Abstract
Phosphatidylinositol 4-phosphate (PI(4)P) is a key regulator of membrane transport required for the formation of transport carriers from the trans-Golgi network (TGN). The molecular mechanisms of PI(4)P signaling in this process are still poorly understood. In a search for PI(4)P effector molecules, we performed a screen for synthetic lethals in a background of reduced PI(4)P and found the gene GGA2. Our analysis uncovered a PI(4)P-dependent recruitment of the clathrin adaptor Gga2p to the TGN during Golgi-to-endosome trafficking. Gga2p recruitment to liposomes is stimulated both by PI(4)P and the small GTPase Arf1p in its active conformation, implicating these two molecules in the recruitment of Gga2p to the TGN, which ultimately controls the formation of clathrin-coated vesicles. PI(4)P binding occurs through a phosphoinositide-binding signature within the N-terminal VHS domain of Gga2p resembling a motif found in other clathrin interacting proteins. These data provide an explanation for the TGN-specific membrane recruitment of Gga2p.
INTRODUCTION
Phosphoinositides (PIs) are signaling molecules regulating key processes in eukaryotic cells including signal transduction, cytoskeleton organization, and membrane transport. Spatially restricted activity of PI-kinases and -phosphatases results in rapid turnover of different PI species. The resulting localized exposure of the corresponding PI headgroups allows for the generation of microdomains on membranes that then serve as sites for the assembly of membrane transport machinery, e.g., coats and their adaptors (De Matteis et al., 2005).
The spatially restricted production of phosphatidylinositol 4-phosphate (PI(4)P) at the Golgi complex is catalyzed by Pik1p in yeast (Hama et al., 1999; Walch-Solimena and Novick, 1999; Audhya et al., 2000), a type III PI 4-kinase conserved up to mammals (Godi et al., 1999; De Matteis et al., 2005). The pool of PI(4)P generated by Pik1p is required for normal Golgi morphology and for membrane transport from the trans-Golgi network (TGN; De Matteis et al., 2005). Like other Golgi mutants, pik1 mutants showed an accumulation of abnormal, ring-like Golgi structures called Berkeley bodies (Hama et al., 1999; Walch-Solimena and Novick, 1999; Audhya et al., 2000). The pik1 mutant analysis uncovered a role of PI(4)P in the formation of TGN vesicles for exocytosis of cargo such as invertase and TGN-to-vacuole transport of cargo like the vacuolar hydrolase carboxypeptidase Y (CPY; Hama et al., 1999; Walch-Solimena and Novick, 1999; Audhya et al., 2000).
Indeed, the TGN represents a major hub within the secretory pathway for the sorting of newly synthesized cargo into the vacuolar protein-sorting (Vps) pathway and for exocytosis. Recent evidence suggests specialized routes for cell surface delivery for the major plasma membrane ATPase Pma1p as well as other plasma membrane and cell wall components as opposed to invertase, which is transported in Vps-dependent secretory vesicles (SVs; Gurunathan et al., 2002; Harsay and Schekman, 2002). The molecular mechanisms underlying protein sorting at the TGN into these distinct exocytosis routes are currently not well understood. In addition, downstream of the TGN-sorting event, the machinery mediating cell surface delivery of proteins traveling via endosomes (e.g., nutrient permeases) is not known. Because PI(4)P affects both vacuolar and cell surface transport identification of PI(4)P effectors is likely to provide an entry point to the dissection of the specific molecular machineries regulating vesicle formation from the TGN.
The first identified effectors of PI(4)P at the Golgi in yeast belong to the family of oxysterol-binding protein (OSBP)-related proteins. The OSBP Kes1p/Osh4p is recruited to the Golgi depending on catalytically active Pik1p (Li et al., 2002) and regulates sterol sensing and transport (Raychaudhuri et al., 2006). Also, the OSBP-related proteins Osh1p/Osh2p require PI(4)P for Golgi association (Levine and Munro, 2001; Roy and Levine, 2004; Yu et al., 2004).
Although clathrin coat complexes in TGN-to-endosome transport have been extensively characterized, possible coats for secretory vesicles remain obscure. GGAs (Golgi-associated, γ-ear containing, ARF-binding proteins) are monomeric clathrin adaptors that regulate transport of TGN cargo destined to vacuoles/lysosomes both in yeast and in mammals. GGAs bind to GTP-bound, active Arf, ubiquitin, cargo containing acidic amino-acid cluster-dileucine motifs and clathrin (Boman, 2001; Bonifacino, 2004; Ghosh and Kornfeld, 2004; Robinson, 2004; Scott et al., 2004). The contribution of Arf-GTP interaction with GGAs to membrane recruitment has remained controversial. Although yeast gga mutants defective in Arf binding have been reported to retain the ability to associate with membranes, such mutants in mammals were found to be predominantly cytosolic (Boman et al., 2002; Takatsu et al., 2002). Because Arf acts at multiple stages of membrane transport, additional determinants are expected to play a role in the specific recruitment of GGAs to the TGN.
In this study, we used synthetic lethal analysis to search for new PI(4)P targets. Our screen resulted in the isolation of Gga2p as new PI(4)P target and uncovered a role of PI(4)P together with Gga2p in a common biosynthetic transport pathway from the TGN for cargo en route to either the vacuole or the cell surface.
MATERIALS AND METHODS
Strains and Media
The genotypes of Saccharomyces cerevisiae strains used in this study are listed in Table S4. Yeast were cultured in YPD, SC drop-out media, or SD minimal media supplemented with the necessary amino acids. Yeasts were transformed using a lithium acetate–based method (Schiestl and Gietz, 1989). Yeast strains were either constructed by tetrad dissection of sporulated diploids or by integration of indicated tagging or disruption cassettes as described previously (Longtine et al., 1998). Disruptions were verified by PCR. Genomic tagging has been confirmed either by Western blotting (VPS10-3xHA) or visually by fluorescence microscopy (SEC7-DsRED).
To create a pik1-101 vps28Δ double mutant, the VPS28 deletion cassette including homologous regions was amplified using genomic DNA of CSY902 as template. The integration cassette was then transformed into CSY544. Yeast strains expressing VPS10-3xHA were constructed by transformation of a PCR-amplified genomic integration construct using pFA6a-3xHA-kanMX6 (CSY210, CSY392) or pFA6a-3xHA-His3MX6 (CSY399, CSY545, CSY900) plasmid as template. A yeast strain expressing SEC7-DsRED as an additional copy in the genome and under the control of the TPI promoter was engineered by transforming the integration plasmid YIplac204-T/C-SEC7-DsRED.T4 (Benjamin Glick, University of Chicago) into NY1175. The plasmid was cut with Bsu36I before transformation. Yeast strains expressing genomically tagged SEC7-DsRED were constructed by transformation of a PCR-amplified genomic integration construct using the pYG42 plasmid as template.
Genetic and DNA Manipulations
SGA (synthetic genetic array) analysis was essentially performed as previously described (Tong et al., 2001). The screen was performed at the permissive temperature of 25°C. The SGA analysis included the generation and evaluation of ∼4800 double mutants of pik1-101 with each of the deletion mutants of the EUROSCARF collection (European Saccharomyces cerevisiae Archives for Functional analysis; http://web-uni-frankfurt.de/fb15/mirko/euroscarf/index.html). Initially, the growth of double mutants in duplicates at 25°C was compared with the growth of the parental strains. The pik1-101 query strain was constructed as described in Supplementary Materials online.
Standard molecular biology techniques were used for DNA manipulations (Sambrook and Russel, 2001). Enzymes used for recombinant DNA techniques were purchased from New England Biolabs (Beverly, MA), Invitrogen (Carlsbad, CA), and USB (Cleveland, OH). PCR was performed according to manufacturer's instructions using Expand High Fidelity PCR System (Roche, Indianapolis, IN) or AmpliTaq (Applied Biosystems, Foster City, CA) DNA polymerase for cloning or diagnostic reactions, respectively.
DNA encoding for the Gga2p VHS (aa 18-170) or VHS-GAT domain (aa 18-327) were amplified from genomic DNA by PCR. The DNA fragment was digested with BamHI and XhoI and ligated into pGEX-6P-1, creating pLD212 or pLD213, respectively. The DNA fragment encoding for VHS-GAT was furthermore ligated into pNP308 creating pLD216. The vector pNP308 contains an ADH promoter and green fluorescent protein (GFP) for N-terminal tagging. To create pLD133, containing glutathione S-transferase (GST)-2xOSBPPH (aa 87-185), two copies of OSBPPH were cloned into pGEX-6P-1 using BamHI and EcoRI. The genomic DNA of GGA1 and GGA2 was amplified by PCR and digested with BamHI and XhoI. The DNA fragment containing GGA1 was then ligated into p415 GALs, creating pLD208. The GGA2 fragment in p416 GALs generated pLD211. To overexpress Gga1pL203Q and Gga2pI207N, the mutant open reading frame (ORF) was amplified by PCR from pAB469 and pAB456 (Boman et al., 2002), respectively, digested with BamHI and XhoI. The DNA fragment encoding for Gga1pL203Q was inserted in p416 GALs, creating pLD231, and the fragment encoding for Gga2pI207N was ligated into p415 GALs giving rise to pLD232.
Site-directed mutagenesis of the GGA2 VHS domain was achieved by overlap extension PCR as described (Ho et al., 1989). Two fragments of GGA2 were amplified from pAB381 in two separate PCR reactions using GGA2 5′ or 3′ flanking oligonucleotides in combination with internal overlapping mismatch oligonucleotides: GGA2_Glu_fw (5′ATGGTATCAAACAATTTGTGAACACTCAAGCTACGAGAATGATATGGGTTATATTAGAGACATGGCCGAATTGTTGAAATATAAGG3′); GGA2_Glu_bw (5′CCTTATATTTCAACAATTCGGCCATGTCTCTAATATAACCCATATCATTCTCGTAGCTTGAGTGTTCACAAATTGTTTGATACCAT3′) with mismatches underlined. The two PCR products were fused in a subsequent primer extension reaction to give rise to full-length GGA2 (K143E, K148E, H158A, R159E) flanked by BamHI and XhoI sites at the 5′ and 3′ ends of the ORF. This PCR product was cloned into pCR2.1-TOPO (Invitrogen) to create pMG7. For expression of GST-Gga2pKKHR-EEAE the mutated GGA2 ORF was cleaved from pMG7 using BamHI/XhoI and subcloned into pGEX-6P-1, resulting in plasmid pMG8. For expression of Gga2pVHS(KKHR-EEAE), DNA encoding the mutated Gga2p VHS domain (aa 18-170) was amplified from pMG7 and subcloned into the BamHI/XhoI sites of pGEX-6P-1, generating pMG10. Plasmid pMG9 for expression of GFP-Gga2pKKHR-EEAE was generated by PCR amplification of the mutated GGA2 ORF from pMG7 to incorporate a SalI site at the 5′ end and cloned into pCR2.1-TOPO. GGA2 (KKHR-EEAE) was then cleaved from plasmid pCR2.1-TOPO using SalI/KpnI and subcloned into the SalI/KpnI sites of pCS136. The plasmid pCS136 (CPYp-GFP-GGA2, pGOGFP, pRS426) was obtained from Chris Stefan and Scott Emr (University of California, San Diego). pMB430 (CPYp-GFP-GGA2, I207N mutation) was constructed from pCS136 using the QuickChange mutagenesis kit (Stratagene, La Jolla, CA). pMB432 [CPYp-GFP-GGA2(KKHR-EEAE)] was obtained by transferring a ClaI/Eco91I fragment from pMG7 to pCS136. For construction of pMB433 [CPYp-GFP-GGA2 (KKHR-EEAE and I207N)], the GGA2 I207N mutation was introduced in pMB432 using the QuickChange mutagenesis kit (Stratagene).
The plasmid pAB381 encoding for GST-Gga2p (pGEX5x-2; Zhdankina et al., 2001), pAB469 encoding for Gga1pL203Q and pAB456 encoding for Gga2pI207N (Boman et al., 2002) were a gift from Patricia Scott (University of Minnesota, Duluth Medical School). The plasmid for the expression of Sec7p-DsReD.T4 (YIplac204-T/C-SEC7-DsRED.T4) was kindly provided by Benjamin Glick. The GFP from the pFA6a-GFP(S65T)-kanMX6 (Longtine et al., 1998) was removed by digest with PacI and AscI, and the sequence encoding for DsRed was inserted giving rise to the plasmid pYG42.
Chemicals and Antibodies
Lipids were purchased from Avanti Polar Lipids (Alabaster, AL), and PIs were obtained from Echelon Biosciences (Salt Lake City, UT). GTPγS was from Sigma (St. Louis, MO) and ECL for chemiluminescence detection was from Amersham Biosciences (Piscataway, NJ). The following commercially available antibodies were used in this study: α-ADH (Chemicon International, Temecula, CA), α-HA (HA.11, affinity-purified, Covance, Madison, WI), α-GST (Protein Expression Facility, MPI-CBG, Dresden, Germany), and goat anti-rabbit antibody conjugated with horseradish peroxidase (Dianova, Hamburg, Germany). The α-Gga2p antibody was a generous gift from Patricia Scott (Zhdankina et al., 2001). For the anti-Arfp antibody we are indebted to Anne Spang (Biozentrum, University of Basel). Complete was from Roche.
Life Cell Imaging
A 5-ml yeast culture was grown to early log phase in YPD or selective media. Then cells were harvested, resuspended in 500 μl residual medium, and observed without fixation under a fluorescence microscope (Axioplan 2, Zeiss or Axioplan 2 MOT, Zeiss, Jena, Germany). If indicated, temperature-sensitive mutants were shifted to the restrictive temperature of 37°C for 1 h.
Subcellular Fractionation
A 200-ml yeast culture was grown at 25°C to an OD600 of 0.5–1.0. A temperature shift was performed for 1 h at 37°C when indicated. Then, 35 ODu of cells were harvested (5 min, 3000 × g) and washed with ice-cold 50 mM potassium phosphate, pH 7.5. Cell lysis and subcellular fractionation was performed as described (Du and Novick, 2001). Modifications and details of the protocol are described in the Supplementary Materials online. Twenty microliters of the lysate were kept as an input sample, and 500 μl of lysate was spun for 45 min at 100,000 × g in a Beckman Optima Ultracentrifuge (TLA120.2 rotor; Fullerton, CA). The S100 was kept, and the P100 pellet was resuspended in 500 μl lysis buffer. Twenty microliters of each fraction in 1× sample buffer were analyzed by SDS-PAGE and immunoblotting. Fractions were quantified using the Image Quant software (Molecular Dynamics, Sunnyvale, CA).
In Vitro Binding Assays for Protein–Lipid Interaction
Liposomes were created, and liposome recruitment assays were performed as described in Baust et al. (2006). See Supplementary Materials online for a more detailed description.
Invertase Assay
The procedure for determination of invertase secretion was performed as described (Nair et al., 1990). Invertase production was induced in YP medium containing 0.1% glucose for 1 h at 37°C. The mean value and SEM of all sets were calculated and are presented in Figure 2.
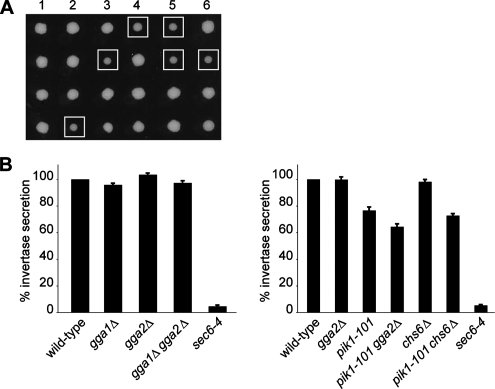
Figure 2.
A pik1-101 gga2Δ double mutant exhibits synthetic genetic and exocytosis defects. (A) pik1-101 mutant cells were crossed to gga2Δ cells, and tetrads were isolated at 25°C. Growth of resulting haploids was monitored after 3 d at 25°C. Squares indicate pik1-101 gga2Δ double mutants, showing a synthetic growth defect. (B) The expression of invertase was derepressed by shifting cells for 1 h into low glucose (0.1%). Left panel, wild-type (YAB200897; invertase secretion set to 100%), gga1Δ (YAB531; 95.8 ± 1.2%), gga2Δ (YAB532; 103.4 ± 1.2%), gga1Δ gga2Δ (YAB538; 97.2 ± 1.6%), and sec6-4 (NY778; 4.6 ± 0.8%); right panel, wild-type (CSY209; invertase secretion set to 100%), gga2Δ (CSY567; 99.9 ± 1.9%), pik1-101(CSY544; 76.7 ± 2.5%), pik1-101gga2Δ (CSY545; 64.4 ± 2.2%), chs6Δ (CSY566; 98.2 ± 1.7%), pik1-101chs6Δ (CSY561; 72.8 ± 1.4%), and sec6-4 (NY778; 5.3% ± 0.5). The exocyst mutant sec6-4 was used as positive control. The experiment was performed at the restrictive temperature of 37°C. Values indicate percentages of secreted invertase (n = 6–15, mean ± SEM). The difference in invertase secretion between the pik1-101 and pik1-101gga2Δ mutants is highly significant (p = 0.003). There is no significant difference (p = 0.18) between pik1-101 and pik1-101chs6Δ. Note that mutants in the same strain background are shown within the same panel with their respective wild type.
Vps10p Stability Assay
Yeast strains were grown overnight in YPD to an OD600 of 0.4–0.7. Twenty ODu were collected, washed with PBS, and transferred to 50 ml SD medium with the appropriate amino acids. Cultures were incubated for 3–4 h at 25°C. For each time point 2.5 ODu were required, and therefore 7.5 ODu were harvested in a 15-ml Falcon tube (10 min, 3,000 × g). Cells were resuspended in 3.75 ml synthetic medium lacking methionine and preincubated at 25°C for 30 min. [35S] methionine (40 μl; 10 mCi/ml; Perkin-Elmer Cetus Life Sciences, Norwalk, CT) was added, and cells were labeled for 10 min. Cells were chased with an excess of unlabeled methionine for the indicated times. At each time point, a 1-ml sample was removed and mixed with 120 μl 50% trichloroacetic acid (TCA) on ice. The conditions for Vps10p-3xHA immunoprecipitation were adapted from Govindan et al. (1995). In each immunoprecipitation, 1.5 μl α-HA antibody (clone HA.11, Covance) has been used.
Electron Microscopy
Yeast cells were cryoimmobilized using an EMPACT2+RTS (Leica Microsystems, Heidelberg, Germany) high-pressure freezer (Manninen et al., 2005). Samples were processed for freeze substitution as described previously (McDonald and Muller-Reichert, 2002). In brief, samples were freeze-substituted at −90°C for 2 d in acetone containing either 1% osmium tetroxide and 0.1% uranyl acetate or 0.01% osmium tetroxide and 0.1% uranyl acetate for morphological analysis and immunolabeling, respectively. The temperature was raised progressively to room temperature over 22 h in an automatic freeze-substitution machine (Leica Microsystems). Samples were embedded in Epon/araldite (morphology) or LR White (immunolabeling). Thin sections (70 nm) were cut using a Leica Ultracut UCT microtome. Sections were collected on Formvar-coated copper grids, poststained with 2% uranyl acetate in 70% methanol followed by aqueous lead citrate, and viewed in a Tecnai 12 (FEI, Eindhoven, The Netherlands) transmission electron microscope operated at 100 kV. Thin sections were labeled using α-Tlg1 antibody diluted in blocking buffer containing 0.8% bovine serum albumin, 0.01% Tween20, and 0.1% fish scale gelatin (Nycomed, Oslo, Norway; Amersham) in PBS. The secondary goat anti-rabbit IgG antibody was coupled to 12-nm colloidal gold (Jackson ImmunoResearch, West Grove, PA). The antibody complex was stabilized with 1% glutaraldehyde in PBS, and the labeled sections were poststained as described.
Bioinformatics Analysis and Structure Prediction
The structures of the yeast VHS domains of Gga1p and Gga2p were predicted using Phyre (Enhanced Genome Annotation using Structural Profiles in the Program 3D-PSSM; Kelley et al., 2000). The multiple sequence alignment of the GGA with Tom1 was done using ClustalX (Multiple sequence alignment with the Clustal series of programs; Chenna et al., 2003). The alignment of Epsin ENTH and CALM ANTH domains with the members of the GGA/Tom families was done manually based on structural alignments provided by the DALI server (http://ekhidna.biocenter.helsinki.fi/dali/; Holm and Sander, 1996). For GenBank accession numbers, see Table S3.
Surface Plasmon Resonance
The binding of recombinant GST-2xOSBPPH, Gga2pVHS, Gga2pVHS-GAT, Gga2pVHS(KKHR-EEAE), and GST proteins to PI(3)P, PI(4)P, and phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2)-containing liposomes was recorded in real time using an surface plasmon resonance (SPR)-based biosensor (BIACORE 2000; Biacore AB, Uppsala, Sweden) at 25°C. Liposomes (0.4 mg/ml) containing 10% PI(3)P, 10% PI(4)P, or 10% PI(4,5)P2 and 70% PC, 20% PE were prepared as described before (Honing et al., 2005). Liposomes containing 80% phosphatidylcholine (PC) and 20% phosphatidylethanolamine (PE) were used for the reference cell. A L1 sensor chip (Biacore) was primed twice with an injection of 20 mM CHAPS for 1 min at a flow rate of 10 μl/min. Subsequently the liposomes in running buffer: 10 mM HEPES (pH 7.4) and 150 mM NaCl were injected at 5 μl/min for 30 min followed by pulse injections of 50 mM NaOH to remove unbound material. This procedure resulted in an increase of the baseline by 7500–8000 RU. All binding experiments with fusion-proteins were performed in running buffer 10 mM HEPES (pH 7.4), 150 mM NaCl at a flow rate of 30 μl/min. GST-2xOSBPPH, Gga2pVHS, and Gga2pVHS-GAT were used at concentrations ranging from 250 nM to 4 μM. Gga2pVHS(KKHR-EEAE) was used at 250 nM to 2 μM. GST showed no binding up to 4 μM. The sensor chip surface was regenerated after each injection with 50 mM NaOH. Subtraction of the unspecific binding to the reference surface coated with PC/PE liposomes was done before the evaluation. Evaluations of steady-state affinity data were performed using BIAevaluation software 4.1. A plot of steady-state binding levels (Req) against analyte concentration was fitted to the general fitting model of steady-state affinity.
RESULTS
Gga2p Is a PI(4)P Effector Candidate
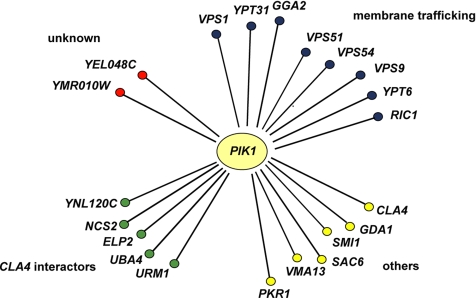
To identify candidate effectors for PI(4)P, we carried out a genome-wide screen for nonessential genes that interact synthetically with the temperature-sensitive pik1-101 mutation. The rationale of the screen was to focus on deletions in those genes that do not show a phenotype by themselves, but would do so in a sensitized background in which PI(4)P production is partially compromised. Therefore, we screened an established library of deletions in nonessential genes (EUROSCARF collection; European Saccharomyces cerevisae Archives for Functional analysis; http://web-uni-frankfurt.de/fb15/mikro/euroscarf/index.html) and analyzed their effect on growth in a pik1-101 mutant background at the permissive temperature. We isolated 86 interacting deletions (Table S1) and focused further on 21 PI(4)P effector candidates that represented the strongest or most interesting genetic interactors. Among them, we found regulators of Golgi-to-endosome transport (GGA2, VPS1, and VPS9), endosome-to-TGN retrieval (VPS51, VPS54, RIC1, and YPT6), and the Golgi small GTPase YPT31 as well as genetic interactors of the p21-activated kinase CLA4 (URM1, ELP2, NCS2, and YNL120C; Figure 1).
Figure 1.
Results of genome-wide synthetic lethality screen of the pik1-101 mutant allele. Genes that have been found in SGA analysis to interact genetically with pik1-101 (synthetic lethal or sick) are represented as nodes. All genetic interactions shown in this scheme have been confirmed by tetrad analysis. Several genetic interactors of CLA4 were found to exhibit synthetic interactions (URM1, ELP2, NCS2, YNL120C, and UBA4). The synthetic genetic interaction with UBA4 was not identified in the genome-wide screen but was found by tetrad analysis in the further course of this study. For a full list of genes isolated in the screen, see Table S1.
Because a number of examples have been reported of clathrin adaptors, which directly interact with phosphoinositides, we first focused on the Golgi clathrin adaptor Gga2p, for which the mechanism of specific targeting to the TGN membrane is still unknown. Double mutants of pik1-101 gga2Δ show a synthetic growth defect, which was confirmed by tetrad analysis (Figure 2A). This synthetic growth phenotype prompted us to ask whether Pik1p and Gga2p show similar phenotypes, and therefore we studied exocytosis of the secretory cargo invertase and monitored the morphology of secretory organelles in the respective mutants.
Mutants of gga2 and pik1 Show Similar Phenotypes
Because we and others have previously reported that pik1 mutants exhibit defects in both surface transport and transport from the TGN to the vacuole via endosomes (Hama et al., 1999; Walch-Solimena and Novick, 1999; Audhya et al., 2000), we investigated whether pik1-101 gga2Δ double mutants show synthetic defects in these pathways. As shown in Figure 2B, surface transport monitored through invertase secretion was reduced from 76.7 ± 2.5% (mean ± SEM) in pik1-101 mutant cells to 64.4 ± 2.2% in the pik1-101 gga2Δ double mutant (p = 0.003). The synthetic effect of this double mutant is specific to gga2Δ because it was not observed in a pik1-101 chs6Δ background (72.8 ± 1.4%, p = 0.18 for the difference between pik1-101 and the pik1-101 chs6Δ double mutant; Figure 2B). Chs6p, which served as a control Golgi mutant in this experiment, is required for transport of the cell wall biosynthetic enzyme Chs3p (chitin synthase III) from the TGN/endosome to the plasma membrane (Trautwein et al., 2006; Wang et al., 2006).
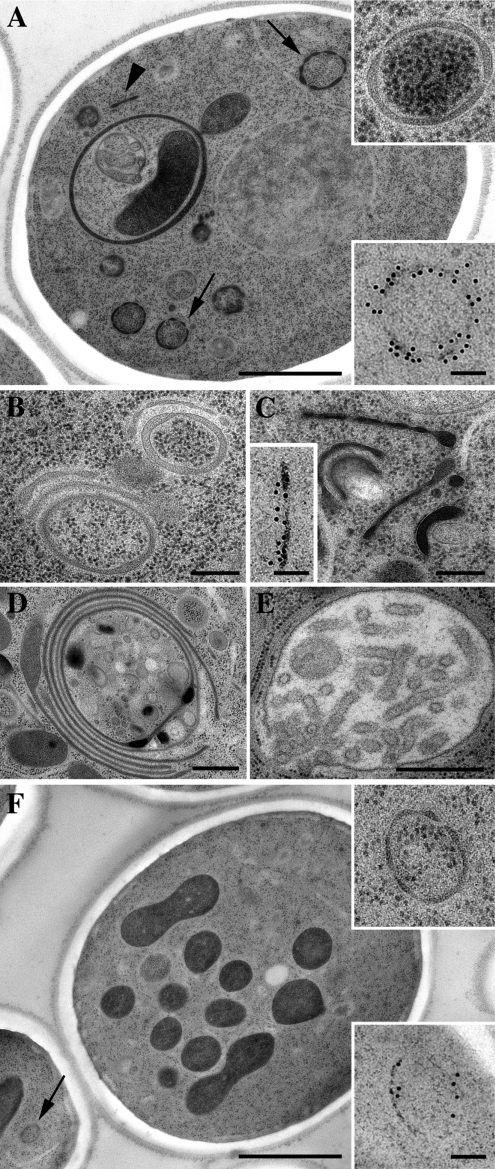
We next compared the organelle morphology of pik1-101 and gga2Δ mutants at the ultrastructural level. Even though gga2Δ mutants accumulated fewer membranes, similar ring-like structures akin to Berkeley bodies were the most prominent membranes both in gga2Δ and pik1-101 mutant cells (Figure 3, A and F; see Table S2 for quantitation). To establish the identity of these structures, we performed immunoelectron microscopy using the late Golgi/endosome marker Tlg1p (Holthuis et al., 1998). Indeed, the ring-like structures in both gga2Δ and pik1-101 mutants correspond to Tlg1p-positive Berkeley bodies, a hallmark of Golgi mutants (Novick et al., 1980). Tubular Tlg1p-positive structures were also found in both mutants (Figure 3, A and C, and Table S2). Moreover, both mutants exhibited a small number of multilayered structures, probably representing further advanced accumulation of Golgi membranes (Figure 3, B, D, and E), and abnormal vacuoles (Figure 3, A and F, and Table S2). In summary, the similar morphology and secretory defects of pik1-101 and gga2Δ mutants raised the possibility of a role of Pik1p and Gga2p at a common step in membrane transport at the TGN.
Figure 3.
Ultrastructural analysis of pik1-101 and gga2Δ mutant cells. The pik1-101 and gga2Δ mutant cells were grown at permissive temperature and processed for electron microscopy. (A–E) pik1-101. (F) gga2Δ. Immunolabeling was with α-Tlg1 antibody. Secondary antibody was coupled to 12-nm colloidal gold. Bar, (A and F) 1 μm; (D and E) 500 nm; (B and C) 250 nm; and (A, C, and F, insets) 100 nm. Note the appearance of ring-like (arrows) and tubular (arrowhead) structures. For quantitation of the results, see Table. S2.
Pik1p Directly Affects the Vacuolar Protein-sorting Pathway at the TGN Exit
We have shown above that Pik1p and Gga2p show similar and synergistic phenotypes and affect TGN-to-endosome trafficking. We then asked whether Pik1p directly regulates vacuolar transport at the TGN exit. To address this question, we investigated the trafficking of the CPY receptor Vps10p, which cycles between TGN and prevacuolar/late endosomes (Bowers and Stevens, 2005). This process can be studied by monitoring the stability of Vps10p. In wild-type cells, pulse-labeled and immunoprecipitated Vps10p is completely stable, and thus only the full-length form is observed. In class E vps mutants (e.g., vps28Δ), however, Vps10p becomes unstable because of trapping of the protein in the abnormal, proteolytically active “class E compartment” (Costaguta et al., 2001). It has been previously reported that in gga1Δ gga2Δ vps28Δ triple mutants, Vps10p is stabilized indicating an inhibition of exit of this protein from the TGN (Costaguta et al., 2001). We asked whether we could observe a similar phenotype in a pik1-101 vps28Δ double mutant. As expected, in the class E mutant vps28Δ, Vps10p was partially in the cleaved, lower molecular weight form, after 30 min and more completely after 60 min. This cleaved product of Vps10p was then not processed any further (Figure 4A). In pik1-101 cells, stability of Vps10p was normal, indicating that the receptor can still recycle from the prevacuolar/late endosome compartment. In the pik1-101 vps28Δ double mutant, Vps10p was stabilized and could be observed in its full-length form for a longer period of time (up to 60 min), compared with the vps28Δ mutant (Figure 4A). This partial rescue of Vps10p from proteolysis in the class E compartment suggests that like in gga1Δ gga2Δ mutants, TGN exit of Vps10p is partially inhibited in the pik1-101 mutant. A pik1-101 gga2Δ vps28Δ triple mutant for comparison was not viable. The observed rescue of Vps10p proteolysis in the class E compartment implies that Pik1p does function in the Vps10p pathway, and, like GGAs, acts at the exit from the TGN.
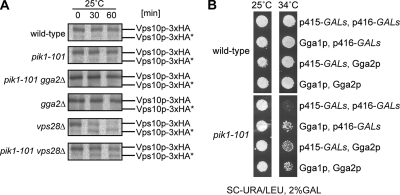
Figure 4.
Pik1p regulates the Vps pathway at the exit from the Golgi. (A) Vps10p transport to endosomes is impaired in pik1-101. Wild-type (CSY392), pik1-101 (CSY391), pik1-101 gga2Δ (CSY398), vps28Δ (CSY900), and pik1-101 vps28Δ (CSY399) containing the CPY receptor Vps10p genomically tagged with 3xHA were grown overnight in YPD, and the next day were shifted for 3–4 h to minimal medium lacking methionine. Proteins were pulse-labeled with 35[S]methionine for 10 min and then chased for 0, 30, and 60 min. Proteins were precipitated with TCA and washed with acetone, and Vps10p-3xHA was immunoprecipitated using an α-HA antibody. Samples were subjected to SDS-PAGE and autoradiography. Vps10–3xHA* indicates the lower protease-resistant form. (B) GGA overexpression suppresses growth defect of pik1-101 at 34°C. Wild-type (NY1211) or pik1-101 (CSY712) cells containing p415 GALs and p416 GALs, control vectors, p415 carrying GALs-GGA1 or p416 carrying GALs-GGA2 as indicated were grown on selective media with 2% raffinose to midlog phase and plated onto selective media with 2% galactose at 25 or 34°C, respectively, for 3 d.
Because similar and synthetic phenotypes were observed in pik1 and gga2 mutants, we hypothesized that both proteins could function together at the same transport step. Overexpression of Gga proteins suppressed the growth defect of pik1-101 cells up to 34°C, further suggesting a role of Pik1p and Gga2p in a common pathway (Figure 4B).
Pik1p Activity Is Required for TGN Localization of Gga2p
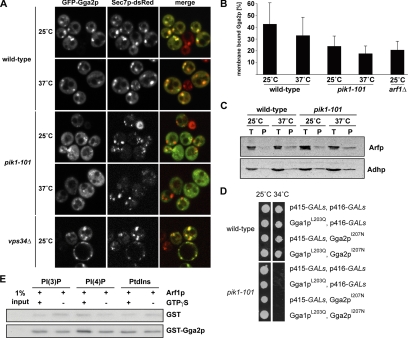
The observed mutant and double mutant phenotypes as well as genetic interactions of gga2 and pik1 might be due to a direct recruitment of Gga2p to the TGN through binding to PI(4)P, the immediate product of Pik1p activity. We therefore decided next to examine the requirement of Pik1p for Golgi localization of Gga2p. As a TGN marker, we used the Arf1p nucleotide exchange factor Sec7p. For visualization of Gga2p we used a plasmid for overexpression of GFP-GGA2, which did not result in suppression of the pik1-101 temperature-sensitive growth defect at 34°C (not shown). In wild-type cells, GFP-Gga2p and Sec7p-DsRed colocalize, and GFP-Gga2p was more prominent at the TGN than in the cytoplasm (Figure 5A). Instead, in pik1-101 cells, GFP-Gga2p is cytosolic, and only residual TGN colocalization was found (Figure 5A). The defect in GFP-Gga2p localization is already apparent at 25°C due to the low activity of the Pik1p mutant protein even at the permissive temperature (Walch-Solimena and Novick, 1999). In contrast to Pik1p, loss of the PI 3-kinase Vps34p (Schu et al., 1993) had no effect on the localization of Gga2p, indicating that PI(3)P and PI(3,5)P2 are not required (Figure 5A). TGN structures do not loose integrity in pik1-101 compared with wild-type cells (Figure 3; Walch-Solimena and Novick, 1999; Audhya et al., 2000), as shown by the Sec7p-dsRed fluorescence in Figure 5A). This excludes the possibility that cytosolic Gga2p localization is due to loss of TGN in pik1-101 mutants. We also controlled for possible changes in Arf1p levels at the membrane, which could be caused by differences in SEC7-DsRED expression but did not find any differences in the strains used for GFP-Gga2p localization (Supplementary Figure 1). We therefore conclude that Gga2p targeting is PI(4)P-dependent. Consistently, pik1-101 mutant cells showed a significant decrease of endogenous Gga2p in Golgi containing membrane fractions (Figure 5B). This decrease was not due to altered fractionation of Arf1p as indicated in Figure 5C.
Figure 5.
Binding of Gga2p to PIs and Arf1p is required for proper localization to the Golgi. (A) Wild-type (CSY349) and vps34Δ cells (CSY911) expressing an additional copy of Sec7p-DsRed under the control of a strong promoter as well as pik1-101 cells (CSY906) containing genomically tagged Sec7p-DsRed were transformed with a plasmid encoding for GFP-Gga2p (pCS136). Cells were grown in selective media to midlog phase, incubated at 25 or 37°C for 1 h, and subsequently analyzed by fluorescence microscopy. In wild-type and vps34Δ cells, Gga2p colocalizes with Sec7p-DsRed, whereas in pik1-101 cells GFP-Gga2p is mostly cytosolic. (B) Subcellular distribution of endogenous Gga2p was evaluated by subcellular fractionation from wild-type (CSY210), pik1-101 (CSY544), and arf1Δ (CSY704) cells. Wild-type, arf1Δ, and pik1-101 mutant cells were grown to midlog phase, homogenized, and then soluble and membrane fractions were separated by 100,000 × g centrifugation of a postnuclear supernatant. Temperature shift to 37°C was performed for 1 h before fractionation. Equal volumes of fractions were loaded and analyzed by SDS-PAGE and immunoblotting. The Image Quant Software was applied for quantification (n = 3, mean ± SD). Less endogenous Gga2p is membrane-bound in pik1-101 cells at both permissive and restrictive temperatures compared with wild type. (C) Equal amounts of subcellular fractions prepared from wild-type (CSY210) and pik1-101 (CSY544) cells as in B were analyzed by SDS-PAGE and Western blots using α-ADH and α-Arf antibodies. (D) Overexpression of gga mutants unable to bind Arf1p does not rescue the growth defect of pik1-101 at 34°C. Wild-type (NY1211) or pik1-101 (CSY712) cells containing p415 GALs and p416 GALs control vectors, or expressing Gga1pL203Q (pLD232) or Gga2pI207N (pLD231) as indicated were grown on selective media with 2% raffinose to midlog phase and plated onto selective media with 2% galactose at 25 or 34°C, respectively, for 3 d. (E) Liposomes containing either 1% PI(3)P, PI(4)P or PtdIns were incubated with purified GST-Gga2p (1.5 μg) or GST (0.5 μg) and purified myristoylated ARF1 in the absence or presence of GTPγS. The liposome-bound fraction of GST-Gga2p was detected using a α-GST antibody. The recruitment of GST-Gga2p to PI(4)P containing liposomes is increased in the presence of active ARF1, indicating a synergistic function of PI(4)P and Arf1p in the recruitment of Gga2p to membranes.
We have previously demonstrated that Pik1p activity is strongly reduced but not completely abolished in the pik1-101 mutant (Walch-Solimena and Novick, 1999). The remaining Gga2p at the Golgi (Figure 5, A and B) might be due to the remaining partial functionality of Pik1p. Alternatively, residual binding of Gga2p to membranes might be due to Arf1p interaction, which on its own is not sufficient for yeast Gga protein localization to the TGN (Boman et al., 2002). It is however necessary, because fractionation of arf1Δ cells did show a significant decrease in membrane-bound Gga2p (Figure 5B), supporting a scenario where Pik1p activity and Arf1p cooperate in Gga2p recruitment to the membranes. Consistent with such a mechanism, we found that the overexpression of Gga's no longer suppressed pik1-101 when Arf-binding deficient mutants Gga1pL203Q or Gga2pI207N (Boman et al., 2002) were used (Figure 5D).
Gga2p Binds Phosphoinositides
The observed Pik1p-dependence of Gga2p localization implied a direct interaction of Gga2p with PI(4)P. We therefore investigated next in a liposome pulldown assay, whether this PI can indeed recruit Gga2p to a lipid bilayer. In the presence of activated Arf1p we observed enhanced recruitment of GST-Gga2p to PI(4)P containing liposomes (Figure 5E). No such increase in recruitment of GST-Gga2p was observed in control liposomes containing PtdIns or in liposomes containing PI(3)P (Figure 5E), which is in agreement with the normal Gga2p localization in vps34Δ cells (Figure 5A). In control experiments, GTPγS without Arf1p had no effect on recruitment of GST-Gga2p to PI(4)P liposomes. Together these results suggest a synergistic role of PI(4)P and Arf1p in the localization of Gga2p to membranes.
The Gga2p VHS Domain Interacts with PI(4)P
Because we have demonstrated PI(4)P-dependent membrane recruitment of Gga2p, we next asked for the structural basis of this interaction. We first compared the sequence of the VHS (Vps27p/Hrs/STAM) domain of Gga2p to ANTH/ENTH domains, which are known to interact with phosphoinositides (Ford et al., 2001; Itoh et al., 2001). This analysis was prompted by the similarity found by structural comparison of ANTH/ENTH with the VHS domain (see Materials and Methods and De Camilli et al., 2002). Indeed, we noticed that the Gga2p VHS domain shows significant sequence homology with ANTH/ENTH domains (Figure 6A). The VHS domain was first identified from sequence comparisons in signal transduction proteins. In mammalian GGAs, the VHS domain binds to cargo proteins through sorting signals of the acidic cluster dileucine family (Bonifacino, 2004; Robinson, 2004). In yeast, the possible cargo interaction of the Gga1p and Gga2p VHS domain has not been clarified yet (Bonifacino, 2004; Bowers and Stevens, 2005).
Figure 6.
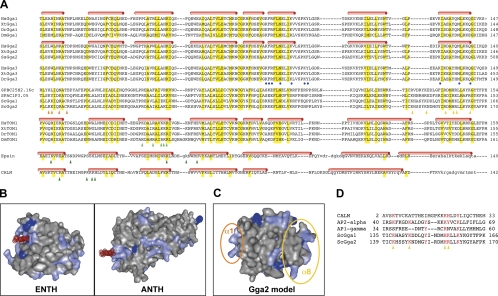
Modeling of a PI-binding motif in the N-terminal VHS domain of Gga2p. The VHS domains of the GGA proteins from different species were analyzed in terms of their potential binding to PI(4)P. (A) Multiple sequence alignment of the GGA VHS domains from fungi, arthropods, and vertebrates with the VHS domain of Target of Myb1 (Tom1) and the ENTH and ANTH domains from Epsin and CALM, respectively. Conserved residues are highlighted in yellow, and secondary structural elements are indicated on top of the one family member containing structural information. Residues that are described to contact Ins(1,4,5)P3 in Epsin and PI(4,5)P2 in CALM are indicated by dark green triangles. The Ins(1,4,5)P3-binding site in Epsin resides mostly within the very N-terminus, which only adopts a helical fold when bound to the ligand (not shown; for details see Ford et al. 2002). Residues thought to be involved in membrane binding of Tom1 are indicated by light green triangles. Residues that are in contact with BACE phosphopeptide in human GGA1 are highlighted by blue circles. The two potential PI(4)P-binding sites in fungal GGAs, which show a strong basic charge on the surface area, are located in helix α1 or helix α8 and are indicated by orange or yellow triangles, respectively. For a list of GenBank accession numbers, see Table S3. (B) Structural display of the Epsin ENTH domain and the CALM ANTH domain complexed with PI(1,4,5)P3 and PI(4,5)P2, respectively. The surface of the protein domains is shown in gray with basic amino acid, including histidine, highlighted in light and dark blue, respectively. The dot surface of the lipid is shown in red. (C) Surface representation of the modeled structure of the VHS domain of yeast Gga2p using the Phyre-server for fold recognition. The two potential sites of interaction of Gga2pVHS with PI(4)P are circled in orange or yellow, respectively. (D) Alignment of the putative phosphoinositide interaction sites of yeast GGAs with vertebrate ANTH and the clathrin adaptors AP1γ and AP2α. The positively charged and aromatic residues contacting the phosphoinositide head group in AP1γ and AP2α are highlighted in red. Note: This site overlaps with the predicted PI-interacting region preceding helix α8 (orange triangles).
How could the Gga2p VHS domain bind to PI(4)P? As a prerequisite of PI binding, a basic patch or groove should be accessible on the surface of the three-dimensional (3D) structure of the VHS domain similar to what has been described for ENTH/ANTH domains (Figure 6B). To explore such potential PI(4)P-binding sites, we modeled the tertiary structure of the VHS domain of yeast Gga2p using 3D-structure information from the GGA2 human homologue (Figure 6C). The resulting 3D model of the Gga2p VHS domain reveals two potential binding sites for PI(4)P (Figure 6C and Ford et al., 2001). One of these potential lipid-binding sites is localized to the last helix, α8 (Figures 6, A and C) and shows similarity to the signature of the PI(4,5)P2-binding site in the CALM ANTH domain (Figure 6D). This site is localized close to the phospho-protein interaction site of vertebrate GGAs (Figure 6A and Shiba et al., 2004). The signature proposed to interact with PI, however, seems not to be conserved in higher eukaryotes.
Interestingly, the proposed phosphoinositide-binding site in the loop preceding helix α8 of yeast Gga's also shows a pattern of charged and aromatic residues that closely resembles the reported phosphoinositide-binding site that was found in cocrystals of the α-subunit of AP2 with InsP6 (Figure 6D and Collins et al., 2002). Similar to AP2α, also AP1γ (Heldwein et al., 2004) uses a set of positively and aromatic residues in the so-called helix2-helix3 corner region to bind to the phospholipids. Introduction of a triple mutation in the conserved lysines in AP2α lead to a failure of membrane recruitment of this adaptor to coated pits (Gaidarov and Keen, 1999). AP1γ mutation in this region prevented normal Golgi targeting of the adaptor, and one specific mutation (R48A) eliminated PI(4)P-dependent enhancement of adaptor recruitment to liposomes (Heldwein et al., 2004). A very similar pattern of conservation can be observed in the yeast Gga sequences (Figure 6D). This similarity additionally hints at loop7 and helix α8 as the site of interaction between Gga2p and PI(4)P. In summary, sequence comparison and 3D modeling suggests that the Gga2p VHS domain mediates membrane interaction via its PI(4)P-binding capacity, similar to the role of the ANTH domain of CALM, the α subunit of AP2 and AP1γ.
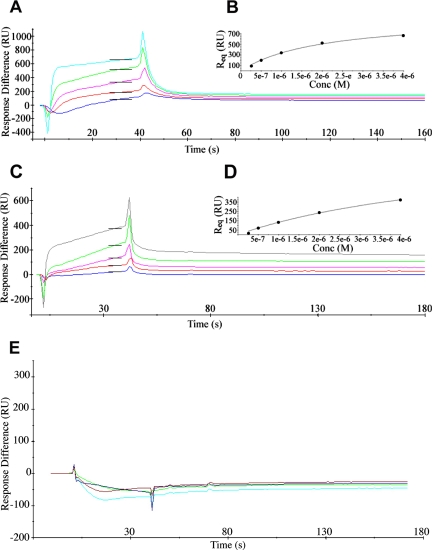
To provide evidence that the Gga2p VHS domain on its own can bind phosphoinositides, we performed an SPR-based binding assay (Honing et al., 2005). The sensor chip was coated with different types of liposomes, and binding of Gga2pVHS or Gga2pVHS-GAT (VHS and the Arf-binding GAT domain) was compared with GST-2xOSBPPH and a GST control (Table 1 and Figure 7, A–D). The pleckstrin homology (PH) domain of OSBP has been previously shown to bind PI(4)P in vitro and in vivo (Levine and Munro, 2002) and was here used as positive control. Monitoring domain binding of Gga2pVHS revealed specific binding (KD = 2 μM) to PI(4)P membranes and weaker binding to membranes containing PI(3)P (KD ≫ 19 μM) or PI(4,5)P2 (KD = 19 μM; Table 1 and Figure 7, A and B). Binding of Gga2pVHS to PI(4)P membranes in this assay is thus in the range of the strongest interactions among the yeast PH domains of the oxysterol-binding proteins Osh1p and Osh2p (Kd = 1–3 μM; Yu et al., 2004). Gga2pVHS-GAT did not show an increase in affinity of PI(4)P binding compared with Gga2pVHS (Table 1 and Figure 7, C and D), suggesting that the VHS domain is mainly responsible for PI binding. To test whether the predicted PI-binding motif is indeed responsible for PI(4)P interaction, we next generated a mutant VHS domain (K143E, K148E, H158A, R159E; KKHR-EEAE). Biosensor chip analysis revealed quantitative loss of PI binding for the isolated mutant VHS domain (Gga2pVHS(KKHR-EEAE); Table 1 and Figure 7E), demonstrating that this signature mediates the interaction. Because this motif is conserved in Gga1p and we have shown that Gga1p like Gga2p overexpression (Figure 4B) can rescue pik1-101, both proteins apparently share PI binding as a common function. This is in agreement with previous work suggesting redundancy of Gga1p/Gga2p function (Boman, 2001). Nevertheless, there might be some degree of specialization of these two proteins because we found by ultrastructure analysis that in gga1Δ mutants mainly the elongated structures accumulated, which were also observed in gga2Δ and pik1-101 at much lower abundance than Berkeley bodies (Supplementary Figure 2 and Table S2).
Table 1.
Estimated Kd values (in M)
| PI(3)P | PI(4)P | PI(4,5)P2 | |
|---|---|---|---|
| GST | No binding | No binding | No binding |
| GST-2 × OSBPPH | (+/−) | 7.4 × 10−7 ± 1.02 × 10−7 Ma | ≫7.4 × 10−7 M |
| Gga2pVHS | ≫1.99 × 10−5 M | 2.0 × 10−6 ± 0.27 × 10−6 Ma | 1.99 × 10−5 ± 0.522 × 10−5 Mb |
| Gga2pVHS-GAT | No binding | 6.28 × 10−6 ± 1.461 × 10−6 Ma | ≫6.28 × 10−6 M |
| Gga2pVHS(KKHR-EEAE) | No binding | No binding | No binding |
a Mean ± SE; n = 5.
b Mean ± SE; n = 4.
Figure 7.
Binding of Gga2pVHS (A), Gga2pVHS-GAT (C) and Gga2pVHS(KKHR-EEAE) (E) to PI(4)P liposomes determined by surface plasmon resonance. Gga2pVHS and Gga2pVHS-GAT were injected at concentrations from 250 nM to 4 μM (bottom to top curves) over immobilized PI(4)P liposomes and over control PC liposomes. The values of control surface were subtracted from the signal. The change in SPR signal during association and dissociation is shown in colored curves. Black bars are report points set on the sensograms in the steady-state region of the curve. (B and D) Plot of steady-state binding levels (Req) against concentrations of Gga2pVHS or Gga2pVHS-GAT and fit to steady-state affinity model. Gga2pVHS(KKHR-EEAE) was used at concentrations from 250 nM to 2 μM and revealed no binding to PI(4)P liposomes.
PI(4)P and Arf1p Cooperate in Gga2p Recruitment to the TGN
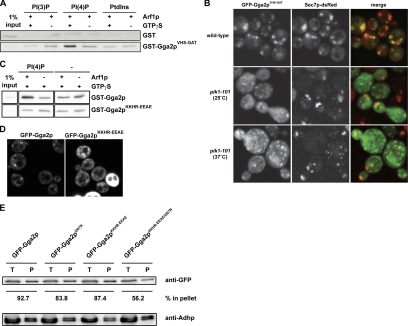
To test for synergy in PI(4)P and Arf1p in liposome recruitment, we performed liposome-pulldown assays. The Gga2pVHS-GAT construct was efficiently recruited by PI(4)P in the presence of activated Arf1p (Figure 8A).
Figure 8.
PI(4)P and Arf1p cooperate in the recruitment of the VHS-GAT domain to the TGN. (A) Liposome pulldown assay was performed as in Figure 5D using GST-Gga2pVHS-GAT (1.5 μg). (B) Wild-type (CSY349) and pik1-101 cells (CSY906) were transformed with a plasmid encoding for GFP-Gga2pVHS-GAT (pLD216). Cells were grown in selective media to midlog phase and subsequently analyzed by fluorescence microscopy. In wild-type cells, the GFP-Gga2pVHS-GAT domain was able to localize to the Golgi, whereas in pik1-101 cells GFP-Gga2pVHS-GAT was mostly cytosolic. (C) Liposome pulldown assay was performed as in Figure 5D using GST-Gga2p and GST-Gga2pKKHR-EEAE (pMG10; 1.5 μg). (D) Localization of GFP-Gga2 and GFP-Gga2pKKHR-EEAE. Wild-type cells (NY1211) expressing GFP-Gga2p (pCS136) or GFP-Gga2pKKHR-EEAE (pMG9) were grown in selective media to midlog phase and subsequently analyzed by fluorescence microscopy. (E) Mutation of the Gga2p Arf-binding domain and VHS domain leads to decreased membrane association of GFP-Gga2p. Wild-type cells (NY604) were transformed with plasmids encoding GFP-Gga2p (pCS136), GFP-Gga2pI207N (pMB430), GFP-Gga2pKKHR-EEAE (pMB432), or GFP-Gga2pKKHR-EEAE,I207N (pMB433). GFP-GGA2 constructs express Gga2p under the control of the CPY promoter. Subcellular fractionations, and quantifications of Western blots were performed as described in Figure 5B. Shown is the result of one representative experiment.
To investigate PI(4)P dependent recruitment of the Gga2p VHS domain to the TGN in vivo, we expressed GFP fusions containing the VHS domain alone (GFP-Gga2pVHS) or VHS-GAT (GFP-Gga2pVHS-GAT; Figure 8B). We then evaluated PI(4)P dependence in wild-type or pik1-101 mutant cells. The VHS domain was not sufficient to target the TGN, and thus GFP-Gga2pVHS was mostly cytosolic (data not shown and Boman et al., 2002). Similarly, the GAT domain alone has been shown to be insufficient for TGN localization (Boman et al., 2002). In contrast, the GFP-Gga2pVHS-GAT domain localized to the TGN, and this targeting was PI(4)P-dependent (Figure 8B). These data demonstrate that Gga2p localization to the TGN is regulated by PI(4)P through the VHS and GAT domains. In further support of this model, specific recruitment of full-length GST-Gga2p to liposome membranes in the pulldown assay was abolished by the KKHR-EEAE mutation (Figure 8C). Similarly, we observed mislocalization of GFP-Gga2pKKHR-EEAE in a wild-type yeast background (Figure 8D). To further test the cooperation of both PI(4)P and Arf1p interaction in GFP-Gga2p membrane recruitment in vivo, we constructed a GFP-Gga2pKKHR-EEAE,I207N double mutant deficient in both PI(4)P and Arf1p interaction (Boman et al., 2002). In subcellular fractionation experiments, a strong additive reduction of membrane-bound GFP-Gga2p was observed in the GFP-Gga2pKKHR-EEAE,I207N double mutant compared with either the GFP-Gga2pKKHR-EEAE or the GFP-Gga2pI207N single mutant (Figure 8E). In agreement with our pik1-101 subcellular fractionation (Figure 5B) and liposome pulldown (Figures 5E and 8, A and C) experiments, these data further imply that Gga2p targeting to the TGN occurs by coincidence detection of two binding sites, provided by PI(4)P and Arf1p and resembling a targeting mode described for other PI-binding domains (Carlton and Cullen, 2005).
DISCUSSION
The formation of clathrin-coated vesicles at the TGN involves monomeric clathrin adaptors of the GGA protein family. Specific targeting of GGAs to the TGN is not fully understood. We identified a novel PI(4)P-dependent mechanism to target Gga2p to TGN exit sites. Gga2p acts as a PI(4)P effector in both TGN-to-vacuole and TGN-to-plasma membrane transport.
Mechanism of GGA Recruitment to TGN Exit Sites
We have demonstrated that PI(4)P and Arf1p cooperate in the specific recruitment of Gga2p to the TGN. Although GGAs are effectors of Arf-GTP, this interaction has been shown to be insufficient for recruitment of Gga proteins to the TGN (Boman et al., 2002). The VHS domain, together with the Arf-GTP–binding GAT domain of GGAs contributes to Golgi localization, and the VHS domain confers specificity to late Golgi localization of this protein (Boman et al., 2002). Our finding of a Pik1p- and PI(4)P-dependent membrane recruitment through the VHS domain provides a mechanism for the specific localization of Gga2p to the TGN.
VHS domains share similarity with ANTH/ENTH domains, which are known to bind PI(4,5)P2 and are found in clathrin adaptor proteins (De Camilli et al., 2002; Legendre-Guillemin et al., 2004). The clathrin adaptor AP2 has also been shown to bind PI(4,5)P2. All three domains, ANTH, ENTH and AP2α, bind PI in a region formed by multiple helices (Balla, 2005). ENTH and ANTH domains differ in their mode of lipid interaction. Although Epsin ENTH forms a basic pocket where the lipid is captured, the ANTH domain of CALM interacts with the lipid in a more superficial manner without burying the PI in a groove-like structure (Ford et al., 2002; Figure 6B). Also in AP2α, PI interacts with a cluster of positive residues on the surface (Collins et al., 2002). PI(4)P binding to the AP1 clathrin adaptor (γ-adaptin) has been suggested to occur through a similar peripheral interaction site (Heldwein et al., 2004).
The structural basis for stereospecificity of the described PI interactions is not fully understood (Balla, 2005). By fold recognition, we uncovered two possible surfaces for interaction with PIs within the Gga2p VHS domain. Either proposed PI-binding site provides a charged surface similar to the ANTH domain of AP180/CALM (Legendre-Guillemin et al., 2004). The two potential PI interaction surfaces of the Gga2p VHS domain, in the α1 and α8 helices, respectively, involve residues that are unique to the fungal subfamilies of the GGAs (Figure 6A). Helix α1 in fungi contains a set of basic residues. Except for Arginine 32, none of the basic residues are conserved throughout all GGA-family members. In case of helix α8, the loop preceding the helix would be involved in PI binding, which is also specific to fungal members of the GGA-family.
Beyond the structural similarity, we found a sequence signature in the predicted PI interaction motif in helix α8 (Figure 6D) with a motif present in both ANTH and AP2α involving the signature (K-X9-KKK-H/Y; Ford et al., 2001; Collins et al., 2002; Lemmon, 2003). The similarity of the yeast Gga proteins with the ANTH consensus has also been noted by Costaguta et al. (2006). We have now shown that mutagenesis of the predicted PI signature in Gga2p results in loss of PI binding, suggesting that this motif is required for PI(4)P interaction.
Consistent with our data, it has recently been reported that dual recognition by PI(4)P and Arf1 is responsible for recruitment of GGAs to the TGN also in the mammalian system (Wang et al., 2007). Variations in the regulation of targeting of yeast versus mammalian GGAs are suggested by the differences in Arf dependence of GGA localization between the two systems: Arf interaction is required for Golgi targeting in mammals, but is not sufficient in yeast (Boman et al., 2002). This view is further supported by the finding that in mammals, the Arf1-binding GAT domain contains the major PI(4)P-binding activity, which seems to be regulated by the VHS domain (Wang et al., 2007). In yeast, we found strong PI(4)P binding by the VHS domain without a major change in a VHS-GAT construct. Thus, the mechanistic details of dual key recruitment of GGAs by PI(4)P and Arf1 seem to differ between the two systems. An additional role for PI(4)P in promoting recognition of ubiquitylated cargo by GGA has been suggested (Wang et al., 2007), adding another level of regulation.
PI(4)P-dependent Clathrin Coat Formation in TGN-to-Endosome Pathway
Recent work has demonstrated that specific cargos traffic through different TGN-to-surface pathways, whereby invertase was sorted into different SVs than Pma1p, and sorting of invertase away from Pma1p was disturbed in mutants affecting the Vps route from the TGN to the endosome (Gurunathan et al., 2002; Harsay and Schekman, 2002). On the basis of these findings, it has been proposed that invertase-containing SVs could be formed at endosomes (Gurunathan et al., 2002; Harsay and Schekman, 2002). The synergistic function of Pik1p/PI(4)P and Gga2p in TGN-to-endosome transport and in surface delivery of invertase (Figure 2) is consistent with the possibility of a shared control for vacuolar and exocytic traffic through endosomes. However, it remains a possible scenario that invertase vesicles bud directly from the TGN but depend on a different subset of TGN proteins than Pma1p vesicles. To resolve this issue, it will now be crucial to determine whether endosomes are indeed an intermediate of exocytosis for a subset of secreted cargo.
We have shown here that Pik1p functions together with Gga2p in TGN-to-late endosome transport. Does Pik1p function also play a role in trafficking between TGN and early endosomes? A number of observations are consistent with such a model. First, Pik1p has also been shown to regulate trafficking of cargos that require recycling through early endosomes, in particular the SNARE Snc1p and chitin synthase Chs3p (data not shown and Sciorra et al., 2005). Second, our study and the work of others (Sciorra et al., 2005) revealed synthetic growth defects of mutants of early endosome-to-TGN recycling with different pik1 mutant alleles. Third, AP-1, a regulator of this retrieval pathway (Valdivia et al., 2002), showed synthetic lethality in an apl4 (AP-1 subunit) pik1 double mutant. This interaction was not identified in our SGA screen but was found by tetrad analysis (data not shown). These findings can be interpreted in different ways. Pik1p and its product PI(4)P could directly participate in TGN-to-endosome trafficking (anterograde or retrograde). Alternatively, the effects on the recycling pathway in pik mutants could be an indirect consequence of impaired TGN-to-late endosome transport, which would then also interfere with early endosome function, e.g., by missorting of late endosome proteins to early endosomes. In favor of the latter hypothesis, a Snc1p recycling defect has also been observed in gga1Δ gga2Δ double mutants (Black and Pelham, 2000). Further studies will be required to distinguish between these different scenarios.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jan Peychl, Jeremy Sanderson, and Kurt Anderson for advice with light microscopy and Susanne Kretschmar for excellent technical assistance with electron microscopy. We are indebted to Yvonne Gloor, Benjamin Glick, Chris Stefan and Scott Emr, Peter Novick (Yale University), Patricia Scott, Annette Boman, Hugh Pelham (MRC Laboratory of Molecular Biology, Cambridge), Anne Spang, and Robert S. Fuller (University of Michigan) for sharing reagents and yeast strains. For helpful discussions and comments on the manuscript, we thank Marcos Gonzalez-Gaitan and Giancarlo Costaguta. This work was supported by the Max Planck Society, by a grant from the Deutsche Forschungsgemeinschaft (SFB 449, A11) to V.H., and in part by Free State of Saxony and EU to T.B.
Abbreviations used:
- CPY
carboxypeptidase Y
- PtdIns
phosphatidylinositol
- PI(3)P
phosphatidylinositol (3)-phosphate
- PI(4)P
phosphatidylinositol (4)-phosphate
- PI(3,5)P2
phosphatidylinositol (3,5)-bisphosphate
- PI(4,5)P2
phosphatidylinositol (4,5)-bisphosphate
- PI
phosphoinositide
- SGA
synthetic genetic array
- SPR
surface plasmon resonance
- SV
secretory vesicle
- TGN
trans-Golgi network.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-10-0937) on February 20, 2008.
REFERENCES
- Audhya A., Foti M., Emr S. D. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. J. Cell Sci. 2005;118:2093–2104. doi: 10.1242/jcs.02387. [DOI] [PubMed] [Google Scholar]
- Baust T., Czupalla C., Krause E., Bourel-Bonnet L., Hoflack B. Proteomic analysis of adaptor protein 1A coats selectively assembled on liposomes. Proc. Natl. Acad. Sci. USA. 2006;103:3159–3164. doi: 10.1073/pnas.0511062103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black M. W., Pelham H. R. A selective transport route from Golgi to late endosomes that requires the yeast GGA proteins. J. Cell Biol. 2000;151:587–600. doi: 10.1083/jcb.151.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman A. L. GGA proteins: new players in the sorting game. J. Cell Sci. 2001;114:3413–3418. doi: 10.1242/jcs.114.19.3413. [DOI] [PubMed] [Google Scholar]
- Boman A. L., Salo P. D., Hauglund M. J., Strand N. L., Rensink S. J., Zhdankina O. ADP-ribosylation factor (ARF) interaction is not sufficient for yeast GGA protein function or localization. Mol. Biol. Cell. 2002;13:3078–3095. doi: 10.1091/mbc.E02-02-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J. S. The GGA proteins: adaptors on the move. Nat. Rev. Mol. Cell Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- Bowers K., Stevens T. H. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Carlton J. G., Cullen P. J. Coincidence detection in phosphoinositide signaling. Trends Cell Biol. 2005;15:540–547. doi: 10.1016/j.tcb.2005.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins B. M., McCoy A. J., Kent H. M., Evans P. R., Owen D. J. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109:523–535. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- Costaguta G., Duncan M. C., Fernandez G. E., Huang G. H., Payne G. S. Distinct roles for TGN/endosome epsin-like adaptors Ent3p and Ent5p. Mol. Biol. Cell. 2006;17:3907–3920. doi: 10.1091/mbc.E06-05-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costaguta G., Stefan C. J., Bensen E. S., Emr S. D., Payne G. S. Yeast Gga coat proteins function with clathrin in Golgi to endosome transport. Mol. Biol. Cell. 2001;12:1885–1896. doi: 10.1091/mbc.12.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Camilli P., Chen H., Hyman J., Panepucci E., Bateman A., Brunger A. T. The ENTH domain. FEBS Lett. 2002;513:11–18. doi: 10.1016/s0014-5793(01)03306-3. [DOI] [PubMed] [Google Scholar]
- De Matteis M. A., Di Campli A., Godi A. The role of the phosphoinositides at the Golgi complex. Biochim. Biophys. Acta. 2005;1744:396–405. doi: 10.1016/j.bbamcr.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Du L. L., Novick P. Yeast rab GTPase-activating protein Gyp1p localizes to the Golgi apparatus and is a negative regulator of Ypt1p. Mol. Biol. Cell. 2001;12:1215–1226. doi: 10.1091/mbc.12.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford M. G., Mills I. G., Peter B. J., Vallis Y., Praefcke G. J., Evans P. R., McMahon H. T. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- Ford M. G., Pearse B. M., Higgins M. K., Vallis Y., Owen D. J., Gibson A., Hopkins C. R., Evans P. R., McMahon H. T. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- Gaidarov I., Keen J. H. Phosphoinositide-AP-2 interactions required for targeting to plasma membrane clathrin-coated pits. J. Cell Biol. 1999;146:755–764. doi: 10.1083/jcb.146.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P., Kornfeld S. The GGA proteins: key players in protein sorting at the trans-Golgi network. Eur. J. Cell Biol. 2004;83:257–262. doi: 10.1078/0171-9335-00374. [DOI] [PubMed] [Google Scholar]
- Godi A., Pertile P., Meyers R., Marra P., Di Tullio G., Iurisci C., Luini A., Corda D., De Matteis M. A. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat. Cell Biol. 1999;1:280–287. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- Govindan B., Bowser R., Novick P. The role of Myo2, a yeast class V myosin, in vesicular transport. J. Cell Biol. 1995;128:1055–1068. doi: 10.1083/jcb.128.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurunathan S., David D., Gerst J. E. Dynamin and clathrin are required for the biogenesis of a distinct class of secretory vesicles in yeast. EMBO J. 2002;21:602–614. doi: 10.1093/emboj/21.4.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H., Schnieders E. A., Thorner J., Takemoto J. Y., DeWald D. B. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:34294–34300. doi: 10.1074/jbc.274.48.34294. [DOI] [PubMed] [Google Scholar]
- Harsay E., Schekman R. A subset of yeast vacuolar protein sorting mutants is blocked in one branch of the exocytic pathway. J. Cell Biol. 2002;156:271–285. doi: 10.1083/jcb.200109077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldwein E. E., Macia E., Wang J., Yin H. L., Kirchhausen T., Harrison S. C. Crystal structure of the clathrin adaptor protein 1 core. Proc. Natl. Acad. Sci. USA. 2004;101:14108–14113. doi: 10.1073/pnas.0406102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- Holthuis J. C., Nichols B. J., Dhruvakumar S., Pelham H. R. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 1998;17:113–126. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honing S., Ricotta D., Krauss M., Spate K., Spolaore B., Motley A., Robinson M., Robinson C., Haucke V., Owen D. J. Phosphatidylinositol-(4,5)-bisphosphate regulates sorting signal recognition by the clathrin-associated adaptor complex AP2. Mol. Cell. 2005;18:519–531. doi: 10.1016/j.molcel.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Itoh T., Koshiba S., Kigawa T., Kikuchi A., Yokoyama S., Takenawa T. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science. 2001;291:1047–1051. doi: 10.1126/science.291.5506.1047. [DOI] [PubMed] [Google Scholar]
- Kelley L. A., MacCallum R. M., Sternberg M. J. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- Legendre-Guillemin V., Wasiak S., Hussain N. K., Angers A., McPherson P. S. ENTH/ANTH proteins and clathrin-mediated membrane budding. J. Cell Sci. 2004;117:9–18. doi: 10.1242/jcs.00928. [DOI] [PubMed] [Google Scholar]
- Lemmon M. A. Phosphoinositide recognition domains. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- Levine T. P., Munro S. Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. Mol. Biol. Cell. 2001;12:1633–1644. doi: 10.1091/mbc.12.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine T. P., Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr. Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- Li X., Rivas M. P., Fang M., Marchena J., Mehrotra B., Chaudhary A., Feng L., Prestwich G. D., Bankaitis V. A. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J. Cell Biol. 2002;157:63–77. doi: 10.1083/jcb.200201037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Manninen A., Verkade P., Le Lay S., Torkko J., Kasper M., Fullekrug J., Simons K. Caveolin-1 is not essential for biosynthetic apical membrane transport. Mol. Cell. Biol. 2005;25:10087–10096. doi: 10.1128/MCB.25.22.10087-10096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald K., Muller-Reichert T. Cryomethods for thin section electron microscopy. Methods Enzymol. 2002;351:96–123. doi: 10.1016/s0076-6879(02)51843-7. [DOI] [PubMed] [Google Scholar]
- Nair J., Muller H., Peterson M., Novick P. Sec2 protein contains a coiled-coil domain essential for vesicular transport and a dispensable carboxy terminal domain. J. Cell Biol. 1990;110:1897–1909. doi: 10.1083/jcb.110.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Field C., Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S., Im Y. J., Hurley J. H., Prinz W. A. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J. Cell Biol. 2006;173:107–119. doi: 10.1083/jcb.200510084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. S. Adaptable adaptors for coated vesicles. Trends Cell Biol. 2004;14:167–174. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Roy A., Levine T. P. Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J. Biol. Chem. 2004;279:44683–44689. doi: 10.1074/jbc.M401583200. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russel D. W. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schiestl R. H., Gietz R. D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schu P. V., Takegawa K., Fry M. J., Stack J. H., Waterfield M. D., Emr S. D. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- Sciorra V. A., Audhya A., Parsons A. B., Segev N., Boone C., Emr S. D. Synthetic genetic array analysis of the PtdIns 4-kinase Pik1p identifies components in a Golgi-specific Ypt31/rab-GTPase signaling pathway. Mol. Biol. Cell. 2005;16:776–793. doi: 10.1091/mbc.E04-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott P. M., et al. GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat. Cell Biol. 2004;6:252–259. doi: 10.1038/ncb1107. [DOI] [PubMed] [Google Scholar]
- Shiba T., Kametaka S., Kawasaki M., Shibata M., Waguri S., Uchiyama Y., Wakatsuki S. Insights into the phosphoregulation of beta-secretase sorting signal by the VHS domain of GGA1. Traffic. 2004;5:437–448. doi: 10.1111/j.1600-0854.2004.00188.x. [DOI] [PubMed] [Google Scholar]
- Takatsu H., Yoshino K., Toda K., Nakayama K. GGA proteins associate with Golgi membranes through interaction between their GGAH domains and ADP-ribosylation factors. Biochem. J. 2002;365:369–378. doi: 10.1042/BJ20020428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Trautwein M., Schindler C., Gauss R., Dengjel J., Hartmann E., Spang A. Arf1p, Chs5p and the ChAPs are required for export of specialized cargo from the Golgi. EMBO J. 2006;25:943–954. doi: 10.1038/sj.emboj.7601007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia R. H., Baggott D., Chuang J. S., Schekman R. W. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev. Cell. 2002;2:283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- Walch-Solimena C., Novick P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat. Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- Wang C. W., Hamamoto S., Orci L., Schekman R. Exomer: a coat complex for transport of select membrane proteins from the trans-Golgi network to the plasma membrane in yeast. J. Cell Biol. 2006;174:973–983. doi: 10.1083/jcb.200605106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Sun H. Q., Macia E., Kirchhausen T., Watson H., Bonifacino J. S., Yin H. L. PI4P promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal. Mol. Biol. Cell. 2007;18:2646–2655. doi: 10.1091/mbc.E06-10-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. W., Mendrola J. M., Audhya A., Singh S., Keleti D., DeWald D. B., Murray D., Emr S. D., Lemmon M. A. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- Zhdankina O., Strand N. L., Redmond J. M., Boman A. L. Yeast GGA proteins interact with GTP-bound Arf and facilitate transport through the Golgi. Yeast. 2001;18:1–18. doi: 10.1002/1097-0061(200101)18:1<1::AID-YEA644>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.