Abstract
Skp1, Cul1, Rbx1, and the FBXO25 protein form a functional ubiquitin ligase complex. Here, we investigate the cellular distribution of FBXO25 and its colocalization with some nuclear proteins by using immunochemical and biochemical approaches. FBXO25 was monitored with affinity-purified antibodies raised against the recombinant fragment spanning residues 2-62 of the FBXO25 sequence. FBXO25 protein was expressed in all mouse tissues tested except striated muscle, as indicated by immunoblot analysis. Confocal analysis revealed that the endogenous FBXO25 was partially concentrated in a novel dot-like nuclear domain that is distinct from clastosomes and other well-characterized structures. These nuclear compartments contain a high concentration of ubiquitin conjugates and at least two other components of the ubiquitin-proteasome system: 20S proteasome and Skp1. We propose to name these compartments FBXO25-associated nuclear domains. Interestingly, inhibition of transcription by actinomycin D or heat-shock treatment drastically affected the nuclear organization of FBXO25-containing structures, indicating that they are dynamic compartments influenced by the transcriptional activity of the cell. Also, we present evidences that an FBXO25-dependent ubiquitin ligase activity prevents aggregation of recombinant polyglutamine-containing huntingtin protein in the nucleus of human embryonic kidney 293 cells, suggesting that this protein can be a target for the nuclear FBXO25 mediated ubiquitination.
INTRODUCTION
Ubiquitin-proteasome system (UPS) controls the abundance of near 80% of all intracellular proteins in eukaryotes (Glickman and Ciechanover, 2002). Proteins destined for degradation by the UPS are first covalently linked to a chain of ubiquitin molecules (ub), which marks them for rapid breakdown to small peptides by the 26S proteasome (Glickman and Ciechanover, 2002). The critical enzymes responsible for attaching ub to protein substrates are the E3 ub-ligases that catalyze the transfer of an activated form of ub from a specific E2 ub-carrier protein to a lysine residue in the substrate (Hershko and Ciechanover, 1998). The E3s are the most numerous and diversified component of the UPS. Three distinct classes of E3 have been identified: the homologous to E6-AP carboxy-terminus domains, really interesting new gene (RING) finger, and U-box domain types (Ardley and Robinson, 2005). The largest class comprises the RING fingers, whose prototype is the SCF that is composed of the invariable components Skp1, Cul1 and Rbx1 (RING finger protein, also named Roc1), and an interchangeable component known as an F-box protein. The F-box protein is the component that contains protein interaction domains for binding the ubiquitination targets (Kipreos and Pagano, 2000; Ang and Harper, 2005; Ardley and Robinson, 2005).
During our studies of structure–function relationship of atrogin-1/FBXO32 and its paralogue FBXO25, we demonstrated that the FBXO25 gene product has the properties of an E3 of the SCF class (Gomes et al., 2001; Maragno et al., 2006). SCF E3s are known to participate in various important cellular processes, but the biological functions of the majority of the F-box proteins, including FBXO25, remain uncharacterized (Jin et al., 2004b). Interestingly, an FBXO25 gene variant has been linked to a genetically inherited cerebral disorder (Hagens et al., 2006). In addition, the level of FBXO25 mRNA is increased in response to interferon β treatment and virus infection, and association of FBXO25 gene with inflammation and tumorigenesis has been proposed (Gorreta et al., 2005; Malathi et al., 2005).
Both Northern blot and reverse transcription-polymerase chain reaction (RT-PCR) studies demonstrated that the predominant site of FBXO25 expression was the central nervous system, although intestine and kidney also showed significant levels of expression (Maragno et al., 2006; Hagens et al., 2006). Few studies have investigated the subcellular localization of F-box proteins, whose results relied on the detection of the respective overexpressed protein as in the case of FBXO25 (Kipreos et al., 2000). Previous research of different groups, including ours, has demonstrated that tagged FBXO25 in cultured cells exhibits a diffuse distribution pattern in the nucleus, and it is excluded from the nucleoli (Hagens et al., 2006; Maragno et al., 2006).
In the present study, we examine the cell cycle dependency of the subcellular localization of endogenous FBXO25 in cultured cells and the expression of the FBXO25 protein in mouse tissues by using immunochemical approaches. Also, we investigate the association of FBXO25 with other subnuclear components and the effects of inhibiting the transcription process on the nuclear distribution of this enzyme. The nuclear ubiquitin ligase activity of FBXO25 was probed using an assay for nuclear aggregation of polyglutamine-containing proteins in cultured cells.
MATERIALS AND METHODS
Materials
Anti-20S (1:600), anti-Skp1 (1:50), anti–β-tubulin (1:2000) mouse monoclonal antibodies (mAbs), and Prolong gold antifade reagent with 4,6-diamidino-2-phenylindole (DAPI) were obtained from Invitrogen (Carlsbad, CA). Anti-hemagglutinin (HA; 1:4000), anti-SC35 (1:100), anti-survival motor neuron protein (1:100), anti-p80-coilin (1:40), anti-B23-nucleophosmin (1:100), anti-FLAG (1:3000), and anti-β-actin (1:3000) mouse mAbs were obtained from Sigma-Aldrich (St. Louis, MO). Anti-promyelocytic leukemia protein (PML; 1:100), anti-glutathione transferase (GST; 1:1000), and anti-green fluorescent protein (GFP; 1:1000) mAbs were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-α-tubulin (1:300) mAb was purchased from GE Healthcare (Little Chalfont, Buckinghamshire, United Kingdom). Mouse anti-polyubiquitinylated proteins (FK2; 1:10,000) was purchased from BIOMOL Research Laboratories (Plymouth Meeting, PA). Secondary antibodies conjugated with Alexa Fluor 488 (1:300) and Alexa Fluor 594 (1:300) were obtained from Invitrogen. Secondary antibodies conjugated with cyanine 3 (1:300) were obtained from Sigma-Aldrich.
Plasmids
The pENTRy-HA-FBXO25-FLAG, pDEST27-HA-FBXO25-FLAG (GST-tagged) GATEWAY constructs were described previously (Maragno et al., 2006). Gateway recombination were used to subclone HA-FBXO25-FLAG (wild type [WT] and ΔF) into pDEST53 (N-terminal enhanced GFP [EGFP] fusion) and pDEST12 (Invitrogen).
Reverse Transcription-Polymerase Chain Reaction
Total RNA was extracted from cells using TRIzol reagent (Invitrogen). Reverse transcription was done with random primers and SuperScript II (Invitrogen). The RT-PCR assay was done as described previously (Maragno et al., 2006). Primers used for huntingtin were HT-F, 5′-ACCCTCGTGACCACCCTGACCTAC-3′ and HT-R, 5′-GGACCATGTGATCGCGCTTCTCGT-3′. Primers used for β-actin were ACT-F, 5′-CTAAGGCCAACCGTGAAAAGA-3′ and ACT-R ACT-R, 5′-ATTGCCGATAGTGATGACCTG-3′.
Production of Affinity-purified Anti-FBXO25 Antibodies
To make GST-tagged fusion with an ∼7-kDa NH2-terminal fragment of FBXO25, cDNA was amplified using IMAGE 4240953 as a template and NT2-F (5′-GGGAATTCCCGTTTCTGGGTCA-3′) and NT2-R (5′-CGGCGGCCGCGGCTGCGTATTCAC-3′) primers; the product was digested with EcoRI and NotI, and it was subcloned into pGEX4T1. This FBXO25 fragment was purified from Escherichia coli DH-5α by using the glutathione-Sepharose affinity matrix, and it was digested with thrombin according to the manufacturer's instructions (GE Healthcare). The polyacrylamide gel band containing ∼150 μg of the thrombin-released fragment of FBXO25 was excised, and it was cut into 1-mm3 pieces, which were finely ground in a mortar before preparing the emulsion with complete Freund's adjuvant. Then, the emulsion was injected into a New Zealand rabbit (Supplemental Figure S1). This initial immunization was followed by booster doses (∼150 μg) of FBXO25 fragment in incomplete Freund's adjuvant given with 3-wk intervals. Serum was obtained and processed using established protocols (Harlow and Lane, 1988). Anti-FBXO25 antibodies were affinity-purified from the serum according to the procedures of Harlow and Lane (1988), by using a Sepharose-matrix (GE Healthcare) onto which the purified FBXO25 fragment had been covalently linked. Bound antibodies were eluted using 100 mM glycine, pH 2.8, and they were used for immunolocalization microscopy and immunoblot studies after appropriate dilution.
Preparation of Nuclear Extracts
The nuclear extract was prepared by a modification of a previously described procedure (Zhou et al., 2004). Briefly, HeLa cells were collected, washed and lysed in buffer A (10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, and 10 mM HEPES, pH 7.9, containing a cocktail of inhibitors). After 15-min incubation on ice, 0.1% Triton X-100 was added to the homogenates, and the tubes were vigorously rocked for 1 min. Then, the homogenate was centrifuged 20,800 × g for 5 min in a microcentrifuge at 4°C. The supernatant fluid (cytoplasmatic extract) was separated. The nuclear pellets were washed once with buffer A, and then they were suspended in 50 μl of buffer B (420 mM NaCl, 0.1 mM EDTA, 0.1 mM EGTA, and 10 mM HEPES, pH 7.9, containing a cocktail of inhibitors) and vigorously vortexed for 30 min. This solution was centrifuged 20,800 × g for 5 min, and the supernatant fluid (nuclear extract-1, N) was separated. The pellet was then solubilized in radioimmunoprecipitation assay (RIPA) buffer (300 mM NaCl, 2% NP-40, 0.1% DOC, 0.2% SDS, and 100 mM Tris-HCl, pH 7.5), sonicated, and centrifuged 20,800 × g for 10 min. The supernatant fluid (nuclear extract-2, NP) was separated, and it was used as a source of protein for the immunoblots.
Western Blotting
For preparation of whole-cell lysates, cells were washed with phosphate-buffered saline (PBS), suspended in 4 volumes of 2× RIPA buffer containing a cocktail of protease and phosphatase inhibitors, and sonicated on ice bath by 40 s. Lysates were then obtained as the supernatant fractions after centrifugation at 20,800 × g for 10 min. Mouse tissue lysates were similarly prepared by freezing the corresponding tissues in liquid nitrogen before grinding with a mortar and pestle and suspending the resulting powder in 2× RIPA buffer containing protease and phosphatase inhibitors (1:4, mass:volume). After sonication and centrifugation as described above, each lysate was recovered as the supernatant fraction. One hundred and fifty micrograms of protein from each lysate was subjected to SDS-polyacrylamide gel electrophoresis (PAGE), transferred onto nitrocellulose membrane and probed with affinity-purified anti-FBXO25 antibodies (1:1500). Horseradish peroxidase-conjugated secondary antibodies were used to detect the primary antibodies. Antibodies were visualized by the enhanced chemiluminescence method (Santa Cruz Biotechnology). Protein concentration in the cell lysates was determined using a Bio-Rad protein assay kit (Bio-Rad, Richmond, CA).
Cell Culture, Synchronization, and Cell Cycle Analysis
For expression of GST/HA/FLAG-, EGFP/HA/FLAG-tagged proteins, HEK293H (Invitrogen) cells were grown in DMEM (Sigma-Aldrich) in 10-cm-diameter dishes supplemented with 10% fetal bovine serum. The plasmid constructs were transfected with into HEK293H cells at 60–80% confluence by using either the calcium phosphate transfection method (Ausubel et al., 1997) or Lipofectamine 2000 (Invitrogen). For the establishment of the cell line, cells were cultured in the presence of Geneticin (Invitrogen) at a concentration of 1 mg/ml. After a period of 2–3 wk, resistant colonies were isolated and tested for the FBXO25 expression. Red fluorescent protein (RFP)-tagged PML-IV plasmids were transfected into HeLa cells using Superfect (QIAGEN, Valencia, CA). Cultured cells were exposed to 5 and 0.05 μg/ml actinomycin D (Sigma-Aldrich) for 2 h, 100 μM α-amanitin (Sigma-Aldrich) for 3 h, and 50 μg/ml dichlororibofuranosylbenzimidazole (DRB; Sigma-Aldrich) for 5 h of to inhibit the transcription. HeLa cells were incubated for 12 h with the proteasomal inhibitor MG132 (5 μM; BostonBiochem, Boston, MA). To induce PML stress, HeLa cells were incubated in 50 μM CdCl2 (Sigma-Aldrich) for 4 h. HeLa cells were synchronized by blocking the cells with thymidine at G1/S as described previously (Stein and Borun, 1972). Cells were treated with 2 mM thymidine (Sigma-Aldrich) for 12 h. Cells were released from the thymidine block by incubation in PBS followed by incubation in serum containing DMEM supplemented with 24 μM deoxycytidine. After 9 h, cells were refed with fresh media containing 2 mM thymidine for 12 h, and subsequently they were released as described above. After release from the double thymidine block, cells were harvested at 1- and 2- to 4-h intervals. Cell cycle distribution was determined by fluorescence-activated cell sorting (FACSORT; BD Biosciences, San Jose, CA). At each time, HeLa mitotic cells extracts were prepared and processed for protein blots as described above. For microscopic studies of mitosis, cells were double-labeled with anti-FBXO25, anti-α-tubulin and costained with DAPI.
Biochemical Partitioning
HeLa cells extracts were prepared in four buffers containing different concentrations of salts and detergents by a modification of a previously described procedure (Platani et al., 2000). HeLa-adhered cells (2 × 10-cm-diameter cell culture dishes) were washed in PBS, centrifuged, and the cell pellet was resuspended and incubated in 1 ml of buffer-1 (10 mM Tris-HCl, pH 7.4, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2, 1 mM EGTA, 0.5% Triton X-100, and 0.2 mg/ml phenylmethylsulfonyl fluoride [PMSF]). This pellet was centrifuged at 20,800 × g for 5 min to produce supernatant 1 and pellet 1. Supernatant 1 was stored, whereas pellet 1 was resuspended and incubated in buffer-2 (10 mM Tris-HCl, pH 7.4, 250 mM KCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, and 0.2 mg/ml PMSF). This pellet was centrifuged at 20,800 × g for 5 min to produce supernatant 2 and pellet 2. Supernatant 2 was stored, and pellet 2 was resuspended in buffer-3 (10 mM Tris-HCl, pH 7.4, 300 mM sucrose, 50 mM NaCl, 3 mM MgCl2, 1 mM EGTA, and 0.5% Triton X-100 with 400 U/ml DNase-I) and incubated at 32°C for 50 min. This pellet was centrifuged at 20,800 × g for 5 min to produce supernatant 3 and pellet 3. Supernatant 3 was stored, and pellet 3 was resuspended in buffer-4 (RIPA) and sonicated. Protein samples from supernatants 1–4 and the pellet from the last extraction were analyzed by SDS-PAGE and immunoblotting.
Immunofluorescence Microscopy
For indirect immunofluorescence, HeLa (CCL-2; American Type Culture Collection, Manassas, VA), HEK293H (Invitrogen), HEK293T (CRL-11268; American Type Culture Collection), COS-7 (CRL-1651; American Type Culture Collection), IMCD (CRL-2123; American Type Culture Collection), LLC-PK1 (CL-101; American Type Culture Collection), and MCI (CRL-1927; American Type Culture Collection) cells were grown on glass coverslips in DMEM supplemented with 10% fetal calf serum. Leydig cells were isolated from Swiss mice as described previously (Costa and Varanda, 2007). The cells were fixed and permeabilized for 10 min at room temperature (RT) with PBS containing 2% paraformaldehyde, 0.3% Triton X-100, and 10 μM taxol, and they were blocked with PBS/2% bovine serum albumin (BSA) containing 5% goat immunoglobulin (Ig)G. Antibodies incubations were performed 1 h at RT in PBS/2% BSA followed by incubation with Alexa 488- and Alexa 594-coupled secondary antibodies (Invitrogen). Coverslips were mounted with Prolong gold antifade mounting medium containing DAPI (Invitrogen). Samples were analyzed with a Leica TCS SP5 laser scanning confocal microscope (Leica Microsystems, Wetzlar, Germany). Preincubation of the FBXO25 fragment with affinity-purified antibodies abolished all signal produced by the antibodies during immunofluorescence. For quantitative analysis, images were examined by confocal microscopy, and FBXO25 associated nuclear domains (FANDs) and clastosomes were counted; the total number of FANDs, clastosomes, and the number of colocalizing dots were counted in 100 cells from randomly chosen fields in each of four independent microscope slides.
In Vivo Incorporation of Bromouridine-Triphosphate (BrUTP)
The in vivo transcription assay was performed as described previously (Chen et al., 2005), with slight modifications. HeLa cells that were grown directly on glass slides were rinsed once with PBS and once with a glycerol buffer (20 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 25% glycerol, 0.5 mM PMSF, and 0.5 mM EGTA). Cells were then permeabilized in the glycerol buffer containing 5 μg/ml digitonin at RT for 5 min. Subsequently, cells were incubated in transcription buffer (50 mM Tris-HCl, pH 7.4, 100 mM KCl, 5 mM MgCl2, 0.5 mM EGTA, 25% glycerol, 1 mM PMSF, 2 mM ATP, 0.5 mM CTP, 0.5 mM GTP, 0.2 mM BrUTP, and 25 U/ml RNAsin [Promega, Madison, WI]) for 10 min at 37°C and 5% CO2. Uridine incorporation was visualized by monoclonal anti-5-bromo-2′-deoxyuridine conjugate to biotin (Invitrogen) followed by incubation with streptavidin conjugate to Alexa 594 (Invitrogen).
Filter Retardation Assay
The filter assay used to detect polyglutamine-containing huntingtin protein aggregates was carry out as described previously (Sittler et al., 1998; Wanker et al., 1999). Briefly, FBXO25WT, or FBXO25ΔF in pDEST12, was cotransfected with of or HA-Skp1, FLAG-Cul1, Myc-Roc1, and EGFP-httEx1-74Q into HEK293T cells at 60–80% confluence by using calcium phosphate. After 24 h, cells were collected, washed, and lysed 30 min on ice in lysis buffer containing 50 mM Tris, pH 8.8, 100 mM NaCl, 5 mM MgCl2, 0.5% NP-40, 1 mM EDTA, and supplemented with a cocktail of protease inhibitors. Total extracts were centrifuged at 20,800 × g for 10 min at 4°C to separate soluble proteins from aggregates. Pellets were washed with PBS and further incubated 1 h at 37°C in a DNase-I buffer (20 mM Tris, pH 8.0, and 15 mM MgCl2) containing 0.5 mg/ml DNase-I. Subsequently, pellets were heated at 95°C for 5 min in 1% SDS, and then they were spotted onto a 0.2-μm pore cellulose acetate membrane (Whatman Schleicher and Schuell, Dassel, Germany) by using a BRL dot-blot filtration unit (Invitrogen). The cellulose membranes were probed with the anti-GFP, and then they were subjected to densitometric scanning by ImageJ software (http://rsb.info.nih.gov/ij/).
RESULTS
Tissue Distribution of FBXO25 Protein in the Mouse
To investigate the tissue distribution and cellular localization of FBXO25, we prepared rabbit polyclonal antibodies to a recombinant NH2-terminal fragment of FBXO25 (residues 2–62; ∼7 kDa) as a source of specific antibody that was purified by affinity chromatography using the cognate FBXO25 fragment as the ligand (Supplemental Figure S2). Antibodies recovered from immune serum and preimmune serum were used in Western blots. The anti-FBXO25 antibodies reacted with a 42-kDa protein of tissue and cell extracts, in good agreement with the predicted molecular weight for the FBXO25 mouse gene product. The distribution and relative expression of FBXO25 protein were examined by Western blotting analysis of some mouse tissue extracts. Figure 1A shows that the antibody recognized an ∼42-kDa protein (lanes 1, 2, and 4) or a doublet ∼42–44 kDa (lanes 3–5) in all tissues tested, except in heart and skeletal muscle. In the liver, an additional protein of ∼55 kDa was detected with equal intensity to that of FBXO25. It remains to be determined whether this protein represents an alternatively spliced form of FBXO25. High levels of FBXO25 expression was detected in testis, spleen and brain and low levels in kidney, liver, and intestine. Preimmune serum did not react with FBXO25 (data not shown). We also used the anti-FBXO25 antibodies to examine FBXO25 protein expression in cell culture lysates by immunoblot analysis. FBXO25 was detected in HEK293T/H, HeLa, COS-7, MCI, IMCD, and LLC-PK1 (Figure 1B). In addition, anti-FBXO25 reacted with a 60-kDa overexpressed GST-tagged FBXO25 (GST-FBXO25WT) protein in HEK293H cells stably transfected (HEK293HFB25-WT-1), in good agreement with the predicted molecular weight for the tagged protein (Figure 1C). The specific FBXO25 bands were no longer detected when the affinity-purified anti-FBXO25 antibodies were preincubated with the FBXO25 fragment (Figure 1C). The Western blots clearly demonstrated that anti-FBXO25 recognized specifically FBXO25 protein in cells and tissue extracts.
Figure 1.
Expression of FBXO25 protein in mouse tissues and cultured cell lines. Approximately 150 μg of protein from the indicated tissues (A) and cultured cells (B) lysates were subjected to SDS-PAGE, transferred onto nitrocellulose membranes, and probed with affinity-purified anti-FBXO25 antibodies (1:1500). Tissue and cell extracts were prepared as described in Materials and Methods. The specificity of the anti-FBXO25 antibodies (C) were ascertained by probing twin blots prepared from SDS-PAGE loaded with two separate sets of cell lysates from HEK293H cells and HEK293H cells stably transfected with GST-FBXO25 (HEK293HFB25-WT-1), with antibodies in the absence and presence of 6 μg/ml recombinant FBXO25 N-terminal fragment. Note that the FBXO25 fragment fully blocked the antibody reaction with both the endogenous and GST-fusioned protein. Ponceau-S staining showed protein loading (middle) and anti-GST antibodies labeling showed the expression of GST-FBXO25 protein in HEK293HFB25-WT-1 cells (bottom).
Subcellular Distribution of FBXO25 in Cultured Cells
The anti-FBXO25 antibodies, whose specificity and selectivity were ascertained by immunoblot analysis, were then used to probe in some detail the localization of FBXO25 within cells. The anti-FBXO25 antibodies labeled HeLa cells predominantly at the nuclei both with a diffuse and with a dot-like pattern, but they did not stain nucleoli (Figure 2, A–C). Thirty to 40% of the HeLa cell population analyzed contained at least one to four brightly labeled dot-like structures in the nucleoplasm, although some cells contained up to 10 dots (Figure 2, A–C). These dot-like structures were heterogeneous in size and shape, and they were randomly located in the interior of the nucleus. Confocal microscopy analysis demonstrated that the staining was present in planes inside the nucleus, not just on the surface. A similar staining pattern was also observed in the COS-7 and HEK293H cells (Figure 2, A–C). Only a small fraction of the LLC-PK1, IMCD, and Leydig cells showed this staining pattern revealed with anti-FBXO25 antibodies (Supplemental Figure S3).
Figure 2.
Mammalian cell nuclei contain domains highly enriched in FBXO25. Affinity-purified antibodies directed against FBXO25 were used to perform immunofluorescence on a variety of mammalian cell types. These included HeLa (A), COS-7 (B), and HEK293H (C). Note that anti-FBXO25 antibodies label both nucleus and cytoplasm, staining of the nucleoplasm is more intense. Observe the presence of bright dot-like structure within the nucleoplasm. DAPI was used to stain nuclei and images were taken by confocal microscopy. Bars, 5 μm.
During the characterization of the anti-FBXO25 antibodies, we found that FBXO25 was partially resistant to detergent extraction. To further examine this retention of FBXO25 in the nucleus during biochemical partitioning, we performed a series of progressively more stringent extractions with or without DNase-I treatment (Figure 3A). The results demonstrated a strong binding of FBXO25 to the nuclear fraction, and they indicated that it was partially resistant to extraction with Triton X-100 (Figure 3A, lane 1). Substantial amounts of the protein were solubilized only in Triton X-100 plus DNase-I (Figure 3A, lanes 3), indicating that FBXO25 is tightly bound to the chromatin.
Figure 3.
Nuclear association of FBXO25. (A) Biochemical partitioning. Whole-cells extracts of controls HeLa cells or cells treated were prepared under the indicated conditions (see Materials and Methods) and analyzed by immunoblotting using anti-FBXO25 antibodies. (B) HeLa cells (T) were fractionated into nucleoplasm (N) and insoluble nuclear pellet (NP) as described in Materials and Methods. Proteins were separated by SDS-PAGE and the corresponding blotted nitrocellulose membranes were probed with anti-FBXO25, anti-nucleophosmin (NPM), and anti-β-tubulin antibodies. (C) An overlay of the differential interference contrast (DIC) microscopy image of HeLa cells and the corresponding FBXO25 immunostaining image generated by affinity-purified anti-FBXO25 antibodies is shown in Ci. Confocal analysis of cells stained for FBXO25 and nuclei/nucleoli with the affinity-purified anti-FBXO25 antibodies and anti-B23 nucleophosmin (anti-NPM) antibodies, respectively, is shown in Cii. FBXO25 is predominantly confined to the nucleus, outside nucleoli structures.
We performed cell fractionation on HeLa cells and nuclear compartments were efficiently enriched as determined by probing the fractions for nucleophosmin and β-tubulin, markers of the nuclear and cytoplasmic fractions, respectively (Figure 3B). The anti-FBXO25 antibodies recognized a protein with molecular mass of ∼42 kDa in Western blot of nuclear fractions (Figure 3B). When examined by confocal microscopy, FBXO25 did not show coincident localization with the nucleolar compartment as determined using antibodies to nucleophosmin as a marker for the nucleoli in HeLa cells (Figure 3C).
To confirm that the dot-like nuclear staining pattern observed with anti-FBXO25 antibodies indeed indicated preferred localization of FBXO25, we transiently expressed exogenous FBXO25 in HEK293H cells. An EGFP fusion with the NH2 terminus of FBXO25 resulted in a nuclear labeling when transiently expressed in HEK293H cells (Figure 4A). The fusion protein showed a pattern similar to that observed when anti-FBXO25 antibodies were used to label untransfected cells, including the formation of the dot-like structures (23 ± 4/100). The tagged FBXO25 did not accumulate in nucleoli, as indicated by immunolabeling for nucleophosmin (Figure 4Aii). The band of the EGFP-FBXO25 fusion protein migrated to the expected size of 66 kDa on the Western blotting membrane (Figure 4B).
Figure 4.
Localization of overexpressed tagged-FBXO25 in HEK293H cells. (A) HEK293H cells transfected with EGFP/HA-FBXO25-FLAG were fixed and immunostained with anti-NPM antibodies. (B) Western blotting analysis of transiently expressed EGFP-FBXO25 protein. Approximately 50 μg of protein from the nontransfected (NT) and transfected cells (T) lysates were subjected to SDS-PAGE, transferred onto nitrocellulose membranes and probed with monoclonal anti-GFP antibodies.
FBXO25 Localizes in a Novel Nuclear Compartment
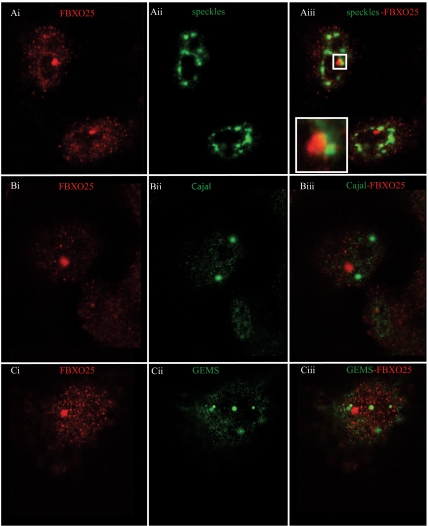
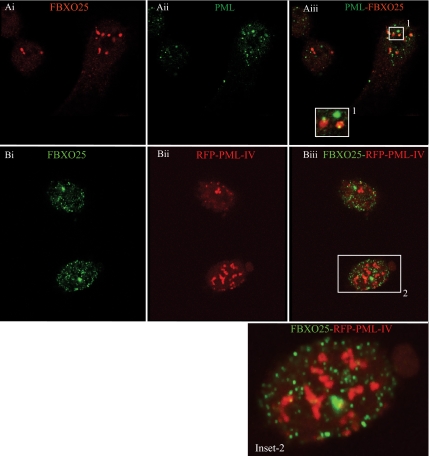
The dot-like structure containing FBXO25 in the nucleoplasm was not readily distinguishable from other known subnuclear compartments. Thus, double-labeling experiments were done using the HeLa cells to determine whether they were coimmunolabeled for both FBXO25 and known markers proteins of other nuclear bodies (Figure 5, A–C). As shown in Figure 5A, no colocalization of FBXO25 with speckles was detected using an antibody specific for the splicing factor SC35 (Figure 5A). However, we found that FBXO25 localize in some dots that were in proximity with SC35 (Figure 5Aiii, box). At a higher magnification (Figure 5Aiii, inset), we observed that in these areas FBXO25 were juxtaposed with SC35, suggesting an interaction of these structures. A double-labeling experiment with anti-p80-coilin monoclonal antibodies (a marker for Cajal bodies) confirmed that there is also no colocalization with FBXO25 (Figure 5B). Furthermore, no colocalization of FBXO25 with Gemini of coiled bodies (GEMS), a nuclear compartment labeled by antibodies against the SMN, was detected (Figure 5C). Another important subnuclear particle that has been studied contains the PML (Zhong et al., 2000). Double labeling using an anti-PML monoclonal antibodies that react against of all isoforms of PML proteins showed that the PML overlap or juxtapose with FBXO25 in some dots (Figure 6Aiii, box). Interestingly, a subset of PML bodies enriched in components of the UPS were suggested to be an important site of protein degradation in the nucleus (Lafarga et al., 2002; Rockel et al., 2005). To assess whether the FBXO25 colocalize with this subset of PML protein, RFP-PML-IV constructs were expressed in HeLa cells, and then they were immunolabeled for FBXO25. As shown in Figure 6B, FBXO25 foci did not show coincident localization with PML-IV bodies. Additionally, cadmium chloride, which is known to disassemble PML bodies (Nefkens et al., 2003) showed no effect on the nuclear distribution of FBXO25-enriched bodies compared with control cells (Supplemental Figure S4). As reported previously (Lafarga et al., 2002), anti-20S antibodies label both the nucleus and the cytoplasm seen as a diffuse pattern, although a few cells contained, in addition, at least one discrete brightly labeled 26S clastosome structure in the nucleoplasm (Figure 7A). In double-labeling experiments, we observed that a fraction of FBXO25 foci of ∼15% accumulated in some of the proteasome-enriched structures that resembled 26S clastosomes (Figure 7A). The remaining 85% of the FBXO25 foci did not colocalize with clastosomes (Figure 7, B and C). However, proteasome inhibitor MG132, which is known to disassemble 26S clastosomes (Lafarga et al., 2002) showed no effect on the nuclear distribution of FBXO25 foci (Supplemental Figure S5). Double-labeling experiments revealed that endogenous FBXO25 and Skp1 colocalize in the brightly labeled nuclear structures (Figure 7D). Interestingly, the relatively abundant Skp1 protein showed an unexpectedly low, diffused staining within the nucleoplasm. As proposed previously (Freed et al., 1999), the weak diffuse staining of Skp1 may be attributed to extraction of this small and very soluble protein during fixation of the cells; it is possible that only the fraction of complexed Skp1 remains bound to cells after fixation to be revealed with anti-Skp1 antibodies. Altogether, we proposed that FBXO25 was associated with a novel subnuclear structure, which we named FAND.
Figure 5.
FBXO25 protein is found in distinct subnuclear domain. Confocal microscopy of cells double stained with affinity-purified anti-FBXO25 antibodies and antibodies against SC35, which label splicing speckles (Ai–Aiii), or against p80-coilin, which label Cajal bodies (Bi–Biii), or against SMN, which label GEMS (Ci–Ciii). The proteins labeled in each panel are indicated in the top left and right of the panel in the relevant color.
Figure 6.
FBXO25 protein is organized in subnuclear structures distinct from PML-IV clastosomes in HeLa cells. (A) Confocal analysis of cells labeled with the FBXO25 and PML antibodies. PML bodies were labeled with monoclonal antibodies that react against of all isoforms of PML proteins (Ai–Aiii). (B) HeLa cells were transiently transfected with RFP-PML isoform IV and immunostained with the FBXO25 antibodies to detect sites of colocalization. FANDs did not colocalize with a large ring-like structure of PML-IV clastosomes (Bi–Biii).
Figure 7.
(A) Double labeling confocal experiments for the detection of FBXO25 and proteasomes in HeLa cells. (A) Proteasomes are accumulated in domains resembling clastosomes (Aiii, inset). (B) Proteasomes show a diffuse nucleoplasmic pattern. (C) Density and colocalization of FANDs and clastosomes in nuclei of HeLa cells as determined by counting 100 cells in each of four microscopic-slides. (D) Confocal analysis of cells double-labeled with antibodies against FBXO25 and Skp1 (Ci–Ciii). The labeled proteins in each panel are indicated in the top left and right of each panel in the respective color.
Effect of Inhibiting RNA Transcription
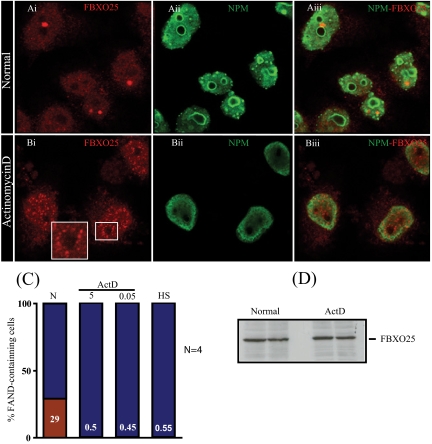
In mammalian cells, the composition and localization of intranuclear bodies respond to changes in transcription, protein phosphorylation, and methylation (Lyonet al., 1997; Shav-Tal et al., 2005; Gary and Clarke, 1998). Inhibition of transcription using actinomycin D (ActD) disrupts Cajal bodies, GEMS, and enlarges speckles (Cioce and Lamond, 2005; Lamond and Spector, 2003; Pellizzoni et al., 2001). To investigate whether the localization of FBXO25 was dependent on active transcription, we initially treated HeLa cells with 5 μg/ml ActD for 2 h, which inhibits transcription by blocking the activities of RNA polymerases I, II, and III. As seen in Figure 8, A and B, ActD treatment caused complete disruption of FANDs. Hardly any cells (0.5 ± 0.2/100) retained FANDs after ActD treatment, compared with 29% (29 ± 2/100) in untreated control cells (Figure 8C). Compared with nuclei in nontreated cells, more diffuse fluorescence could be seen, suggesting that FBXO25 was reorganized from this FBXO25-containing compartment as a consequence of the ActD treatment. An interesting finding is that FBXO25 formed perinucleolar structures in HeLa cells treated with ActD (Figure 8Biii). Identical results were obtained with COS-7 cells, which showed FBXO25-specific dots without treatment, and diffuse nuclear distribution of FBXO25 after treatment with ActD (data not shown).
Figure 8.
Inhibition of RNA polymerases using actinomycin D disrupts FBXO25-associated nuclear domains. Confocal microscopy of labeled FBXO25 in untreated and ActD-treated (5 μg/ml for 2 h) HeLa cells are shown. Without ActD treatment, FANDs were found in the nucleoplasm (Ai–Aiii). In the presence of ActD, the majority of endogenous (Bi–Biii) FANDs disappeared (B). Note that FBXO25 occurs in perinucleolar structures in cells treated with ActD (Bi, inset). (C) Proportion of cells containing FANDs was estimated. After treatment with ActD, the proportion of cells containing FANDs (red) significantly differs from the control population. Four separate experiments were performed, and ∼100 cells were analyzed for drug treatment. CR, untreated; 0.5, ActD, 0.5 μg/ml; ActD 0.05, 0.05 μg/ml; and HS, heat shock. (D) Western blotting analysis of FBXO25 from protein extracts of cells at time points as shown in A. ActD does not alter FBXO25 subcellular levels.
To explore this further, we treated HeLa cells with α-amanitin (50 μg/ml for 5 h) or DRB (100 μM for 3 h), which block only RNA polymerase II (Weinmann et al., 1975; Granick, 1975). Unexpectedly, FANDs did not disassemble (data not shown). However, when ActD was added to the HeLa cells plates at a concentration of 0.05 μg/ml for 2 h, sufficient to inhibit only polymerase I (Perry and Kelley, 1970), FANDs were severely disrupted in the entire all population (0.45 ± 0.2/100; Figure 8C). Together, these results suggest that the distribution of the domain depend on RNA polymerase I activity.
No evident change in the amount of the FBXO25 protein was observed by Western blotting analysis of extracts from HeLa cells subjected to short exposure to ActD (Figure 8D), suggesting that the disappearance of FANDs was due to relocalization of FBXO25 rather than to its nuclear degradation. In parallel experiment we showed that endogenous nucleophosmin was relocalized from the nucleoli to nucleoplasm after transcription inhibition (Figure 8B). In addition, we tested whether ActD affected the biochemical state of FBXO25. HeLa cells were first extracted with 0.5% Triton X-100, followed by treatment with 250 mM KCl and subsequent DNase-I digestion. Protein blotting analysis using anti-FBXO25 revealed ActD does not alter the solubility or subnuclear partitioning of FBXO25 (Figure 3B). Finally, we demonstrated by pull-down experiments that ActD treatment does not impede the interaction between FBXO25 and Skp1 (Supplemental Figure S6).
The above-mentioned experiments suggested a correlation between transcriptional state and the localization of FANDs within the nucleoplasm. In an attempt to strengthen this correlation, experiments in which transcription was inhibited by heat shock (Yost and Lindquist, 1986; Bond, 1988) were performed, a condition that causes redistribution of different subnuclear structures (Zeng et al., 1997; Chiodi et al., 2000). It was observed that both heat-shock treatment at 42°C for 1 h and incubation with 5 or 0.05 μg/ml ActD for 2 h caused nearly complete disruption of FANDs in HeLa cells nuclei (Figure 8C).
FANDs Are Dispersed during the Cell Cycle
We observed significant variability in the incidence and quantity of FANDs among asynchronous cells, suggesting that these structures might assemble and disassemble in coordination with the cell cycle. To investigate this further, we synchronized HeLa cells by using the double thymidine block. As expected, FACS analysis revealed a marked accumulation of HeLa cells in the G1/S phase of the cell cycle after thymidine treatment (Supplemental Figure S7). Cells were double-labeled with anti-FBXO25 and anti-α-tubulin antibodies, and they are costained with DAPI to reveal by confocal microscopy any distributional relationship that might exist between FBXO25, chromatin, and spindles throughout mitosis. There seems to be a sharp transition in the assemblage of FANDs, because they became undetectable as soon as the cells entered S phase (Figure 9). From prophase through metaphase, FBXO25 is diffusely localized in the nucleoplasm (Figures 9 and 10). FANDs reappear in late telophase and disappear again in S phase (Figures 9 and 10). In prophase, FBXO25 was restricted to the remaining nonchromosomal nuclear space (Figure 10). From metaphase through telophase, FBXO25 showed no accumulation with condensed chromosomes or association with the mitotic spindle or other relevant structures. Interestingly, we detected no significant change in the amount of FBXO25 protein at different stages of the cell cycle (Figure 9B). These studies demonstrate that endogenous FANDs are regulated during the cell cycle and that their appearance correlates with the onset of transcriptional activity. To our knowledge, this is the first evidence that a subnuclear structure is G1/telophase specific.
Figure 9.
Distribution of FANDs throughout cell division. (A) Three separate experiments were performed, and ∼100 cells were analyzed for each mitotic phase. The percentages of cells containing FANDs are indicated (red). (B) Western blotting of FBXO25 protein levels during the cell cycle. HeLa cell lysates were prepared by harvested cells in RIPA buffer at the indicated time points after release from the thymidine arrest. RIPA cell extracts (60 μg/lane) were processed for protein blots. The membrane was stripped and reprobed with an anti-β-actin antibodies.
Figure 10.
FANDs are dispersed throughout the nucleoplasm during mitosis. HeLa cells released from a double thymidine block at the G1/S were immunolabeled using antibodies to the FBXO25 (red), costained with α-tubulin (green) and DAPI (blue) to identify the mitotic phase. Merged images are shown in the last column.
FANDs Are Major Sites of Ubiquitination but Not of Transcription
As described previously (Lafarga et al., 2002; Janer et al., 2006), ub-conjugates concentrate in immunochemically well-defined nuclear structures, a pattern also observed in FBXO25-containning nuclear domains (Figure 11A). Remarkably, double-labeling experiments in HeLa cells revealed that FANDs colocalized perfectly with some of the polyubiquitinated protein-enriched nuclear structures (Figure 11A). To our knowledge, there has been no previous evidence of an E3 ligase localized in a major focal site of ubiquitination.
Figure 11.
FANDs are major focal sites of ubiquitination but not transcription. (A) Ubiquitin-conjugates were labeled with antibody FK2, specific for conjugated ubiquitin but not for free ubiquitin (Fujimuro et al., 1994). The localization of FBXO25 (red) and ubiquitin-conjugates (green) are shown. (B) The transcription sites in HeLa cells were labeled using in vivo Br-UTP incorporation, followed by detection with anti-BrdUTP. No labeling was observed in control labeling in the absence of Br-UTP (data not shown). The localization of FBXO25 (green) and BrRNA (red) are shown.
Next, we investigated whether FANDs were transcriptionally active using in vivo Br-UTP incorporation. HeLa cells were permeabilized and incubated with BrUTP. The nascent Br-transcripts were labeled with an anti-BrdUTP biotin-conjugated antibody and visualized with Alexa Fluor 594 streptavidin. Double labeling with anti-FBXO25 antibodies revealed that the major Br-UTP–labeled domains never overlapped with FANDs (Figure 11B). The specificity of this labeling was tested in experiments performed without Br-UTP incorporation (data not shown).
SCFFBXO25 Prevents Polyglutamine Amyloid Fiber Formation
Several studies have associated UPS with the degradation of intranuclear inclusions formed by deposits of aggregated protein, and they have demonstrated an enhancement of neurodegeneration by further impairment of the UPS (Goldberg, 2003; Rubinsztein, 2006). Recent evidence suggests that PML-IV clastosomes recruit soluble polyglutamine (polyQ)-containing proteins and promote their degradation by proteasome-dependent proteolysis, thus preventing the aggregate formation (Janer et al., 2006).
To further explore the possibility of FBXO25 being involved in preventing polyQ-containing proteins aggregation, we expressed wild-type huntingtin (htt) exon-1 (Ex1) with 103Q glutamines fused to EGFP (EGFP-httEx1-103Q) in HEK293T cells, and we processed them for immunofluorescence confocal microscopy analysis. The results showed that FBXO25 colocalized with EGFP-httEx1–103Q aggregates largely in the intranuclear region (Figure 12A). Additionally, we analyzed the effect of overexpression of FBXO25 on the nuclear aggregation of polyQ-containing proteins in cultured cells. We expressed in HEK293T cells EGFP-httEx1-74Q, Skp1, Cul1, and Roc1 in combination with full-length WT or mutant version of FBXO25 in which the F-box had been deleted (ΔF). The FBXO25ΔF protein cannot interact with Skp1 and thus with other components of the SCF E3 complex. Full-length FBXO25, but not the FBXO25ΔF protein, strongly reduced the level of aggregated EGFP-httEx1-74Q in the filter retardation assay (Figure 12B). Importantly, coexpression of full-length wild-type or mutant version of FBXO25 had no effect on expression of EGFP-httEx1-74Q mRNA (Figure 12E). Also, we observed that the mutant FBXO25ΔF was capable of reaching the nucleus (Supplemental Figure S8). To confirm the involvement of a functional SCF in mediating reduction of nuclear aggregation of polyQ-containing proteins, we expressed in HEK293T cells EGFP-httEx1-74Q, FBXO25WT, Skp1, and Roc1 in parallel with similar combination of proteins in which Cul1 was replaced by mutant that lacked the N-terminal domain (Cul1DN). The Cul1DN protein interacts with Skp1, but not with Roc1; thus, it inhibits the function of Cul1-containing SCF complexes (Wu et al., 2000). As shown in Figure 12F, the combination containing Cul1DN resulted in significant increase of polyQ-containing proteins aggregates trapped in the cellulose membrane in comparison with the aggregates formed in the presence of full-length wild-type (Cul1WT).
Figure 12.
FBXO25 prevents aggregation of polyQ-containing proteins. (A) HeLa cells transfected with EGFP-httEx1-103Q were fixed and labeled with anti-FBXO25 antibodies. Images were taken by confocal microscopy, and the labeled proteins are indicated on top of each panel with the respective color. (B) Slot-blot (S.B) filter retardation assays of polyQ aggregates formed in HEK293T cells cotransfected as indicated in B (top). Results shown correspond to two of four sets of independently cotransfected cells. The EGFP-httEx1-74Q protein-containing aggregates were detected with anti-GFP antibodies. (C) Western blotting (W.B) analysis of cell lysates of one set of HEK293T cells cotransfected as indicated in B. The different forms of FBXO25 protein, indicated by arrows, were revealed with ati-FBXO25 antibodies. Input: lysates of control, WT and ΔF cells. (D) Densitometric analysis of the slot-blot membranes prepared with samples of lysates from all four sets of cotransfected cells indicated in B. Results are expressed as percentage of aggregated protein in the control samples. (E) Detection of EGFP-httEx1-74Q mRNAs by RT-PCR from HEK293T cells cotransfected as indicated in E (top). Coexpression of full-length wild-type or mutant version of FBXO25 had no effect on expression of EGFP-httEx1-74Q mRNA. (F) S.B filter retardation assays of polyQ aggregates formed in HEK293T cells that were transfected with either Cul1WT or Cul1DN and EGFP-httEx1-74Q, FBXO25WT, Skp1, and Roc1. Results shown correspond to two of four sets of independently cotransfected cells. (G) W.B analysis of cell lysates of one set of HEK293T cells cotransfected as indicated in F. The different forms of FLAG-Cul1 protein, indicated by arrows, were revealed with anti-FLAG antibodies. Input, lysates of Cul1WT and Cul1DN cells. (H) Densitometric analysis of the slot-blot membranes prepared with samples of lysates from all four sets of cotransfected cells indicated in F. Results are expressed as percentage of aggregated protein in the Cul1DN.
DISCUSSION
The eukaryotic nucleus is a highly organized, membrane-enclosed organelle that is composed of numerous subnuclear compartments (Handwerger and Gall, 2006; Carmo-Fonseca et al., 2000). Investigation into the nuclear functions related to these structures has increased recently as it has become clear that these membraneless compartments are not just storage spaces but rather highly dynamic entities that can exchange their constituent molecules with the nucleoplasm and/or cytoplasm in response to a variety of stimuli. The functions of the various nuclear compartments have been attributed, in part, to their molecular components and arrays. For example, Cajal bodies are enriched in U7 small nuclear ribonucleoproteins, the protein coilin, and many other factors involved in the biogenesis of nuclear RNA (Handwerger and Gall, 2006). Clastosomes are compartments enriched in PML-IV, 19S and 20S proteasomes, ubiquitin, and substrates of proteasomes involved in the proteolysis of a variety of nuclear proteins (Lafarga et al., 2002; Rockel et al., 2005; Janer et al., 2006). Here, we found that FBXO25 localized to a new class of subnuclear structure, the FAND, that is distinct from clastosomes and other subnuclear compartments and that does not harbor focal sites of transcription.
Immunochemical visualization of FBXO25 in cells indicated that the protein is found in the nucleoplasm, either diffusely spread or arranged in dot-like structures, the FANDs. It should be mentioned that FBXO25 protein was not immunochemically detected in nucleoli. FANDs have distinct localization and morphology relative to other known subnuclear domains such as splicing speckles, Cajal bodies, GEMS, and PML bodies. We observed that a fraction of FANDs colocalize with structures resembling clastosomes. It is unclear whether FANDs are functionally related to clastosomes, but they certainly have distinct properties and composition as we have summarized in Table 1. There are other less well-characterized subnuclear bodies that were not investigated here; despite their diverse morphologic appearance compared with FANDs, at present we cannot disregard the possibility that some proteins are shared between these subnuclear domains.
Table 1.
Comparison between properties of clastosomes and FANDs
Subnuclear compartments and their components are exquisitely sensitive to the transcriptional state of the cell. Remodeling of subnuclear bodies and relocalization of nucleoplasmic proteins occur under physiological circumstances and in certain diseases that involve transcriptional shutdown, which can be mimicked by drug-induced transcriptional arrest (Vera et al., 1993; Malatestaet al., 2000; Gonda et al., 2003). For example, ActD treatment disrupts Cajal bodies and GEMS and relocates nucleophosmin (Raska et al., 1990; Yung et al., 1990; Pellizzoni et al., 2001). Our studies showed that FANDs were completely dispersed and that the FBXO25 redistributed within the nucleosplasm after ActD treatment, indicating that they are dynamic compartments influenced by the transcriptional activity of the cell. This effect of ActD at low dosage is usually considered to be not related to DNA damage (Ljungmanet al., 1999). Additionally, a similar reorganization of FANDs was observed when transcription was experimentally arrested by heat-shock treatment, indicating that the reorganization of FANDs was specifically dependent on inhibition of transcription. We have found that the disruption of FANDs caused by the transcriptional inhibition was accompanied by rearrangement of FBXO25 proteins into discrete perinucleolar structures. Resembling those of some of the ultrastructural components of functionally active nucleolus that are specifically committed with different steps of ribosome biosynthesis, and whose compositions vary over the cell cycle (Van Gansen and Schram, 1972; Stahl et al., 1991; Hyttel et al., 2000).
Our immunofluorescence analyses showed that endogenous Skp1, an adaptor protein of the SCF complex known to interact with F-box proteins, colocalizes and accumulates in FANDs. The possibility that the Skp1-FBXO25 subcomplex or SCFFBXO25 complex is kept in FANDs in the active state, being released as inactive proteins to the nucleoplasm upon transcriptional inactivation is not supported by the results of pull-down experiments because it was shown that ActD treatment does not change the interaction between FBXO25 and Skp1. Thus, the FBXO25 dispersed in the nucleoplasm may still be in a complexed form, whose catalytic role as an E3 ub-ligase is restricted by its accessibility to the ubiquitination targets, a hypothesis that emphasizes the functional significance of the compartmentalization brought about spatial domains in controlling nuclear phenomena.
Also, it can be speculated that the observed nuclear reorganization of FANDs that ensues after treatment of cells with ActD would be accompanied by regulation of the FBXO25 activity. It is well established that another E3 ub-ligase, Mdm2, is regulated by the ribosomal proteins L11, L5, and L13 in response to the ribosomal biogenesis stress caused by ActD (Lohrum et al., 2003; Jin et al., 2004a; Dai and Lu, 2004). These L proteins associate with Mdm2 and inhibit its activity, causing stabilization and activation of the p53 tumor suppressor protein among other effects. The interaction between Mdm2 and each of these L ribosomal proteins is enhanced selectively by inhibition of the activity of RNA polymerase I. Similarly, it is possible that perturbations in rRNA synthesis or ribosome assembly in response to nucleolar stress might result in the release of unknown ribosomal component(s) that could bind FBXO25 and modulate its localization and interaction with the ubiquitination targets.
During mitosis, mammalian cell nuclei go through major structural and functional alterations such as repression of the transcriptional machinery, and redistribution of subnuclear domains such as nucleoli and Cajal bodies (Gottesfeld and Forbes, 1997; Cioce and Lamond, 2005). Observation of HeLa cells after thymidine arrest under confocal microscopy indicated that FANDs were completely dispersed in the nuclei from S phase until the end of telophase, reappearing as cells complete mitosis. Interestingly, a parallel analysis of the FBXO25 showed that the levels of this protein were not significantly affected throughout the cell cycle. The fact that FANDs disassembles at the S phase and reassembles at late telophase in the nuclei of daughter cells supports the view that FANDs are dependent upon the transcriptional status of the cell. It is well documented that during cell division both rRNA synthesis and ribosome assembly are halted (Gottesfeld and Forbes, 1997). As cells enter G1, the concomitant reactivation of RNA synthesis and reorganization of FANDs corroborates the aforementioned notion that FAND organization during mitosis, and possibly FBXO25 regulation as well, follows the inherent fluctuation in RNA synthesis or ribosome assembly in normal cells just as observed in ActD-treated cells. However, the finding that FANDs are completely dispersed at S/G2 was unexpected. It is known that polymerase I activity is elevated in S/G2, suggesting that additional stimuli for the relocalization of FANDs might exist.
The anti-FBXO25 antibodies used in this study did not cross-react with atrogin-1, a protein with which FBXO25 shares a high degree of sequence identity. As expected, immunoblot analyses showed clear distinction between the tissue distribution of these proteins in mice in which FBXO25 is widely expressed, whereas atrogin-1 expression is largely restricted to striated muscle. Interestingly, skeletal muscle and heart showed no significant reactivity with anti-FBXO25 antibodies. Western blot analyses using anti-FBXO25 antibodies revealed a protein as doublet bands in various tissues and cultured cells, suggesting that there may be alternatively spliced forms of mouse FBXO25. In agreement with this observation, it has been shown that in humans there are at least three FBXO25 isoforms (Hagens et al., 2006). Our biochemical data provide evidence that FANDs are predominantly present in the nucleus. Also, we observed that FBXO25 was only partially extracted from adherent HeLa cells upon treatment with salt/detergent mixtures, digestion of DNA with DNase-I; however, caused complete solubilization of FBXO25, indicating that the fraction of the protein that is refractory to detergent extraction is probably chromatin associated.
The results presented here have highlighted some functional similarities between FANDs and PML-IV clastosomes that may contribute to the understanding of polyQ disorders because both subnuclear structures accumulate polyQ-containing proteins aggregates in cultured cell assay for studying the mechanism of these diseases. Also, some of our results provided experimental evidence that overexpressed SCFFBXO25 prevented aggregation of polyQ-containing proteins in cultured cells prone to develop the abnormal accumulation of these proteins. Thus, it will be now of interest to ascertain whether FBXO25 are also found in the neuronal inclusions that characterize Huntington's disease (HD) patients. The observation that only full-length but not F-box–deleted FBXO25, which is inactive, reduced the level of polyQ-containing protein aggregation reinforces the hypothesis that ubiquitin ligase activity of the SCFFBXO25 was needed for the decrease of the aggregation. It remains to be determined whether SCFFBXO25 directly binds to and ubiquitinates the polyQ-containing proteins or another protein associated with polyQ-containing proteins.
In summary, the major conclusions from this study are that a protein that participates in ubiquitination reactions, FBXO25, is localized in a novel subnuclear compartment, the FAND, which is disrupted by inhibition of transcription with subsequent relocation of FBXO25. Our results also indicated that FAND is a dynamic structure capable of rapidly adapting its architecture and probably its ub-ligase activity. In addition to providing new insight into the subcellular localization of FBXO25, our findings also suggest that FANDs recruit polyQ-containing proteins and prevent their accumulation in the nucleus, supporting the notion that FBXO25 is a functional E3 ligase and that FANDs are competent sites of polyubiquitination in the nucleus.
Supplementary Material
ACKNOWLEDGMENTS
We specially thank Dr. Roy E. Larson (Faculty of Medicine of Ribeirão Preto, University of São Paulo [FMRP-USP], São Paulo, Brazil) and Drs. Alfred L. Goldberg and Andreas Schild (Harvard Medical School, Boston, MA) for helpful discussion in the preparation of this article. Confocal microscopy was performed in the Laboratório de Microscopia Confocal da FMRP-USP with technical assistance from Márcia S.Z. Graeff. We thank Dr. Dulce E. Casarini (UNIFESP, Brazil) for providing MCI and IMCD cell lines. We thank Dr. Annie Sittler (Institut National de la Santé et de la Recherche Médicale, Neurologie et Thérapeutique Expérimentale, Paris, France) for providing the RFP-PML IV construct. We thank Dr. David C. Rubinsztein (Cambridge Institute for Medical Research, Addenbrooke's Hospital, Cambridge, United Kingdom) for providing the EGFP-httEx1-74Q construct. We thank Dr. Zhen-Qiang Pan (Mount Sinai School of Medicine, New York City, NY) for providing the Flag-CUL1 (1-452) construct. We are grateful to Ligia S. Antonio from Dr. Wamberto A. Varanda (FMRP-USP) for providing the mouse Leydig cells. We are also grateful to Odete A.B. Cunha and Lucia Sakagute for excellent technical support. FACS analysis was performed with technical assistance from Walter M. Turato (FMRP-USP). This study was supported by grants from the Fundação de Amparo a Pesquisa do Estado de São Paulo (Fundação de Amparo à Pesquisa do Estado de São Paulo [FAPESP] 03/08055-7 and 06/58140-9) and Fundação de Apoio ao Ensino, Pesquisa e Assistência. During these studies, A.O.M., A.L.G.C.M., F.R.T., and M.M.A.B. were recipients of FAPESP fellowships; S. Yokoo was fellow from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior.
Abbreviations used:
- ActD
actinomycin D
- FAND
FBXO25-associated nuclear domain
- htt
Huntingtin
- PML
promyelocytic leukemia protein
- polyQ
polyglutamine
- ub
ubiquitin
- UPS
ubiquitin-proteasome system.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-08-0815) on February 20, 2008.
REFERENCES
- Ang X. L., Harper J. W. Interwoven ubiquitination oscillators and control of cell cycle transitions. Oncogene. 2005;17:2860–2870. doi: 10.1126/stke.2422004pe31. [DOI] [PubMed] [Google Scholar]
- Ardley H. C., Robinson P. A. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D, Seidman J. G., Smith J. A., Struhl K. Short Protocols in Molecular Biology. 3rd ed. New York: John Wiley & Sons; 1997. [Google Scholar]
- Bond U. Heat shock but not other stress inducers leads to the disruption of a sub-set of snRNPs and inhibition of in vitro splicing in HeLa cells. EMBO J. 1988;7:3509–3518. doi: 10.1002/j.1460-2075.1988.tb03227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M., Mendes-Soares L., Campos I. To be or not to be in the nucleolus. Nat. Cell Biol. 2000;2:E107–E112. doi: 10.1038/35014078. [DOI] [PubMed] [Google Scholar]
- Chen D., Dundr M., Wang C., Leung A., Lamond A., Misteli T., Huang S. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J. Cell Biol. 2005;168:41–54. doi: 10.1083/jcb.200407182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiodi I., Biggiogera M., Denegri M., Corioni M., Weighardt F., Cobianchi F., Riva S., Biamonti G. Structure and dynamics of hnRNP-labelled nuclear bodies induced by stress treatments. J. Cell Sci. 2000;113:4043–4053. doi: 10.1242/jcs.113.22.4043. [DOI] [PubMed] [Google Scholar]
- Cioce M., Lamond A. I. Cajal bodies: a long history of discovery. Annu. Rev. Cell Dev. Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- Costa R. R., Varanda W. A. Intracellular calcium changes in mice Leydig cells are dependent on calcium entry through T-type calcium channels. J. Physiol. 2007;585:339–349. doi: 10.1113/jphysiol.2007.137950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M. S., Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- Freed E., Lacey K. R., Huie P., Lyapina S. A., Deshaies R. J., Stearns T., Jackson P. K. Components of an SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication cycle. Genes Dev. 1999;13:2242–2257. doi: 10.1101/gad.13.17.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimuro M., Sawada H., Yokosawa H. Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 1994;349:173–180. doi: 10.1016/0014-5793(94)00647-4. [DOI] [PubMed] [Google Scholar]
- Gary J. D., Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- Glickman M. H., Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Gomes M. D., Lecker S. H., Jagoe R. T., Navon A., Goldberg A. L. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. USA. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda K., Fowler J., Katoku-Kikyo N., Haroldson J., Wudel J., Kikyo N. Reversible disassembly of somatic nucleoli by the germ cell proteins FRGY2a and FRGY2b. Nat. Cell Biol. 2003;5:05–210. doi: 10.1038/ncb939. [DOI] [PubMed] [Google Scholar]
- Gorreta F., Runfola T. P., VanMeter A. J., Barzaghi D., Chandhoke V., Del Giacco L. Identification of thioredoxin reductase 1-regulated genes using small interference RNA and cDNA microarray. Cancer Biol. Ther. 2005;4:1079–1088. doi: 10.4161/cbt.4.10.1987. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Forbes D. J. Mitotic repression of the transcriptional machinery. Trends Biochem. Trends Biochem. Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- Granick D. Nucleolar necklaces in chick embryo fibroblast cells. I. Formation of necklaces by dichlororibobenzimidazole and other adenosine analogues that decrease RNA synthesis and degrade preribosomes. J. Cell Biol. 1975;65:389–417. doi: 10.1083/jcb.65.2.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagens O., Minina E., Schweiger S., Ropers H. H., Kalscheuer V. Characterization of FBX25, encoding a novel brain-expressed F-box protein. Biochim. Biophys. Acta. 2006;1760:110–118. doi: 10.1016/j.bbagen.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Handwerger K. E., Gall J. G. Subnuclear organelles: new insights into form and function. Trends Cell Biol. 2006;16:19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Harlow E., Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hyttel P., Laurincik J., Rosenkranz C., Rath D., Niemann H., Ochs R. L., Schellander K. Nucleolar proteins and ultrastructure in preimplantation porcine embryos developed in vivo. Biol. Reprod. 2000;63:1848–1856. doi: 10.1095/biolreprod63.6.1848. [DOI] [PubMed] [Google Scholar]
- Janer A., Martin E., Muriel M. P., Latouche M., Fujigasaki H., Ruberg M., Brice A., Trottier Y., Sittler A. PML clastosomes prevent nuclear accumulation of mutant ataxin-7 and other polyglutamine proteins. J. Cell Biol. 2006;174:65–76. doi: 10.1083/jcb.200511045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin A., Itahana K., O'Keefe K., Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 2004a;24:7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Cardozo T., Lovering R. C., Elledge S. J., Pagano M., Harper J. W. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004b;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos E. T., Pagano M. The F-box protein family. Genome Biol. 2000;1:1–7. doi: 10.1186/gb-2000-1-5-reviews3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafarga M., Berciano M. T., Pena E., Mayo I., Castano J. G., Bohmann D., Rodrigues J. P., Tavanez J. P., Carmo-Fonseca M. Clastosome: a subtype of nuclear body enriched in 19S and 20S proteasomes, ubiquitin, and protein substrates of proteasome. Mol. Biol. Cell. 2002;13:2771–2782. doi: 10.1091/mbc.E02-03-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A. I., Spector D. L. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Ljungman M., Zhang F., Chen F., Rainbow A. J., McKay B. C. Inhibition of RNA polymerase II as a trigger for the p53 response. Oncogene. 1999;18:583–592. doi: 10.1038/sj.onc.1202356. [DOI] [PubMed] [Google Scholar]
- Lohrum M. A., Ludwig R. L., Kubbutat M. H., Hanlon M., Vousden K. H. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- Lyon C. E., Bohmann K., Sleeman J., Lamond A. I. Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp. Cell Res. 1997;230:84–93. doi: 10.1006/excr.1996.3380. [DOI] [PubMed] [Google Scholar]
- Malatesta M., Gazzanelli G., Battistelli S., Martin T. E., Amalric F., Fakan S. Nucleoli undergo structural and molecular modifications during hibernation. Chromosoma. 2000;109:506–513. doi: 10.1007/s004120000102. [DOI] [PubMed] [Google Scholar]
- Malathi K., Paranjape J. M., Bulanova E., Shim M., Guenther-Johnson J. M., Faber P. W., Eling T. E., Williams B. R., Silverman R. H. A transcriptional signaling pathway in the IFN system mediated by 2′–5′-oligoadenylate activation of RNase L. Proc. Natl. Acad. Sci. USA. 2005;102:14533–14538. doi: 10.1073/pnas.0507551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragno A. L., Baqui M. M., Gomes M. D. FBXO25, an F-box protein homologue of atrogin-1, is not induced in atrophying muscle. Biochim. Biophys. Acta. 2006;1760:966–972. doi: 10.1016/j.bbagen.2006.03.020. [DOI] [PubMed] [Google Scholar]
- Nefkens I., Negorev D. G., Ishov A. M., Michaelson J. S., Yeh E. T., Tanguay R. M., Muller W. E., Maul G. G. Heat shock and Cd2+ exposure regulate PML and Daxx release from ND10 by independent mechanisms that modify the induction of heat-shock proteins 70 and 25 differently. J. Cell Sci. 2003;116:513–524. doi: 10.1242/jcs.00253. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Baccon J., Charroux B., Dreyfuss G. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol. 2001;11:1079–1088. doi: 10.1016/s0960-9822(01)00316-5. [DOI] [PubMed] [Google Scholar]
- Perry R. P., Kelley D. E. Persistent synthesis of 5S RNA when production of 28S and 18S ribosomal RNA is inhibited by low doses of actinomycin D. J. Cell. Physiol. 1970;76:127–140. doi: 10.1002/jcp.1040720311. [DOI] [PubMed] [Google Scholar]
- Platani M., Goldberg I., Swedlow J. R., Lamond A. I. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J. Cell Biol. 2000;151:1561–1574. doi: 10.1083/jcb.151.7.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I., Ochs R. L., Andrade L. E., Chan E. K., Burlingame R., Peebles C., Gruol D. Association between the nucleolus and the coiled body. J. Struct. Biol. 1990;104:120–127. doi: 10.1016/1047-8477(90)90066-l. [DOI] [PubMed] [Google Scholar]
- Rockel T. D., Stuhlmann D., von Mikecz A. Proteasomes degrade proteins in focal subdomains of the human cell nucleus. Cell Sci. 2005;118:5231–5242. doi: 10.1242/jcs.02642. [DOI] [PubMed] [Google Scholar]
- Rubinsztein D. C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y., Blechman J., Darzacq X., Montagna C., Dye B. T., Patton J. G., Singer R. H., Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittler A., Walter S., Wedemeyer N., Hasenbank R., Scherzinger E., Eickhoff H., Bates G. P., Lehrach H., Wanker E. E. SH3GL3 associates with the Huntingtin exon 1 protein and promotes the formation of polyglncontaining protein aggregates. Mol. Cell. 1998;2:427–436. doi: 10.1016/s1097-2765(00)80142-2. [DOI] [PubMed] [Google Scholar]
- Stahl A., Wachtler F., Hartung M., Devictor M., Schofer C., Mosgoller W., de Lanversin A., Fouet C., Schwarzacher H. G. Nucleoli, nucleolar chromosomes and ribosomal genes in the human spermatocyte. Chromosoma. 1991;101:231–244. doi: 10.1007/BF00365155. [DOI] [PubMed] [Google Scholar]
- Stein G. S., Borun T. W. The synthesis of acidic chromosomal proteins during the cell cycle of HeLa S-3 cells. I. The accelerated accumulation of acidic residual nuclear protein before the initiation of DNA replication. J. Cell Biol. 1972;52:292–307. doi: 10.1083/jcb.52.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gansen P., Schram A. Evolution of the nucleoli during oogenesis in Xenopus laevis studied by electron microscopy. J. Cell Sci. 1972;10:339–367. doi: 10.1242/jcs.10.2.339. [DOI] [PubMed] [Google Scholar]
- Vera M. I., Norambuena L., Alvarez M., Figueroa J., Molina A., Leon G., Krauskopf M. Reprogramming of nucleolar gene expression during the acclimatization of the carp. Cell. Mol. Biol. Res. 1993;39:665–674. [PubMed] [Google Scholar]
- Wanker E. E., Scherzinger E., Heiser V., Sittler A., Eickhoff H., Lehrach H. Membrane filter assay for detection of amyloid-like polyglutamine-containing protein aggregates. Methods Enzymol. 1999;309:375–386. doi: 10.1016/s0076-6879(99)09026-6. [DOI] [PubMed] [Google Scholar]
- Weinmann R., Raskas H. J., Roeder R. G. The transcriptional role of host DNA-dependent RNA polymerases in adenovirus-infected KB cells. Cold Spring Harb. Symp. Quant. Biol. 1975;34:495–500. doi: 10.1101/sqb.1974.039.01.061. [DOI] [PubMed] [Google Scholar]
- Wu K., Fuchs S. Y., Chen A., Tan P., Gomez C., Ronai Z., Pan Z. Q. The SCF(HOS/β-TRCP)-ROC1 E3 ubiquitin ligase utilizes two distinct domains within CUL1 for substrate targeting and ubiquitin ligation. Mol. Cell. Biol. 2000;20:1382–1393. doi: 10.1128/mcb.20.4.1382-1393.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost H. J., Lindquist S. RNA splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell. 1986;25:185–193. doi: 10.1016/0092-8674(86)90382-x. [DOI] [PubMed] [Google Scholar]
- Yung B. Y., Bor A. M., Chan P. K. Short exposure to actinomycin D induces “reversible” translocation of protein B23 as well as “reversible” inhibition of cell growth and RNA synthesis in HeLa cells. Cancer Res. 1990;50:5987–5991. [PubMed] [Google Scholar]
- Zeng C., Kim E., Warren S. L., Berget S. M. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 1997;16:1401–1412. doi: 10.1093/emboj/16.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Salomoni P., Pandolfi P. P. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2000;2:E85–E90. doi: 10.1038/35010583. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Tang F. D., Mao G. G., Bian R. L. Effect of alpha-pinene on nuclear translocation of NF-kappa B in THP-1 cells. Acta Pharmacol. Sin. 2004;25:480–484. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.