type. (E) Amphid cilia bundle morphology was (top and bottom) grossly abnormal in tza-1;nph-4 double mutants. The head region contained few cilia axonemes and lacked typical bundle arrangement. (F) tza-1;nph-4 double mutants exhibited stunted phasmid cilia that were observed (top) slightly or (bottom) severely mispositioned. (G and H) nph-4;tza-2 double mutants exhibited ciliary defects similar to those in tza-1;nph-4 double mutants, including (arrowhead) laterally oriented phasmid cilia in comparison to wild type.
Table 3.
Phasmid cilia length statistics
| Genotype | n | Length (μM)a | p values compared with |
||||
|---|---|---|---|---|---|---|---|
| N2 | xbx-7 | tza-1 | nph-4 | tza-2 | |||
| Wild type | 33 | 7.04 ± 0.09 | |||||
| xbx-7(tm2705) | 27 | 7.03 ± 0.15 | 0.486 | ||||
| tza-1(tm2452) | 40 | 6.97 ± 0.13 | 0.3478 | ||||
| tza-2(ok2092) | 36 | 7.15 ± 0.09 | 0.1957 | ||||
| nph-4(tm925) | 24 | 6.87 ± 0.27 | 0.2766 | ||||
| xbx-7;nph-4 | 38 | 4.39 ± 0.24 | <0.0001* | <0.0001* | <0.0001* | ||
| tza-1;nph-4 | 33 | 2.88 ± 0.20 | <0.0001* | <0.0001* | <0.0001* | ||
| nph-4;tza-2 | 33 | 2.88 ± 0.15 | <0.0001* | <0.0001* | <0.0001* | ||
| nph-1;xbx-7 | 53 | 6.68 ± 0.17 | 0.0346* | 0.0644 | |||
| nph-1;tza-1 | 30 | 3.43 ± 0.26 | <0.0001* | <0.0001* | |||
| nph-1;tza-2 | 58 | 3.96 ± 0.23 | <0.0001* | <0.0001* | |||
a Values are mean ± SEM.
* p < 0.05 was deemed significant.
More severe ciliary defects were evident in tza-1;nph-4 and nph-4;tza-2 double mutant worms expressing the cilia marker CHE-13::YFP. The amphid cilia bundles of these worms failed to properly form, and the few cilia in the head that were observed appeared shortened, were sometimes misplaced, and were at times oriented in aberrant directions (Figure 8, E and G). Phasmid cilia of these double mutant worms were almost always shortened, mispositioned, and occasionally oriented laterally in comparison to wild type (Figure 8, F and H; Table 3).
Because NPH-4 is required for NPH-1 localization to the transition zones, we were interested in determining whether the ciliary defects observed in the B9 gene;nph-4 double mutants were due directly to compromised function of the mutant NPH-4 protein or were caused instead by the loss of NPH-1 from the transition zones. To address this question, we generated nph-1;xbx-7, nph-1;tza-1, and nph-1;tza-2 double mutant worms expressing the cilia marker CHE-13::YFP. Remarkably, these worms exhibited ciliary defects closely resembling those observed in the B9 gene;nph-4 double mutants (Figure 9). Much like xbx-7;nph-4 double mutants, nph-1;xbx-7 double mutants had the least severe ciliary defects of the three strains. Interestingly, the amphid cilia bundles of nph-1;xbx-7 double mutants were nearly indistinguishable from wild type, apart from rare instances of single cilia being positioned away from the bundle (Figure 9, A and C). However, more distinct defects were observed in the phasmid neurons which most often expressed cilia of normal length and positioning but occasionally appeared both shortened and mispositioned (Figure 9D; Table 3). Likewise, the phenotype of nph-1;tza-1 double mutants mimicked that of tza-1;nph-4 double mutants; fewer amphid cilia were observed and those present were disorganized and had shortened or no ciliary axonemes. (Figure 9E). In the phasmid neurons of the tail, cilia were both short and mispositioned relative to wild type (Figure 9F; Table 3). Interestingly, the phenotype of nph-1;tza-2 double mutants differed somewhat from nph-4;tza-2 double mutants; unlike nph-4;tza-2 worms, the amphid cilia of nph-1;tza-2 double mutants retained some semblance of organized bundles although shortened and mispositioned cilia were still observed (Figure 9G). Phasmid cilia in these worms exhibited typical defects observed in the other B9 gene;nph gene double mutants (Figure 9H; Table 3).
Figure 9.
B9 gene;nph-1 double mutants exhibit cilia morphology and positioning defects. Cilia were analyzed in nph-1;xbx-7, nph-1;tza-1, and nph-1;tza-2 double mutant worms expressing CHE-13::YFP as a cilia/transition zone marker. (A) A representative amphid cilia bundle in the head of an N2 wild-type worm. (B) A representative pair of phasmid cilia in the tail of an N2 wild-type worm. These cilia are positioned near the cuticle of the worm where they are exposed to the external environment. See Figure 3A for schematic representation of the N2 wild-type amphid and phasmid cilia bundles. (C) Amphid cilia bundle morphology was largely unaltered in nph-1;xbx-7 double mutant background. nph-1;xbx-7 double mutants occasionally exhibited cilia positioned (arrowhead) posterior to the rest of the bundle but otherwise resembled wild-type worms. (D) Phasmid cilia in nph-1;xbx-7 double mutants most often appeared (top) normal but in some cases were observed (bottom) shortened and mispositioned relative to wild type. (E) Amphid cilia bundle morphology was (top and bottom) grossly abnormal in nph-1;tza-1 double mutants. The head region contained (arrowhead) few cilia, consisted mostly of (arrow) transition zones lacking axonemes, and had no discernable amphid bundle arrangement. (F) nph-1;tza-1 double mutants exhibited stunted phasmid cilia that were observed (top) slightly or (bottom) severely mispositioned and often oriented laterally. (G) nph-1;tza-2 double mutants exhibited amphid ciliary defects more severe than nph-1;xbx-7 double mutants but less severe than nph-1;tza-1 double mutants. Although not perfectly arranged, (top and bottom) some bundling of multiple cilia was typically observed among (arrowhead) shortened and mispositioned cilia and (arrowhead) transition zones lacking axonemes. (H) In contrast to the relatively mild defects observed in the amphid bundles, the phenotype of phasmid cilia in nph-1;tza-2 double mutant worms mimicked the severity of defects seen in the phasmid cilia of nph-1;tza-1 double mutant worms.
Dendrites of Ciliated Sensory Neurons Are Malformed in B9 Gene;nph Gene Double Mutants But Associated Sheath Glia Are Intact
To further analyze the altered morphology of ciliated sensory neurons in B9 gene;nph gene double mutants, we generated transgenic mutant worms that expressed the transcriptional xbx-7::YFP fusion, which enabled us to visualize the dendrites of the neurons. In the xbx-7;nph-4 double mutants, the dendrites of the phasmid neurons in the tail were sometimes not properly extended compared with the phasmid dendrites of xbx-7(tm2705) single mutant worms (Figure 10, A and B). Predictably, tza-1;nph-4, and nph-4;tza-2 double mutants expressing transcriptional xbx-7::YFP fusion displayed severely shortened phasmid dendrites in comparison to tza-1(tm2452) and tza-2(ok2092) single mutant worms (Figure 10, C–F). These data are reflective of our previous observations in B9 gene;nph gene double mutants that phasmid cilia were positioned at random locations between the cell bodies of the neurons and the cuticle opening where the cilia are normally found.
Figure 10.
The dendrites of ciliated sensory neurons in B9 gene;nph gene double mutants are malformed although the surrounding sheath cells are intact. (A–F) Transgenic B9 gene single mutants and B9 gene;nph-4 double mutants were generated that expressed the xbx-7::YFP transcriptional fusion to visualize the ciliated sensory neurons. (A, C, and E) Expression of xbx-7::YFP revealed that the (arrow) phasmid dendrites are properly extended from the (arrowhead) cell bodies in the background of single B9 gene mutations. (B, D, and F) The phasmid dendrites of B9 gene;nph-4 double mutant worms were shortened at variable lengths and sometimes (arrow) misdirected. (G and H) Morphological analysis of (left) amphid and (right) phasmid sheath cells in wild-type and tza-1;nph-4 double mutant worms. Transgenic worms were generated that coexpressed the tza-1::DsRed2 transcriptional fusion to visualize the ciliated sensory neurons and the f16f9.3::GFP transcriptional fusion to visualize the associated sheath cells. (G) In wild-type worms, the amphid and phasmid sheath cells fully surrounded the dendrites of the ciliated sensory neurons at (arrows) their distal tips where the cilia are formed. (H) Sheath cell morphology was unaffected in tza-1;nph-4 double mutants. The amphid sheath cells were properly extended to surround the distal tips of the associated neurons. Likewise, phasmid sheath cells in the tail were appeared normally extended while the phasmid neuron dendrites were (arrows) shortened and clearly dissociated from (arrowhead) the distal ends of the sheath cells.
In wild-type C. elegans, the ciliated sensory neurons in both the head and tail of the worm are associated with sheath glia cells that surround the cilia at the distal tips of the dendrites. Immediately adjacent to the transition zones, the dendrites form physical attachments with the surrounding sheath cell. This point of attachment is a form of an adherens junction known as a belt junction due to its ring-like appearance (Perkins et al., 1986). To determine whether the B9 gene;nph gene double mutants had altered sheath cell morphology in addition to malformed dendrites of the ciliated sensory neurons, we generated transgenic wild-type and mutant worms that expressed the tza-1::DsRed2 transcriptional fusion as a ciliated sensory neuron marker and the f16f9.3::GFP transcriptional fusion, which specifically stains amphid and phasmid sheath cells (gift of Dr. S. Shaham, The Rockefeller University and Shaham, unpublished results). In both the head and tail of tza-1;nph-4 double mutant worms, sheath cell morphology was indistinguishable from wild type (Figure 10, G and H). Although the distal tips of the amphid neuron dendrites in tza-1;nph-4 double mutants were properly surrounded and could potentially still be physically attached, it was evident in the tails of the double mutants that the shortened phasmid neuron dendrites were no longer associated with the distal ends of the sheath cells. Together, these data suggest that the B9 and NPH proteins function redundantly as part of a mechanism required for dendrite patterning and attachment to sheath cells and for ciliogenesis.
DISCUSSION
Ciliopathies in humans include the phenotypes seen in syndromes such as MKS, NPHP, JBTS, and BBS where most of the genes identified encode proteins that localize to cilia or basal bodies. One of the common phenotypes associated with defects in cilia signaling is the formation of renal cysts. This is seen for the cilia-localized proteins polycystin 1 and polycystin 2, which are affected in PKD. Mutations in the PKD genes do not disrupt cilia structure but do result in the loss of a flow-regulated mechano-sensitive calcium signal, and in the case of polycystin-1 altered AP-1 pathway activation and increased mTOR activity (Low et al., 2006; Shillingford et al., 2006). Proteins involved in the cystic renal pathology in NPHP patients localize to the base of cilia; however, the roles of many of the NPHP proteins are largely unknown. Mutations in nph-1 and nph-4 in C. elegans cause defects in morphology of a subset of cilia on a few amphid neurons in the hermaphrodite and in sensory neurons that are specific to the male (Jauregui et al., 2008) and result in several phenotypes typically associated with disruption of cilia signaling pathways (Jauregui and Barr, 2005; Winkelbauer et al., 2005). It is uncertain whether these phenotypes are due to loss of a specific signaling pathway mediated by the NPH proteins or due to ultrastructural ciliary defects that may disrupt multiple pathways. In contrast to PKD- and NPHP-associated proteins, evidence suggests that MKS1 and mecklin, which are involved in renal cyst formation in MKS, affect cell polarization and centriole migration and severely disrupt cilia assembly, at least based on siRNA knockdown data (Dawe et al., 2007b). An intriguing finding that is emerging with regards to MKS, NPHP, and JBTS is that genes associated with one of these syndromes are being identified as responsible for the phenotypes in one or both of the other disorders. These data argue that the syndromes represent a spectrum of the same underlying defect and that the resulting phenotypes will be dictated by the nature of the mutation occurring in these genes. However, other than their colocalization to cilia or basal bodies, there is currently very little known about how proteins associated with MKS, NPHP, or JBTS are functionally related or how mutations in one of the genes may affect the localization and/or activity of another.
To begin addressing these questions, we are using C. elegans as a model system to elucidate the functional relationships between cilia and proteins associated with human cystic kidney disease syndromes. Here we analyzed the connection between the MKS1 homolog XBX-7 and its related B9 protein family members and the NPHP1 and NPHP4 homologues, NPH-1 and NPH-4. Intriguingly, we demonstrate that all three B9 genes in C. elegans are coregulated by the same transcription factor in ciliated sensory neurons and that the B9 proteins localize to the transition zones at the base of the sensory cilia. Similar localization to basal bodies has been reported for the XBX-7 homolog in mammalian cells, suggesting conserved function (Dawe et al., 2007b). However, siRNA-mediated knockdown of MKS1 expression in mammalian cells or RNAi knockdown of the TZA-1 homolog in P. tetraurelia suggest these proteins are required for cilia assembly or formation (Dawe et al., 2007b; Ponsard et al., 2007). Additionally, during review of this manuscript a conditional mutant in the tza-1 mouse homolog was described. These mutant mice had absent or truncated cilia and systemic phenotypes typically associated with disruption of cilia in mammals (Town et al., 2008). In contrast, our analysis of single, double, or triple mutations in the B9 genes in C. elegans did not reveal cilia morphology defects unless these mutations were combined with mutations in either nph-1 or nph-4. It is plausible that the B9 gene single mutants exhibit ultrastructural abnormalities in cilia or subsets of cilia that would not be detectable by the methods used in this analysis. Alternatively, the discrepancy could be due in part to the nature of the alleles we analyzed. xbx-7(tm2705) and tza-1(tm2452) both result in internal deletions that do not cause frame shifts. Thus, the proteins encoded by xbx-7(tm2705) and tza-1(tm2452) may retain partial function with respect to their potential roles in ciliogenesis. In the case of the xbx-7(tm2705) mutants, this could explain the consistently milder phenotype relative to that seen in the other B9 gene mutants when crossed with an nph gene mutant. Although the tza-1(tm2452) mutation is an internal in-frame deletion, we show that this mutation results in loss of both XBX-7 and TZA-2 from the transition zones, which would thus be expected to acerbate the phenotype as was observed. In contrast, the tza-2(ok2092) allele results in deletion of most of the protein including the B9 domain and is thought to represent a null mutation.
Our genetic analyses in C. elegans indicate that the B9 proteins form part of a complex at the transition zone. This is supported by data showing that XBX-7 localization was dependent on the presence of both TZA-1 and TZA-2, and TZA-2 required only TZA-1 for proper localization. In contrast, disruption of TZA-2 or XBX-7 did not alter localization of TZA-1. We interpret these data to indicate that TZA-1 functions early in the assembly of a complex containing the B9 proteins followed by TZA-2 and subsequently by XBX-7. It is unknown whether the loss of fluorescent signal from the transition zones was due to the unincorporated proteins being diffused throughout the neurons or alternatively due to destabilization and degradation of these proteins. However, using a yeast two-hybrid assay, we observed a positive interaction between TZA-1 and TZA-2, suggesting at least in the case of TZA-2 that the protein was mislocalized due to failure in assembly into the complex at the transition zone. In contrast, we did not observe a physical interaction between XBX-7 and the other B9 proteins, suggesting that there is an unknown factor that is responsible for anchoring XBX-7 in the complex. Whether there is a similar hierarchy of complex assembly with regards to the mammalian proteins remains to be determined.
Because the NPHP protein homologues NPH-1 and NPH-4 are part of a complex that localizes similarly to the B9 proteins at the transition zone (Winkelbauer et al., 2005), and mutations in the NPHP proteins result in cystic renal disease in humans and mice, we explored the possibility that the C. elegans B9 and NPH proteins were functionally related or perhaps components of the same complex. However, our analysis of the localization of NPH-1 and NPH-4 in the B9 gene mutants revealed there were no effects on their localization to the transition zones nor were the B9 proteins affected by mutations in nph-1 or nph-4. Thus, there does not appear to be a direct link between the B9 proteins and the NPH proteins with regards to complex formation at the base of cilia.
Cilia mutants in C. elegans exhibit numerous behavioral and sensory defects including abnormalities in chemotaxis, osmotic avoidance deficiencies, altered dauer formation, and a marked reduction in foraging activity in the presence of food. In the B9 gene mutants, we did not observe defects in osmotic avoidance, chemotaxis, or dauer formation, in agreement with the presence of overtly normal cilia morphology. However, both the tza-1(tm2452) and tza-2(ok2092) mutants did exhibit subtle yet significant reductions in foraging behavior, which was not evident in the xbx-7(tm2705) mutants. Interestingly, we also found that nph-4(tm925) mutants had defects in foraging activity that was significantly more severe than either tza-1(tm2452) or tza-2(ok2092) mutants. These observations led us to evaluate whether the foraging behavior defects in the B9 gene mutants were due to disruption of the same pathway affected in the nph-4(tm925) mutants. We analyzed this in B9 gene;nph-4 double mutants and found that mutations affecting both the B9 proteins and NPH-4 resulted in a markedly more severe dwelling phenotype than either mutation alone. This phenotype was similar to that seen in worms completely lacking cilia, suggesting redundant functions of the B9 proteins and NPH-4 at the base of cilia.
The B9 gene;nph-4 double mutants were also unable to normally dye-fill. This was due to abnormalities in bundling, orientation, or length of the cilia along with gross cilia positioning defects caused by dendrite malformation. Additionally, we observed the same altered morphology in B9 gene;nph-1 double mutants, suggesting that because NPH-4 is critical for the proper localization of NPH-1 to the transition zones, the defects observed in the B9 gene;nph-4 double mutants can be attributed to the loss of NPH-1. The complex phenotype observed in these worms is unique among mutants defective in dye-filling. daf-19 mutant worms exhibit phasmid neuron dendrites of varying lengths similar to those seen in the B9 gene;nph gene double mutants, but daf-19 mutants lack all ciliary and transition zone structures (Swoboda et al., 2000). Notably, centrioles positioned at the distal tips of the dendrites are detected by electron microscopy in daf-19 mutants (Perkins et al., 1986). Because each of the B9 and nephrocystin genes are strongly regulated by the DAF-19 transcription factor and are therefore not expressed in daf-19 mutant worms, it seems likely that the B9 proteins and nephrocystins will not be critical for positioning the centrioles at the distal tips of the dendrites. Disruption of the forkhead domain transcription factor gene fkh-2 results in shortened dendrites and cilia morphology defects, but this phenotype is specific to AWB neurons alone (Mukhopadhyay et al., 2007). B9 gene;nph gene double mutants may most closely resemble mec-8 mutants in which amphid cilia fail to fully penetrate the sheath glia, are sometimes misoriented laterally, and do not fasciculate at the sheath/socket channel (Perkins et al., 1986). However, gross dendrite morphology defects have not been reported in mec-8 mutants. Although we observed grossly normal sheath cells in B9 gene;nph gene double mutants, we have not directly determined whether these cells cooperate with the socket cells to properly form the channels through which the cilia normally project. It is likely, however, that the channels do properly form since the B9 gene;nph gene mutants are still able to uptake DiI to some extent, indicating that on occasion some cilia are correctly formed and exposed to the external environment.
The additive effects seen in the B9 gene;nph gene double mutants indicate that the B9 proteins and NPH proteins likely function redundantly to coordinate how cilia and dendrites will form. It is possible that the phenotypes we see in these double mutants are related to those observed when the B9 proteins are singularly disrupted in other biological systems. Together, our findings provide insights into the allelic nature of the genes that can be involved in multiple syndromes and more importantly indicate that the mammalian homologues of the B9 domain proteins TZA-1 and TZA-2 are strong candidates as loci involved in the pathology of MKS, NPHP, or JBTS patients for which the underlying genetic defects have not yet been identified.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Drs. M. Barr, C. Haycraft, N. Katsanis, and M. Leroux for valuable discussions and critical reading of the manuscript. We thank A. Tousson of the UAB Imaging Facility and J. Lehman for assistance in imaging. We thank J. Davenport for assistance in statistical analysis. We thank M. Croyle and V. Roper for technical assistance. The C. elegans Genome Sequencing Consortium provided sequence information, and the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health, provided some of the C. elegans strains used in this study. We thank the C. elegans Knockout Consortium and the National BioResource Project in Japan for the xbx-7(tm2705), tza-1(tm2452), and tza-2(ok2092) deletion mutants. This work was supported by National Institutes of Health R01 DK65655 to B.K.Y. and sponsored by the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory (ORNL), managed by UT-Battelle, LLC, for the U.S. Department of Energy under Contract No. DE-AC05-00OR22725 to E.J.M. Additional support was provided by the P30 DK074038 UAB Recessive Polycystic Kidney Disease Research and Translational Core Center.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-10-1070) on March 12, 2008.
REFERENCES
- Alexiev B. A., Lin X., Sun C. C., Brenner D. S. Meckel-Gruber syndrome: pathologic manifestations, minimal diagnostic criteria, and differential diagnosis. Arch. Pathol. Lab. Med. 2006;130:1236–1238. doi: 10.5858/2006-130-1236-MS. [DOI] [PubMed] [Google Scholar]
- Apfeld J., Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402:804–809. doi: 10.1038/45544. [DOI] [PubMed] [Google Scholar]
- Arts H. H., et al. Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat. Genet. 2007;39:882–888. doi: 10.1038/ng2069. [DOI] [PubMed] [Google Scholar]
- Attanasio M., et al. Loss of GLIS2 causes nephronophthisis in humans and mice by increased apoptosis and fibrosis. Nat. Genet. 2007;39:1018–1024. doi: 10.1038/ng2072. [DOI] [PubMed] [Google Scholar]
- Baala L., et al. Pleiotropic effects of CEP290 (NPHP6) mutations extend to Meckel syndrome. Am. J. Hum. Genet. 2007a;81:170–179. doi: 10.1086/519494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baala L., et al. The Meckel-Gruber syndrome gene, MKS3, is mutated in Joubert syndrome. Am. J. Hum. Genet. 2007b;80:186–194. doi: 10.1086/510499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano J. L., Mitsuma N., Beales P. L., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics. Hum. Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Barr M. M., DeModena J., Braun D., Nguyen C. Q., Hall D. H., Sternberg P. W. The Caenorhabditis elegans autosomal dominant polycystic kidney disease gene homologs lov-1 and pkd-2 act in the same pathway. Curr. Biol. 2001;11:1341–1346. doi: 10.1016/s0960-9822(01)00423-7. [DOI] [PubMed] [Google Scholar]
- Beales P. L., et al. IFT80, which encodes a conserved intraflagellar transport protein, is mutated in Jeune asphyxiating thoracic dystrophy. Nat. Genet. 2007;39:727–729. doi: 10.1038/ng2038. [DOI] [PubMed] [Google Scholar]
- Blacque O. E., Leroux M. R. Bardet-Biedl syndrome: an emerging pathomechanism of intracellular transport. Cell Mol. Life Sci. 2006;63:2145–2161. doi: 10.1007/s00018-006-6180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque O. E., et al. Functional genomics of the cilium, a sensory organelle. Curr. Biol. 2005;15:935–941. doi: 10.1016/j.cub.2005.04.059. [DOI] [PubMed] [Google Scholar]
- Blacque O. E., et al. Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev. 2004;18:1630–1642. doi: 10.1101/gad.1194004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. P. Meckel syndrome: genetics, perinatal findings, and differential diagnosis. Taiwan J. Obstet. Gynecol. 2007;46:9–14. doi: 10.1016/S1028-4559(08)60100-X. [DOI] [PubMed] [Google Scholar]
- Cole D. G., Diener D. R., Himelblau A. L., Beech P. L., Fuster J. C., Rosenbaum J. L. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet J., Spike C. A., Lundquist E. A., Shaw J. E., Herman R. K. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics. 1998;148:187–200. doi: 10.1093/genetics/148.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culotti J. G., Russell R. L. Osmotic avoidance defective mutants of the nematode Caenorhabditis elegans. Genetics. 1978;90:243–256. doi: 10.1093/genetics/90.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport J. R., Watts A. J., Roper V. C., Croyle M. J., van Groen T., Wyss J. M., Nagy T. R., Kesterson R. A., Yoder B. K. Disruption of intraflagellar transport in adult mice leads to obesity and slow-onset cystic kidney disease. Curr. Biol. 2007;17:1586–1594. doi: 10.1016/j.cub.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe H. R., Farr H., Gull K. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J. Cell Sci. 2007a;120:7–15. doi: 10.1242/jcs.03305. [DOI] [PubMed] [Google Scholar]
- Dawe H. R., et al. The Meckel-Gruber Syndrome proteins MKS1 and meckelin interact and are required for primary cilium formation. Hum. Mol. Genet. 2007b;16:173–186. doi: 10.1093/hmg/ddl459. [DOI] [PubMed] [Google Scholar]
- Deane J. A., Cole D. G., Seeley E. S., Diener D. R., Rosenbaum J. L. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr. Biol. 2001;11:1586–1590. doi: 10.1016/s0960-9822(01)00484-5. [DOI] [PubMed] [Google Scholar]
- Delous M., et al. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat. Genet. 2007;39:875–881. doi: 10.1038/ng2039. [DOI] [PubMed] [Google Scholar]
- Dusenbery D. B., Sheridan R. E., Russell R. L. Chemotaxis-defective mutants of the nematode Caenorhabditis elegans. Genetics. 1975;80:297–309. doi: 10.1093/genetics/80.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimenko E., Bubb K., Mak H. Y., Holzman T., Leroux M. R., Ruvkun G., Thomas J. H., Swoboda P. Analysis of xbx genes in C. elegans. Development. 2005;132:1923–1934. doi: 10.1242/dev.01775. [DOI] [PubMed] [Google Scholar]
- Fliegauf M., et al. Nephrocystin specifically localizes to the transition zone of renal and respiratory cilia and photoreceptor connecting cilia. J. Am. Soc. Nephrol. 2006;17:2424–2433. doi: 10.1681/ASN.2005121351. [DOI] [PubMed] [Google Scholar]
- Fujiwara M., Sengupta P., McIntire S. L. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron. 2002;36:1091–1102. doi: 10.1016/s0896-6273(02)01093-0. [DOI] [PubMed] [Google Scholar]
- Haycraft C. J., Banizs B., Aydin-Son Y., Zhang Q., Michaud E. J., Yoder B. K. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft C. J., Schafer J. C., Zhang Q., Taulman P. D., Yoder B. K. Identification of CHE-13, a novel intraflagellar transport protein required for cilia formation. Exp. Cell Res. 2003;284:251–263. doi: 10.1016/s0014-4827(02)00089-7. [DOI] [PubMed] [Google Scholar]
- Haycraft C. J., Swoboda P., Taulman P. D., Thomas J. H., Yoder B. K. The C. elegans homolog of the murine cystic kidney disease gene Tg737 functions in a ciliogenic pathway and is disrupted in osm-5 mutant worms. Development. 2001;128:1493–1505. doi: 10.1242/dev.128.9.1493. [DOI] [PubMed] [Google Scholar]
- Hildebrandt F., Otto E. Cilia and centrosomes: a unifying pathogenic concept for cystic kidney disease? Nat. Rev. Genet. 2005;6:928–940. doi: 10.1038/nrg1727. [DOI] [PubMed] [Google Scholar]
- Hildebrandt F., Zhou W. Nephronophthisis-associated ciliopathies. J. Am. Soc. Nephrol. 2007;18:1855–1871. doi: 10.1681/ASN.2006121344. [DOI] [PubMed] [Google Scholar]
- Jauregui A. R., Barr M. M. Functional characterization of the C. elegans nephrocystins NPHP-1 and NPHP-4 and their role in cilia and male sensory behaviors. Exp. Cell Res. 2005;305:333–342. doi: 10.1016/j.yexcr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Jauregui A. R., Nguyen K.C.Q., Hall D. H., Barr M. M. The C. elegans nephrocystins act as global modifiers of cilium structure. J. Cell Biol. 2008;180:973–988. doi: 10.1083/jcb.200707090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaddour R., et al. Spectrum of MKS1 and MKS3 mutations in Meckel syndrome: a genotype-phenotype correlation. Mutation in brief #960. Online. Hum. Mutat. 2007;28:523–524. doi: 10.1002/humu.9489. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Gengyo-Ando K., Ishihara T., Katsura I., Mitani S. IFT-81 and IFT-74 are required for intraflagellar transport in C. elegans. Genes Cells. 2007;12:593–602. doi: 10.1111/j.1365-2443.2007.01076.x. [DOI] [PubMed] [Google Scholar]
- Kozminski K. G., Johnson K. A., Forscher P., Rosenbaum J. L. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K. S., Sternberg P. W. Sensory regulation of male mating behavior in Caenorhabditis elegans. Neuron. 1995;14:79–89. doi: 10.1016/0896-6273(95)90242-2. [DOI] [PubMed] [Google Scholar]
- Low S. H., Vasanth S., Larson C. H., Mukherjee S., Sharma N., Kinter M. T., Kane M. E., Obara T., Weimbs T. Polycystin-1, STAT6, and P100 function in a pathway that transduces ciliary mechanosensation and is activated in polycystic kidney disease. Dev. Cell. 2006;10:57–69. doi: 10.1016/j.devcel.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Marshall W. F. What is the function of centrioles? J. Cell Biochem. 2007;100:916–922. doi: 10.1002/jcb.21117. [DOI] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet G., et al. The gene mutated in juvenile nephronophthisis type 4 encodes a novel protein that interacts with nephrocystin. Nat. Genet. 2002;32:300–305. doi: 10.1038/ng996. [DOI] [PubMed] [Google Scholar]
- Mollet G., Silbermann F., Delous M., Salomon R., Antignac C., Saunier S. Characterization of the nephrocystin/nephrocystin-4 complex and subcellular localization of nephrocystin-4 to primary cilia and centrosomes. Hum. Mol. Genet. 2005;14:645–656. doi: 10.1093/hmg/ddi061. [DOI] [PubMed] [Google Scholar]
- Morgan D., Eley L., Sayer J., Strachan T., Yates L. M., Craighead A. S., Goodship J. A. Expression analyses and interaction with the anaphase promoting complex protein Apc2 suggest a role for inversin in primary cilia and involvement in the cell cycle. Hum. Mol. Genet. 2002;11:3345–3350. doi: 10.1093/hmg/11.26.3345. [DOI] [PubMed] [Google Scholar]
- Moyer J. H., Lee-Tischler M. J., Kwon H. Y., Schrick J. J., Avner E. D., Sweeney W. E., Godfrey V. L., Cacheiro N. L., Wilkinson J. E., Woychik R. P. Candidate gene associated with a mutation causing recessive polycystic kidney disease in mice. Science. 1994;264:1329–1333. doi: 10.1126/science.8191288. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S., Lu Y., Qin H., Lanjuin A., Shaham S., Sengupta P. Distinct IFT mechanisms contribute to the generation of ciliary structural diversity in C. elegans. EMBO J. 2007;26:2966–2980. doi: 10.1038/sj.emboj.7601717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama T., Toh Y., Ohshima Y., Koga M. The dyf-3 gene encodes a novel protein required for sensory cilium formation in Caenorhabditis elegans. J. Mol. Biol. 2005;346:677–687. doi: 10.1016/j.jmb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Murcia N. S., Richards W. G., Yoder B. K., Mucenski M. L., Dunlap J. R., Woychik R. P. The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left-right axis determination. Development. 2000;127:2347–2355. doi: 10.1242/dev.127.11.2347. [DOI] [PubMed] [Google Scholar]
- Orozco J. T., Wedaman K. P., Signor D., Brown H., Rose L., Scholey J. M. Movement of motor and cargo along cilia. Nature. 1999;398:674. doi: 10.1038/19448. [DOI] [PubMed] [Google Scholar]
- Otto E., et al. A gene mutated in nephronophthisis and retinitis pigmentosa encodes a novel protein, nephroretinin, conserved in evolution. Am. J. Hum. Genet. 2002;71:1161–1167. doi: 10.1086/344395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto E. A., et al. Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat. Genet. 2003;34:413–420. doi: 10.1038/ng1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto E. A., et al. Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nat. Genet. 2005;37:282–288. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- Ou G., Koga M., Blacque O. E., Murayama T., Ohshima Y., Schafer J. C., Li C., Yoder B. K., Leroux M. R., Scholey J. M. Sensory ciliogenesis in Caenorhabditis elegans: assignment of IFT components into distinct modules based on transport and phenotypic profiles. Mol. Biol. Cell. 2007;18:1554–1569. doi: 10.1091/mbc.E06-09-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M. A., Bennett C. L., Eckert M. L., Dobyns W. B., Gleeson J. G., Shaw D. W., McDonald R., Eddy A., Chance P. F., Glass I. A. The NPHP1 gene deletion associated with juvenile nephronophthisis is present in a subset of individuals with Joubert syndrome. Am. J. Hum. Genet. 2004;75:82–91. doi: 10.1086/421846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Dickert B. L., Vucica Y., Seeley E. S., Rosenbaum J. L., Witman G. B., Cole D. G. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene Tg737, are required for assembly of cilia and flagella. J. Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L. A., Hedgecock E. M., Thomson J. N., Culotti J. G. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Piperno G., Siuda E., Henderson S., Segil M., Vaananen H., Sassaroli M. Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J. Cell Biol. 1998;143:1591–1601. doi: 10.1083/jcb.143.6.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponsard C., Skowron-Zwarg M., Seltzer V., Perret E., Gallinger J., Fisch C., Dupuis-Williams P., Caruso N., Middendorp S., Tournier F. Identification of ICIS-1, a new protein involved in cilia stability. Front. Biosci. 2007;12:1661–1669. doi: 10.2741/2178. [DOI] [PubMed] [Google Scholar]
- Porter M. E., Bower R., Knott J. A., Byrd P., Dentler W. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell. 1999;10:693–712. doi: 10.1091/mbc.10.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle D. L., Swanson M. M., Albert P. S. Interacting genes in nematode dauer larva formation. Nature. 1981;290:668–671. doi: 10.1038/290668a0. [DOI] [PubMed] [Google Scholar]
- Roepman R., Letteboer S. J., Arts H. H., van Beersum S. E., Lu X., Krieger E., Ferreira P. A., Cremers F. P. Interaction of nephrocystin-4 and RPGRIP1 is disrupted by nephronophthisis or Leber congenital amaurosis-associated mutations. Proc. Natl. Acad. Sci. USA. 2005;102:18520–18525. doi: 10.1073/pnas.0505774102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J., Witman G. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sayer J. A., et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat. Genet. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- Schafer J. C., Haycraft C. J., Thomas J. H., Yoder B. K., Swoboda P. XBX-1 encodes a dynein light intermediate chain required for retrograde intraflagellar transport and cilia assembly in Caenorhabditis elegans. Mol. Biol. Cell. 2003;14:2057–2070. doi: 10.1091/mbc.E02-10-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey J. M., Ou G., Snow J., Gunnarson A. Intraflagellar transport motors in Caenorhabditis elegans neurons. Biochem. Soc. Trans. 2004;32:682–684. doi: 10.1042/BST0320682. [DOI] [PubMed] [Google Scholar]
- Shillingford J. M., et al. The mTOR pathway is regulated by polycystin-1, and its inhibition reverses renal cystogenesis in polycystic kidney disease. Proc. Natl. Acad. Sci. USA. 2006;103:5466–5471. doi: 10.1073/pnas.0509694103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith U. M., et al. The transmembrane protein meckelin (MKS3) is mutated in Meckel-Gruber syndrome and the wpk rat. Nat. Genet. 2006;38:191–196. doi: 10.1038/ng1713. [DOI] [PubMed] [Google Scholar]
- Starich T. A., Herman R. K., Kari C. K., Yeh W. H., Schackwitz W. S., Schuyler M. W., Collet J., Thomas J. H., Riddle D. L. Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics. 1995;139:171–188. doi: 10.1093/genetics/139.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda P., Adler H. T., Thomas J. H. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol. Cell. 2000;5:411–421. doi: 10.1016/s1097-2765(00)80436-0. [DOI] [PubMed] [Google Scholar]
- Taulman P. D., Haycraft C. J., Balkovetz D. F., Yoder B. K. Polaris, a protein involved in left-right axis patterning, localizes to basal bodies and cilia. Mol. Biol. Cell. 2001;12:589–599. doi: 10.1091/mbc.12.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Town T., et al. The stumpy gene is required for mammalian ciliogenesis. Proc. Natl. Acad. Sci. USA. 2008;105:2853–2858. doi: 10.1073/pnas.0712385105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Pan J., Snell W. J. Intraflagellar transport particles participate directly in cilium-generated signaling in Chlamydomonas. Cell. 2006;125:549–562. doi: 10.1016/j.cell.2006.02.044. [DOI] [PubMed] [Google Scholar]
- Ward S., Thomson N., White J. G., Brenner S. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans. J. Comp. Neurol. 1975;160:313–337. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- Ware R. W., Clark D., Crossland K., Russell R. L. The nerve ring of the nematode Caenorhabditis elegans: sensory input and motor out. J. Comp. Neurol. 1975;162:71–110. [Google Scholar]
- Winkelbauer M. E., Schafer J. C., Haycraft C. J., Swoboda P., Yoder B. K. The C. elegans homologs of nephrocystin-1 and nephrocystin-4 are cilia transition zone proteins involved in chemosensory perception. J. Cell Sci. 2005;118:5575–5587. doi: 10.1242/jcs.02665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.