Abstract
Three different types of autophagy—macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA)—contribute to degradation of intracellular components in lysosomes in mammalian cells. Although some level of basal macroautophagy and CMA activities has been described in different cell types and tissues, these two pathways are maximally activated under stress conditions. Activation of these two pathways is often sequential, suggesting the existence of some level of cross-talk between both stress-related autophagic pathways. In this work, we analyze the consequences of blockage of macroautophagy on CMA activity. Using mouse embryonic fibroblasts deficient in Atg5, an autophagy-related protein required for autophagosome formation, we have found that blockage of macroautophagy leads to up-regulation of CMA, even under basal conditions. Interestingly, different mechanisms contribute to the observed changes in CMA-related proteins and the consequent activation of CMA during basal and stress conditions in these macroautophagy-deficient cells. This work supports a direct cross-talk between these two forms of autophagy, and it identifies changes in the lysosomal compartment that underlie the basis for the communication between both autophagic pathways.
INTRODUCTION
Autophagy is a catabolic process by which intracellular components are delivered to lysosomes for degradation by their resident hydrolases. Three different types of autophagy have been described in mammalian cells depending on the mechanisms used for the delivery of cargo to lysosomes: macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA) (Cuervo, 2004; Mizushima, 2005; Yorimitsu and Klionsky, 2005; Dice, 2007). In microautophagy, the lysosomal membrane invaginates or tubulates to engulf whole regions of the cytosol, but its components and regulation are poorly understood in mammals (Marzella et al., 1981; Mortimore et al., 1988).
Substrates for macroautophagy are complete areas of the cytosol sequestered by a limiting membrane that seals to form a double membrane vesicle or autophagosome (Mizushima et al., 2002; Ohsumi and Mizushima, 2004). The trapped cargo is degraded after the autophagosome fuses with lysosomes, which provide the degradative enzymes. A complex set of genes, known as ATG or autophagy-related genes participate in different steps of macroautophagy (Klionsky et al., 2003). Formation of autophagic vacuoles is mediated by two different conjugation events, a protein-to-protein (to form the Atg5-12-16 complex) and a protein-to-lipid conjugation (to form the Atg8 [LC3 in mammals]-phosphatidyl ethanolamine complex) (Ohsumi and Mizushima, 2004). A kinase nucleation complex, which includes vps34, a class III phosphoinositide 3-kinase (PI3K), and beclin-1 (Liang et al., 1999), also contributes to autophagosome formation. Induction of autophagy is negatively regulated by a second kinase complex, with mammalian target of rapamycin (mTOR) as its main component (Blommaart et al., 1995). Inhibitors of mTOR, such as rapamycin, are widely used as macroautophagy activators. Both the conjugation cascades and the class III PI3K complex are essential for autophagy, because formation of autophagosomes can be blocked with class III PI3K inhibitors (i.e., 3-methyladenine) (Seglen and Gordon, 1982) or by knocking down Atg5, -7, or -12 (the Atgs involved in conjugation) (Komatsu et al., 2005; Hara et al., 2006; Komatsu et al., 2006). In fact, Atg5 or Atg7 null mice die a few hours after birth because of the essential role of macroautophagy, at least partially, as a source of nutrients until the beginning of the lactation period (Kuma et al., 2004). Mice conditionally knocked-out in brain, liver, or heart for any of these two proteins have revealed the involvement of macroautophagy in normal cellular homeostasis and in the clearance of altered proteins, which accumulated in the targeted tissues in these animals (Komatsu et al., 2005, 2006; Hara et al., 2006; Nakai et al., 2007). Although macroautophagy was mainly considered a stress-induced form of autophagy, the studies in these conditionally knocked-out mice strikingly show that constitutive turnover of cytosolic contents by macroautophagy is indispensable for many organs.
In CMA, selective cytosolic proteins are recognized by a chaperone/cochaperone complex that delivers them to the lysosomal surface (Massey et al., 2006b; Dice, 2007). After interacting with the lysosome-associated membrane protein (LAMP) type 2A, which acts as a lysosomal receptor for CMA (Cuervo and Dice, 1996), the substrate protein unfolds, and, assisted by a luminal chaperone, it is translocated directly through the membrane into the lysosomal lumen, where it is rapidly degraded.
Macroautophagy and CMA activity are maximally activated under stress conditions such as starvation, oxidative stress, or conditions leading to enhanced protein misfolding (Cuervo et al., 1995; Kiffin et al., 2004; Mizushima et al., 2004; Ravikumar et al., 2004; Iwata et al., 2005; Scherz-Shouval et al., 2007). Despite both pathways being activated by stressors, a sequential rather than simultaneous activation of these two stress-related forms of autophagy often occurs in most of these conditions. The intrinsic peculiarities of each of these pathways may be behind this coordinated mechanism of activation. Thus, for example, in liver and confluent fibroblasts in culture, starvation stimulates both macroautophagy and CMA. However, whereas macroautophagy is activated during the first hours of starvation, providing a first peak of free amino acids for the synthesis of essential proteins, macroautophagy-dependent proteolysis is rarely observed beyond 4–6 h of starvation in these cells (Fuertes et al., 2003; Finn et al., 2005; Finn and Dice, 2005; Massey et al., 2006a). Instead, at that time, CMA activity progressively increases, reaching a plateau of maximal activity at 20 h after serum removal (in fibroblasts in culture) or 3 d of starvation (in rodent livers) (Dice et al., 1986; Wing et al., 1991; Cuervo et al., 1995; Massey et al., 2006a). This delayed activation of CMA provides a second peak of amino acids to maintain synthesis of essential proteins through the selective degradation of nonessential proteins. A similar simultaneous activation but in reverse order has been described in certain protein conformational disorders (Ravikumar et al., 2002; Cuervo et al., 2004; Iwata et al., 2005). Certain soluble forms of misfolded proteins, bearing the CMA-targeting motif, can be readily degraded via CMA; however, because the misfolded proteins organize in multimeric complexes, they block the CMA translocation machinery and they can no longer be degraded through this pathway (Cuervo et al., 2004). Up-regulation of macroautophagy under these conditions removes protein aggregates (Ravikumar et al., 2002; Iwata et al., 2005). Although the mechanisms that result in activation of macroautophagy in this setting are not clear, and it is likely that blockage in other proteolytic pathways contributes to this activation (Iwata et al., 2005), we have shown experimentally that blockage of CMA in cultured cells results in similar constitutive activation of macroautophagy (Massey et al., 2006a). Although macroautophagy cannot compensate for all CMA functions (cells with impaired CMA are more susceptible to numerous stressors), it is able to maintain proper protein degradation and cell survival under normal conditions (Massey et al., 2006a). Understanding the possible compensatory mechanisms among the different proteolytic systems is of relevance as failure of different autophagic pathways occurs in aging and in severe human pathologies (reviewed in Cuervo, 2004; Shintani and Klionsky, 2004; Rubinsztein et al., 2005; Nixon, 2006; Martinez-Vicente and Cuervo, 2007). Up-regulation of some autophagic pathways after the failure of a particular form of autophagy could be the basis for future therapeutic interventions to preserve normal cellular function in these pathologies.
To better characterize the cross-talk between macroautophagy and CMA, we have analyzed in this work the effect of blockage of macroautophagy on CMA. Using both pharmacological inhibition of macroautophagy and fibroblasts from mouse embryos deficient in Atg5 (Atg5−/− cells) (Kuma et al., 2004), we have found up-regulation of CMA after nutritional stress and also under normal nutritional conditions. Interestingly, different mechanisms contribute to the observed changes in CMA-related proteins and the consequent activation of CMA during basal and stress conditions. This work supports a direct cross-talk between these two forms of autophagy, and it identifies changes in the lysosomal compartment that underlie the basis for the communication between both autophagic pathways.
MATERIALS AND METHODS
Animals and Cells
Male Wistar rats (200–250 g) fed or fasted for the indicated times were used. Where indicated, vinblastine (5 mg/100g body weight) was i.p. injected 2 h before sacrifice. Immortalized mouse embryonic fibroblasts (MEFs) derived from wild-type (WT) and ATG5−/− mice (two different clones each) (Kuma et al., 2004) were maintained in DMEM (Sigma-Aldrich, St. Louis, MO) in the presence of 10% fetal bovine serum (FBS) (Mediatech, Herdon, VA). Except where indicated, all results shown correspond to the mean value of the two different clones from each group. Mouse fibroblasts (NIH3T3) were from the American Type Culture Collection (Manassas, VA), and they were maintained in the same media but supplemented with 10% newborn calf serum (NCS) (Atlanta Biologicals, Lawrenceville, GA). To deprive cells from serum, plates were extensively washed with Hank's balanced salts solution (Invitrogen, Carlsbad, CA), and fresh medium without serum was added. Cells were transfected with the cDNA for Atg5 in frame with three hemagglutinin (HA) sequences subcloned in the pCI-neo mammalian expression vector by using Lipofectamine (Invitrogen).
Chemicals
Sources of chemicals and antibodies were as described previously (Kiffin et al., 2004; Kuma et al., 2004; Kaushik et al., 2006; Massey et al., 2006a). [14C]Formaldehyde was from PerkinElmer Life and Analytical Sciences (Boston, MA), and 3-methyladenine, trichloroacetic acid, Filipin and fluorescent substrates were from Sigma-Aldrich. The Amplex Red Cholesterol Assay kit was from Invitrogen. Antibodies against the cytosolic tail of rat and mouse LAMP-2A, -2B, and -2C were prepared in our laboratory (Cuervo and Dice, 1996; Massey et al., 2006a; Zhang and Cuervo, unpublished data). The antibody against mouse LAMP-1 (1D4B) was from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA), antibodies against flotillin-1 and HA-tag were from BD Biosciences Transduction Laboratories (San Diego, CA), and the antibodies against heat-shock cognate (hsc)70 (SPA-810) and heat-shock protein (hsp)90 (SPA-845) were from Nventa Biopharmaceuticals (San Diego, CA).
Isolation of Lysosomes
Lysosomes from cultured cells were isolated from a light mitochondrial-lysosomal fraction in Percoll/metrizamide discontinuous gradients as described previously (Storrie and Madden, 1990). Two separate lysosomal fractions with different activities for CMA were isolated from rat liver from a light mitochondrial-lysosomal fraction through a discontinuous metrizamide gradient as described previously (Cuervo et al., 1997). β-Hexosaminidase (EC 3.2.1.52) latency was measured as an index for the integrity of the lysosomal membrane after isolation (Storrie and Madden, 1990). Preparations with >10% broken lysosomes were systematically discarded. Lysosomal matrices and membranes were isolated after hypotonic shock of the lysosomal fraction followed by centrifugation as described previously (Ohsumi et al., 1983). Where indicated, regions of different resistance to detergent solubilization were isolated from lysosomal membranes through flotation in a sucrose density gradient as described previously (Kaushik et al., 2006). Briefly, lysosomal membranes (150 μg of protein) from mouse cultured fibroblasts were incubated with 1% Triton-X114 in 150 mM NaCl, 50 mM Tris-HCl, and 5 mM EDTA, pH 7.4, on ice for 30 min, and then they were subjected to centrifugation in a stepwise discontinuous sucrose gradient (5–40%). A visible translucent band in the 15% sucrose region (previously identified as the detergent-resistant (DR) part of the lysosomal membrane), and all the fractions below that band (detergent-soluble; DS) were concentrated by precipitation with trichloroacetic acid (TCA) and then subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblot.
Intracellular Protein Degradation
Confluent cells were incubated with [3H]leucine (2 μCi/ml) for 48 h at 37°C, and then they were extensively washed and maintained in either complete (10% FBS or 10% NCS) or serum-deprived medium containing an excess of unlabeled leucine (2.8 mM), to prevent reutilization of radiolabeled leucine (Auteri et al., 1983). Where indicated, 10 mM 3-methyladenine or 20 mM ammonium chloride was added to the incubation medium during the chase period. Aliquots of the medium taken at different times were precipitated with TCA, and proteolysis was calculated as the amount of initial acid precipitable radioactivity (protein) transformed to acid soluble (peptides and amino acids) at each time point (Auteri et al., 1983). Total radioactivity incorporated into cellular proteins was determined in duplicate samples as the amount of acid-precipitable radioactivity in labeled cells immediately after washing. In HA-Atg5–transfected cells, label was added 1 d before transfection and maintained throughout the transfection procedure to start the chase 12 h after transfection (when maximal levels of HA-Atg5 were detected).
Uptake and Degradation of Substrate Proteins by Isolated Lysosomes
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH; EC 1.2.1.13) was radiolabeled with [14C]formaldehyde by reductive methylation (Jentoft and Dearborn, 1983) to a specific activity of 1.5 × 108 dpm/nmol. Degradation of radiolabeled proteins by isolated intact or broken lysosomes was measured as described previously (Terlecky and Dice, 1993; Cuervo et al., 1997). Briefly, lysosomes were incubated with the radiolabeled protein in 3-(N-morpholino)propanesulfonic acid (MOPS) buffer (10 mM MOPS, pH 7.3, 0.3 M sucrose, 1 mM dithiothreitol, and 5.4 μM cysteine) for 30 min at 37°C. Reactions were stopped with 10% TCA and filtered through a Millipore Multiscreen Assay System (Millipore, Billerica, MA) using a 0.45 μm pore filter. Radioactivity in the flow through and in the filter was converted to disintegration per minute in a WinSpectral 1414 liquid scintillation analyzer (PerkinElmer Wallac, Gaithersburg, MD) by correcting for quenching using an external standard. Proteolysis was expressed in percentage as described above. Degradation of proteins by broken lysosomes was measured as described above, but incubations were performed for 15 min and in 1 mM dithiothreitol supplemented water to maintain the final pH close to that in the lysosomal lumen. Degradation of radiolabeled cytosolic pool of proteins by isolated intact or broken lysosomes was measured similarly.
Uptake of GAPDH by isolated lysosomes was analyzed as described previously (Cuervo et al., 1997). Briefly, freshly isolated lysosomes from NIH3T3 cells were incubated with GAPDH in MOPS buffer at 37°C for 20 min. Where indicated, lysosomes were preincubated with a cocktail of protease inhibitors for 10 min at 0°C. Lysosomes were collected by centrifugation, washed with MOPS buffer, and subjected to SDS-PAGE and immunoblot for GAPDH. Uptake was calculated from densitometric analysis by subtracting the amount of GAPDH associated with lysosomes in the presence (protein bound to the lysosomal membrane and taken up by lysosomes) and absence (protein bound to the lysosomal membrane) of protease inhibitors.
Immunocytochemical Staining
Immunofluorescence studies of cultured cells were performed following conventional procedures (Cuervo and Dice, 2000b; Kiffin et al., 2004; Kaushik et al., 2006). Cells grown on coverslips until subconfluent and kept in the presence or absence of serum for 16 h were fixed with 3% formaldehyde solution, blocked, and then incubated with the primary and corresponding fluorescein isothiocyanate (FITC)- or cyanine 5-conjugated secondary antibody, or 0.5 mg/ml Filipin as described previously (Cuervo and Dice, 2000b; Kiffin et al., 2004; Kaushik et al., 2006). To detect the hsc70 associated to lysosomes, cells were fixed with methanol instead, which removes cytosolic proteins and thus facilitates visualization of the lysosome associated hsc70. Mounting medium contained 4′, 6-diamidino-2-phenylindole (DAPI) stain to highlight the cellular nucleus. Images were acquired with an Axiovert 200 fluorescence microscope (Carl Zeiss, Thornwood, NY) and subjected to deconvolution with the manufacture's software. Quantification was performed using ImageJ software (National Institutes of Health, Bethesda, MD). The mean distance of lysosomes from the nucleus was calculated in 10 different cells for each condition per experiment by tracing lines (6 in each cell) from the nuclear membrane to the most distal fluorescent vesicle (lysosome). The distribution of fluorescent vesicles along each line was calculated with the measure function of the ImageJ (http://rsb.info.nih.gov/ij/) program. Distances and number of vesicles at each distance for each cell were averaged, and mean values were calculated from the individual distributions in 10 cells per condition. Percentage of colocalization was calculated using the JACoP plug-in in ImageJ software. Number of particles was calculated using the analyze particles function of the ImageJ program. All digital microscopic images were prepared using Adobe Photoshop 6.0 software (Adobe Systems, Mountain View, CA).
mRNA Quantification
Total RNA was extracted from cultured cells using the RNeasy Protect Mini kit (QIAGEN, Valencia, CA) following the manufacturer's indications, and stored at −80°C until use. The first-strand cDNA was synthesized from 1 μg of the total RNA with the SuperScript II RNase H reverse transcriptase (Invitrogen) and oligo(dT)12-18 primers. Actin and a region of the exon 8 of LAMP-A were amplified with specific primers (LAMP-2A, 5′-GCAGTGCAGA TGAAGACAAC-3′, 5′-AGTATGATGGCGCTTGAGAC-3′; and actin, 5′-AAGGACTCCTATAGTGGGTGACGA-3′, 5′-ATCTTC TCCATGTCGTCCCAGTTG-3′) by using the SYBR Green polymerase chain reaction (PCR) kit (PE Biosystems, Warrington, United Kingdom). Amplification of the LAMP-2A and actin DNA products (120 and 108 base pairs, respectively) was measured in real time in a SmartCycler (Cepheid, Sunnyvale, CA). For all amplifications, the presence of a single amplified product was verified by agarose gel electrophoresis, and by analysis of the melting curves of the reverse transcription-PCR reaction. The expression level of LAMP-2A in different samples was normalized with respect to those of actin in the same samples. Differences between samples were calculated based on the differences in the number of cycles required to reach a threshold fluorescence intensity level. We did not need to correct for fragment length because the size of the amplified fragments was very similar.
Proteasome Activity
The 20S proteasome chymotrypsin-like, trypsin-like, and peptidylglutamyl peptide hydrolase activities were measured against the fluorogenic synthetic peptides N-succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin, BZ-Val-Gly-Arg-7-amido-4-methylcoumarin, and N-benzyloxy-carbonyl-Leu-Leu-Glu-2-naphtylamide, respectively, following standard procedures (Shibatani et al., 1996).
Viability Measurement
Viability of the cells was measured by incubating cells with 1 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) in DMEM for 1 h at 37°C. The formazan product formed was solubilized with 1-propanol, and its absorbance was measured at 560 nm. Cell viability after HA-Atg5 cDNA transient transfection was calculated as the percentage of the absorbance in mock-transfected cells.
Measurement of Intralysosomal pH
The intralysosomal pH was measured using FITC-dextran fluorescence as described previously (Ohkuma et al., 1982). Briefly, cells were incubated with 1 mg/ml FITC-dextran for 12 h, and then lysosomes were isolated by the standard procedures and the FITC fluorescence associated to isolated lysosomes was determined in a spectrofluorometer at 25°C at 495 nm (pH-sensitive fluorescence) and 450-nm (pH-insensitive fluorescence) excitation and 550-nm emission wavelengths. Double standard curves were plotted by calculating the ratio of fluorescence intensities at 495 nm/450 nm for 2 mg of FITC-dextran at different pH values and in different buffers and by comparing the fluorescence intensities, at 495-nm excitation wavelengths, in intact lysosomes and after disruption of lysosomes with 0.1% Triton X-100. Intralysosomal pH in the isolated lysosomes was calculated with these curves from their ratio of the 495/450-nm fluorescence intensities, after subtracting the background fluorescence.
Measurement of LAMP-2A Degradation
Rates of degradation of LAMP-2A in the isolated lysosomes were determined by immunoblot by using a specific antibody against the cytosolic tail of LAMP-2A as described previously (Cuervo and Dice, 2000a). Briefly, isolated lysosomes were incubated in MOPS buffer at 37°C, and at different times aliquots were removed and subjected to SDS-PAGE and immunoblot for LAMP-2A.
General Methods
Protein concentration was determined by the Lowry method (Lowry et al., 1951) by using bovine serum albumin as a standard. After SDS-PAGE (Laemmli, 1970) gels were transferred to nitrocellulose membranes by using a Mini-TransBlot SD wet transfer cell (Bio-Rad, Richmond, VA). Immunoblotting of the nitrocellulose membranes was performed following standard procedures (Towbin et al., 1979). The proteins recognized by the specific antibodies were visualized by chemiluminescence methods (RenaissanceR; PerkinElmer Life and Analytical Sciences). Densitometric quantification of the immunoblotted membranes was performed with an Image Analyzer System (Inotech S-100; Inotech, Sunnyvale, CA). Quantification of intracellular cholesterol was done using the Amplex Red Cholesterol Assay kit (Invitrogen) as described previously (Eskelinen et al., 2004). The Student's t test was used for statistical analyses.
RESULTS
Pharmacological Blockage of Macroautophagy Activates CMA
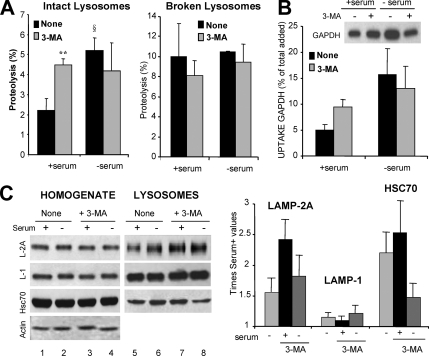
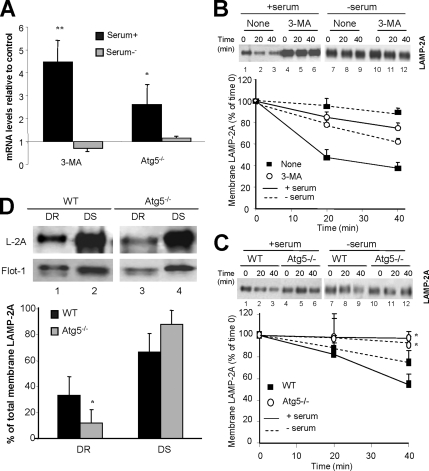
We have previously described that blockage of CMA results in constitutive activation of macroautophagy in mouse fibroblasts in culture (Massey et al., 2006a). To check whether the reciprocal was true, we treated cultured mouse fibroblasts with 3-methyladenine (3-MA), a well-characterized class III PI3K inhibitor that blocks autophagosome formation, and we analyzed CMA activity. The most direct method to measure CMA activity is by analyzing the rates of translocation and degradation of cytosolic proteins into isolated lysosomes (Klionsky et al., 2007). As described previously (Terlecky et al., 1992; Cuervo et al., 1995), lysosomes isolated from cells grown in the absence of serum showed significantly higher ability to take up and degrade a pool of substrate proteins (Figure 1A; note that in these experiments cells were starved for 6 h, as this is the time during which formation of autophagosomes in response to serum removal has been described in these cells, but CMA does not reach maximal activity until 12–18 h after serum removal). Treatment with 3-MA did not modify levels of CMA activity in response to serum removal. In contrast, we found significantly higher CMA activity in lysosomes isolated from 3-MA-treated cells when they were maintained in the presence of serum, reaching levels close to those detected upon serum removal (Figure 1A, left). The observed differences did not result from changes in the proteolytic activity of lysosomes upon 3-MA treatment, because they were no longer present when we compared proteolytic rates in lysosomes in which the lysosomal membrane had previously been disrupted (Figure 1A, right). The similar proteolytic ability detected in lysosomes isolated from treated and untreated cells maintained in the absence of serum, suggested that the lack of a stimulatory effect of 3-MA on CMA during starvation was probably not due to changes in the lysosomal lumen induced by 3-MA (i.e., increase in lysosomal pH described in other cell types). We further confirmed the direct effect of 3-MA treatment on CMA by using GAPDH, a well characterized CMA substrate, in a different in vitro assay that allows us to monitor lysosomal uptake separately from degradation (Figure 1B). Uptake of GAPDH was significantly higher in lysosomes isolated from 3-MA treated cells compared with untreated cells, but only when maintained in the presence of serum.
Figure 1.
Pharmacological inhibition of macroautophagy up-regulates CMA. NIH3T3 mouse fibroblasts were maintained in culture in the presence (+ serum) or absence of serum (− serum) without additions (None) or in media supplemented with 10 mM 3-MA. After 6 h, lysosomes were isolated as described under Materials and Methods. (A) CMA activity. Intact isolated lysosomes (left) or lysosomes disrupted by hypotonic shock (right) were incubated in an isotonic medium or in water, respectively, with a 14C-labeled pool of cytosolic proteins for 30 min at 37°C. Reactions were stopped by addition of TCA, and proteolysis was calculated as the percentage of acid precipitable radioactivity transformed to acid soluble at the end of the incubation. Values are the mean + SE of three different experiments with triplicate samples (**p < 0.001, differences with untreated cells; §p < 0.01, differences with serum supplemented cells). (B) Lysosomal uptake of CMA substrates. The isolated lysosomes, pretreated or not with a cocktail of protease inhibitors, were incubated with 10 μg of GAPDH for 20 min at 37°C. At the end of the incubation, lysosomes were collected by centrifugation and the amount of GAPDH taken up by lysosomes was calculated as described under Materials and Methods. Inset shows a representative immunoblot of the association of GAPDH to lysosomes (lysosomes treated with protease inhibitors). Values are expressed as percentage of the total amount of GAPDH added into the incubation media and are mean + SE of four different experiments. (C) Levels of CMA components. Total cellular homogenates and lysosomes (50 μg of protein) were subjected to SDS-PAGE and immunoblot for the indicated proteins. The densitometric quantification of two immunoblots from different experiments is shown on the right. Values are mean + SE, and they are expressed as -fold increase the value in untreated cells maintained in media supplemented with serum, which was given an arbitrary value of 1. Actin is shown as a loading control in homogenates.
Activation of CMA is often mediated by an increase in the lysosomal levels of LAMP-2A, the CMA receptor, and of hsc70, the chaperone that participates in substrate targeting, unfolding, and translocation (Agarraberes et al., 1997; Cuervo and Dice, 2000b). In agreement with our functional data, levels of both proteins were significantly higher in lysosomes isolated from cells maintained in the presence of serum during 3-MA treatment (Figure 1C). In serum-deprived cells, treatment with 3-MA reduced the amount of both proteins in lysosomes, which probably accounts for the lower CMA activity in response to serum removal detected in these cells compared with untreated cells.
These results support that treatment with 3-MA, a well-characterized inhibitor of macroautophagy, leads to up-regulation of CMA under basal conditions, but it is not able to increase the activity of this pathway beyond the increase observed during nutritional deprivation.
Changes in Lysosomal Protein Degradation in Atg5−/− Fibroblasts
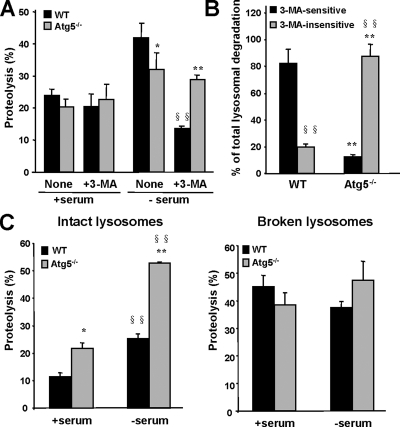
To directly link the changes in CMA observed after 3-MA treatment to its inhibitory effect on macroautophagy, we then used embryonic fibroblasts from a genetic mouse model with defective macroautophagy (knocked out for the essential autophagy gene ATG5 [Atg5−/− MEFs); Kuma et al., 2004). Total rates of degradation of long-lived proteins were similar in wild-type and Atg5−/− MEFs when maintained in the presence of serum, but as expected, they become significantly lower in Atg5−/− cells after serum removal (Figure 2A; except where indicated, the results shown in this work correspond to the mean values obtained for two different wild-type and two Atg5−/− clones). Despite the well-reported blockage of macroautophagy in the Atg5−/− cells, we still observed an increase in long-lived protein degradation in response to serum removal in the Atg5−/− fibroblasts (1.5-fold increase compared with 2.2-fold increase in wild type). This increase in protein degradation is unlikely to result from up-regulated activity of the ubiquitin/proteasome system in Atg5−/− cells, because we did not find significant differences in the three major proteolytic activities of the 26S proteasome in cellular extracts from wild-type and Atg5−/− cells (Supplemental Figure 1A). Furthermore, levels of polyubiquitinated proteins, the main substrates of the ubiquitin/proteasome system, were similar in both groups of cells (Supplemental Figure 1B). These results support that macroautophagy blockage does not have direct consequences on proteasome-dependent degradation, and they are in agreement with the normal activity of this proteolytic system described in brain and liver of conditional Atg5 and Atg7 knockout mice (Komatsu et al., 2005; Komatsu et al., 2006).
Figure 2.
Increased CMA activity in Atg5−/− cells. (A) Rates of long-lived proteins degradation. MEFs from WT and Atg5−/− mice were labeled with [3H]leucine for 48 h at 37°C, and then they were incubated with an excess of unlabeled leucine, in the presence (+ serum) or absence (− serum) of serum and supplemented or not with 10 mM 3-MA. Proteolysis at 20 h was calculated as described under Materials and Methods. Values are expressed as percentage and are the mean + SE of three different experiments with two different WT and Atg5−/− clones each. (B) Type of lysosomal degradation. The total amount of lysosomal-dependent degradation was calculated in experiments as the ones described in A by adding 20 mM ammonium chloride. The percentage of lysosomal-dependent degradation (ammonium chloride inhibited) sensitive or insensitive to 3-MA is plotted. Values are the mean + SE of three different experiments (*p < 0.01; **, §§p < 0.001; *, differences with wild-type cells; §, differences with untreated cells). (C) Intact lysosomes isolated from the same cells (left) or lysosomes disrupted by a hypotonic shock (right) were incubated in an isotonic medium or in water, respectively, with [14C]GAPDH for 30 min at 37°C. Reactions were stopped by addition of TCA acid, and GAPDH proteolysis was calculated as the percentage of acid precipitable radioactivity transformed to acid soluble at the end of the incubation. Values are expressed as percentage, and they are the mean + SE of three different experiments with two different WT and Atg5−/− clones each (*p < 0.01; **, §§p < 0.001; *, differences with wild-type cells; §, differences with serum supplemented cells).
The residual proteolytic activity detected after serum removal in Atg5−/− cells, was still occurring for the most part in lysosomes, because it could be inhibited by ammonium chloride (a well-characterized lysosomal inhibitor), but as expected, in contrast to wild-type cells where 3-MA blocked ∼75% of the lysosomal degradation, the degradation of long-lived protein in Atg5−/− cells was practically unmodified after 3-MA treatment (Figure 2A). In fact, when considering only lysosomal protein degradation (sensitive to ammonium chloride), ∼90% of the total lysosomal degradation was 3-MA–insensitive in Atg5−/− cells after serum removal (Figure 2B). These results support that different lysosomal pathway(s) are activated in these cells during serum removal to compensate for the lack of macroautophagy.
Both microautophagy and CMA can contribute to the observed residual degradation of long-lived proteins in lysosomes. To directly determine the contribution of CMA to protein degradation in Atg5−/− cells, we used the translocation assays described above, with lysosomes isolated from cells maintained in the presence or absence of serum. The purity (<0.01% mitochondria and cytosolic enzymes activity) and the integrity of the isolated lysosomes (as determined by the release of lysosomal enzymes [β-hexosaminidase] into the incubation media) was comparable in both groups (4 and 3.5% broken lysosomes in wild-type and Atg5−/−, right after isolation). We found, however, higher enrichment of lysosomal enzymes (β-hexosaminidase and β-N-acetyl-glucosaminidase, 2.6 and 3.1 times higher, respectively) in the lysosomes isolated from the Atg5−/− cells compared with wild type, which we attributed to a decrease in the content of nonlysosomal proteins (cargo) delivered to lysosomes via macroautophagy (absent in these cells). Our protocol is optimized to isolate lysosomes with high activity for CMA (enriched in hsc70), which represent ∼30% of total lysosomes in fibroblasts. To determine whether lack of fusion with autophagosomes could modify the ability of these CMA-active lysosomes to pellet or to migrate in the density gradient during the isolation procedures, we analyzed the distribution of lysosomal enzymes at the different steps required for the isolation of lysosomes. Whereas in wild-type cells most of the lysosomal enzymatic activity is detected in the 5–10% interface of the metrizamide gradient, we found 15% of the total β-hexosaminidase activity in the gradient in the higher density region in Atg5−/− cells (17–10% interface), where most mitochondria are detected under normal conditions. Immunoblot of that region revealed the presence of LAMP-1 and LAMP-2 but not of hsc70 in this fraction, supporting that lack of macroautophagy altered the physical properties of a subset of intracellular lysosomes but not of those active for CMA (data not shown). Different reasons could account for the change in density of that lysosomal pool, including problems with cholesterol trafficking. Alterations in traffic of cholesterol and intracellular accumulation of lipids have been earlier reported in embryonic fibroblasts derived from the LAMP-2 knockout mice, which also show impaired macroautophagy (Eskelinen et al., 2004). However, in contrast to that mouse model, total intracellular levels of unesterified cholesterol were significantly lower in the Atg5−/− MEFs compared with wild-type cells (Supplemental Figure 2A). Analysis of Filipin-stained cells also revealed changes in cholesterol distribution (more intense staining at the plasma membrane and lesser number but enlarged size of the compartments positive for Filipin) in the Atg5−/− cells (Supplemental Figure 2B). The reasons and cellular consequences of these changes in cholesterol content remain unknown, but they did not seem to affect the purity or stability of the population of lysosomes more active for CMA, which allowed us to proceed to the analysis of CMA activity in lysosomes isolated from these cells.
Atg5−/− Fibroblasts Have Increased Rates of Chaperone-Mediated Autophagy
When we compared the degradation of GAPDH, a well-characterized CMA substrate, we found that GAPDH was degraded more efficiently by lysosomes isolated from the Atg5−/− cell clones (Figure 2C). Interestingly, the increased ability to degrade GAPDH was observed not only in lysosomes isolated from serum-deprived cells (when CMA is activated) but also from cells maintained in the presence of serum.
To determine whether the higher degradation of GAPDH in Atg5−/− cells was a consequence of higher proteolytic activity or of increased rate of uptake through the lysosomal membrane, we compared the degradation of GAPDH by lysosomes in which the lysosomal membrane had been previously disrupted (Figure 2D). Under these conditions, we did not find significant differences in the proteolysis of GAPDH between both groups of cells, suggesting that, in fact, the differences observed with intact lysosomes were a result of increased uptake of the protein through the lysosomal membrane via CMA. As further support that the effect of 3-MA on CMA observed in macroautophagy competent cells (Figure 1, A and B) was mainly through the inhibitory effect of this compound on macroautophagy, we did not find changes in CMA or in the proteolytic activity of lysosomes isolated from Atg5−/− cells treated with 3-MA (Supplemental Figure 3).
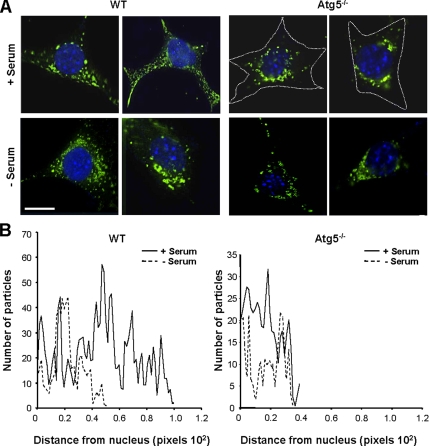
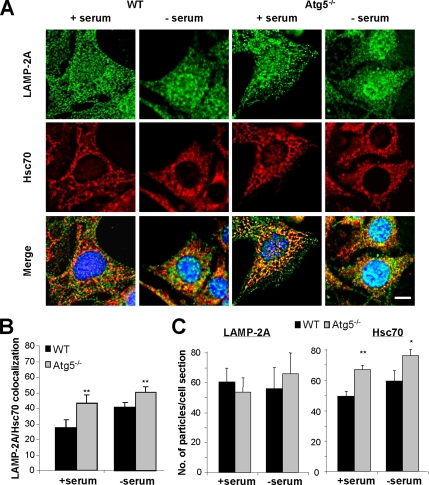
We further verified the activation of CMA in Atg5−/− cells in culture by specifically analyzing the patterns of distribution of LAMP-2A–enriched lysosomes in these cells. As we have previously described in other cell types (Agarraberes et al., 1997; Kiffin et al., 2004; Cuervo and Dice, 2000b), activation of CMA after serum removal in wild-type embryonic mouse fibroblasts associated to the relocation of CMA-active lysosomes (LAMP-2A–enriched) toward the perinuclear region (Figure 3, left). However, in the Atg5−/− cells, CMA-active lysosomes were preferentially located in the perinuclear region even when cells were maintained in the presence of serum (Figure 3, right). Similar perinuclear location was observed in serum-supplemented wild-type cells when macroautophagy was pharmacologically inhibited by treatment with 3-MA (Supplemental Figure 4).
Figure 3.
Relocation of CMA-active lysosomes toward the perinuclear region in Atg5−/− cells. MEFs from WT and Atg5−/− mice were maintained in the presence or absence of serum. After 12 h, cells were fixed and subjected to immunofluorescence for LAMP-2A. Mounting media contains DAPI to highlight the cell nuclei. (A) Representative cells for each condition. Discontinuous line marks the cellular perimeter in Atg5−/− cells. Bar, 5 μm. (B) Quantification of the distribution of lysosomes (LAMP-2A–positive puncta) with respect to the nucleus in wild-type (left) and Atg5−/− (right) MEFs under basal conditions and after serum removal. Values are mean of the quantification of 10 cells per condition in two independent experiments with two different WT and Atg5−/− clones each. Mean cell size of Atg5−/− cells was 1.3 + 0.2 times bigger than wild-type cells.
Both lines of evidence, the enhanced lysosomal ability to take up CMA substrates and the perinuclear location of CMA-active lysosomes in Atg5−/− cells maintained in serum-supplemented media, support constitutive activation of CMA in cells with impaired macroautophagy.
Up-Regulation of CMA Components in Lysosomes from Atg5−/− Cells
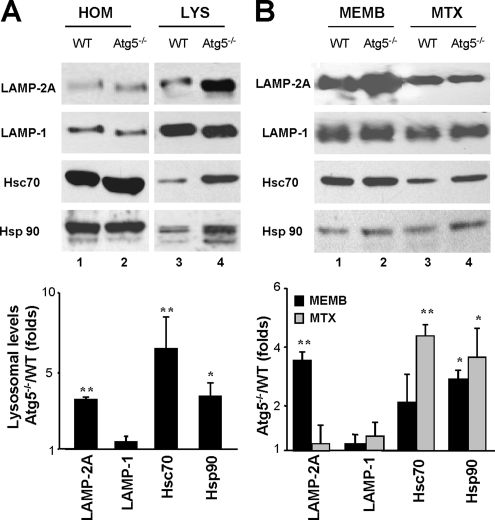
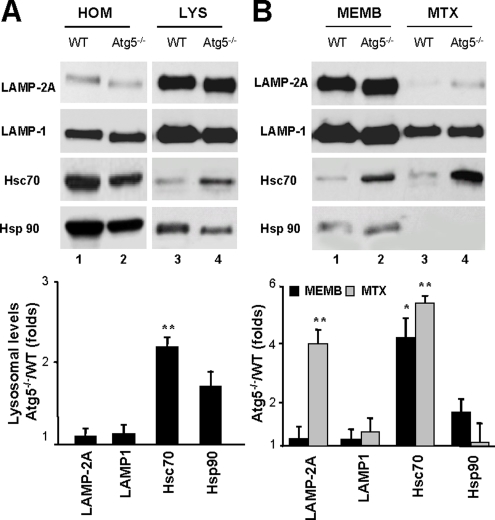
To determine the molecular changes in the lysosomal compartment that lead to CMA up-regulation in fibroblasts with impaired macroautophagy, we first compared the levels of LAMP-2A and other lysosomal membrane proteins in lysosomes isolated from wild-type and Atg5−/− fibroblasts maintained in the absence (Figure 4) or presence of serum (Figure 5). Although total cellular levels of LAMP-2A were similar in both groups of cells (Figure 4A), levels of LAMP-2A were significantly higher in lysosomes from Atg5−/− cells upon serum removal compared with wild-type MEFs (Figure 4A). The levels of the other LAMP-2 variants and other lysosomal membrane proteins remained unchanged under these conditions (Supplemental Figure 5). The higher levels of LAMP-2A in lysosomes from serum-deprived Atg5−/− MEFs were mostly due to an increase in the content of this protein at the lysosomal membrane (Figure 4B). In contrast, the slight increase in LAMP-2A content in lysosomes from serum-supplemented Atg5−/− MEFs was attributable to higher levels of LAMP-2A at the lysosomal lumen (Figure 5B). Several groups including ours have identified the presence of full size LAMP-2A in the lysosomal lumen (Jadot et al., 1996; Cuervo and Dice, 2000b). Part of this protein originates from the mobilization of LAMP-2A from the membrane into the lumen as CMA substrates are translocated. It is thus possible that the higher LAMP-2A luminal content in lysosomes from Atg5−/− cells under basal conditions is a consequence of the enhanced basal CMA in these cells.
Figure 4.
Changes in CMA lysosomal components in Atg5−/− cells during nutritional stress. (A) Homogenate (HOM) and lysosomes (LYS) isolated from WT and Atg5−/− mouse embryonic fibroblasts maintained in media not supplemented with serum for 16 h were subjected to SDS-PAGE and immunoblotted for the indicated proteins. (B) Membranes (MEMB) and matrices (MTX) of lysosomes isolated under the same conditions were separated after hypotonic shock and centrifugation, and then they were processed as described in A. Bottom of A and B, densitometric quantification of three to four immunoblots. Values are mean + SE, and they are expressed as -fold increase in lysosomes isolated from Atg5−/− cells compared with wild-type. Wild-type value is assigned an arbitrary value of 1 (*p < 0.01, **p < 0.001 compared with WT cells).
Figure 5.
Changes in CMA lysosomal components in Atg5−/− cells under basal conditions. (A) Homogenate (HOM) and lysosomes (LYS) isolated from wild-type and Atg5−/− mouse embryonic fibroblasts maintained in the presence of serum were subjected to SDS-PAGE and immunoblotted for the indicated proteins. (B) Lysosomal membranes (MEMB) and matrices (MTX) isolated after hypotonic shock and centrifugation of intact lysosomes were processed as described in A. Bottom of A and B, densitometric quantification of two to three immunoblots. Values are expressed as -fold increase in lysosomes isolated from Atg5−/− cells compared with wild type, which was assigned an arbitrary value of 1 (*p < 0.01, **p < 0.001 compared with WT cells).
Intracellular levels of the subset of chaperones that participate in CMA were comparable in wild-type and Atg5−/− MEFs. However, as described in other conditions with increased CMA activity (i.e., mild oxidative stress or exposure to toxic compounds; Cuervo et al., 1995; Kiffin et al., 2004), levels of hsc70 and hsp90 associated to lysosomes were significantly higher in lysosomes from serum-deprived Atg5−/− cells (Figure 4A). Increased levels of hsc70 were also detected in lysosomes from serum-supplemented Atg5−/− MEFs (Figure 5). Both the lysosome membrane-associated hsc70 and more markedly, the luminal form of hsc70 were augmented in the lysosomes from Atg5−/− MEFs. Although the lumenal hsp90 does not participate directly in substrate uptake, during maximal CMA activation, a portion of lysosomal hsp90 localizes in the lysosomal lumen, and it seems important to confer stability to LAMP-2A (Bandyopadhyay and Cuervo, unpublished data). Lysosomes from Atg5−/− MEFs displayed higher levels of this luminal hsp90 than lysosomes from wild-type cells under serum-deprivation conditions (Figure 4B).
We next determined possible changes in the contribution of CMA active lysosomes to the total lysosomal pool in Atg5−/− MEFs. Both lysosomes active and inactive for CMA contain LAMP-2A in their membrane, but only those containing hsc70 in their lumen are competent for CMA (Cuervo et al., 1997). Using immunofluorescence in cultured cells maintained in the presence or absence of serum, we found that the number of vesicular compartments positive for both proteins was higher in Atg5−/− MEFs (higher percentage of colocalization of puncta) (Figure 6, A and B). Quantification of the total number of vesicles positive for each protein revealed a significant increase in the number of hsc70 positive vesicles in Atg5−/− MEFs compared with wild-type cells, but no significant changes in the total number of LAMP-2A–positive vesicles (Figure 6, A and C) or of acid vesicular compartments (stained by lysosensor; Supplemental Figure 6). These results support that the increase in CMA in cells with impaired macroautophagy does not result from a global expansion of the lysosomal compartment in these cells, but from an increase in the percentage of total lysosomes that contain hsc70 and that are consequently able to perform CMA.
Figure 6.
Changes in lysosomal supbopulations in Atg5−/− cells. (A) WT and ATG5−/− MEFs maintained in the presence or absence of serum for 16 h, as indicated, were methanol fixed (to eliminate soluble cytosolic proteins), blocked, and processed for double immunofluorescence with antibodies against LAMP-2A (green) and Hsc70 (red). Merged images of both channels are shown at the bottom. Bar, 5 μm. (B) Quantification of the fraction of LAMP-2A colocalizing with Hsc70 (lysosomes competent for CMA) in each condition. (C) Average number of puncta positive for LAMP-2A (left) and Hsc70 (right) per cell section in each condition. Values are mean of the quantification of 20 cells per condition in two independent experiments (*p < 0.05, **p < 0.001 compared with WT cells).
Based on our observations, we conclude that both an increase in CMA components per lysosome (in LAMP-2A) and in the total number of lysosomes with CMA capability (total number of LAMP-2A/hsc70 lysosomes) contribute to the observed higher CMA activity in Atg5−/− cells.
Mechanism for CMA Activation during Macroautophagy Blockage
Our results suggest that the increased ability of lysosomes from 3-MA–treated and Atg5−/− cells to directly take up soluble proteins via CMA is probably in part attained through the increase in the levels of protein translocation complexes in the membrane of these lysosomes. Levels of LAMP-2A at the lysosomal membrane are rate limiting for CMA, and they are subjected to very tight regulation (Cuervo and Dice, 2000b). Two different mechanisms contribute to regulate LAMP-2A levels at the lysosomal membrane depending on the type of stimuli. Activation of CMA in conditions of mild-oxidative stress requires de novo synthesis of LAMP-2A (Kiffin et al., 2004), whereas during prolonged nutritional starvation, the observed increase in levels of LAMP-2A at the lysosomal membrane results from a decrease in the rates of degradation of this protein (Cuervo and Dice, 2000a).
Both transcriptional up-regulation of the LAMP-2 gene (Figure 7A) and reduced membrane degradation of LAMP-2A (Figure 7B) contributed to the observed increase in levels of LAMP-2A in cells treated with 3-MA under basal conditions. In serum-deprived cells, treatment with 3-MA did not significantly change levels of LAMP-2A mRNA. The absence of transcriptional changes for LAMP-2A, along with the slightly accelerated rates of LAMP-2A degradation at the lysosomal membrane observed in 3-MA–treated cells after serum removal (Figure 7B) could explain the reduced CMA activity observed under those conditions.
Figure 7.
Mechanisms for CMA up-regulation in macroautophagy-impaired cells. (A) Transcriptional changes of the LAMP-2 gene. Semiquantitative real-time PCR was used to compare mRNA expression levels for LAMP-2A in total mRNA isolated from NIH3T3 cells treated or not with 3-MA and from wild-type and Atg5−/− mouse embryonic fibroblasts maintained in the presence (+serum) or absence (−serum) of serum. Values were corrected for actin amplification in each sample, and they are expressed as times the values in control cells (untreated NIH3T3 cells or wild-type MEFs), which were giving an arbitrary value of 1. Values are mean + SE from four different experiments (*p < 0.01 and **p < 0.001 compared with WT cells). (B and C) Lysosomal degradation of LAMP-2A. Lysosomes isolated from untreated (none) or 3-MA–treated NIH3T3 cells maintained in the presence or absence of serum for 6 h (B) and from WT or Atg5−/− mouse embryonic fibroblasts maintained in media with or without serum for 16 h (C) were incubated at 37°C in isotonic media in the absence of substrate proteins, supplemented with dithiothreitol and 1 mM CaCl2. The levels of LAMP-2A in aliquots taken at the indicated times were determined by immunoblot with the antibody against the cytosolic tail of LAMP-2A. A representative immunoblot (top) and the densitometric quantification of two to four immunoblots from different experiments (bottom) are shown. Values are expressed as percentage of LAMP-2A detected at time 0. (D) Association of LAMP-2A to lipid microdomains. Lysosomes isolated from WT and Atg5−/− mouse embryonic fibroblasts maintained in media supplemented with serum were subjected to extraction with Triton X-114. DR and DS regions were separated by flotation in sucrose density gradients as described under Materials and Methods, and then they were subjected to SDS-PAGE and immunoblot for LAMP-2A (L-2A) and flotillin-1 (Flot-1). A representative immunoblot (top) and the densitometric quantification of three immunoblots from different experiments (bottom) are shown. Values are expressed as percentage of total LAMP-2A at the lysosomal membrane present in each fraction.
Although more moderated, we also observed increased levels of LAMP-2A mRNA in Atg5−/− cells when compared with wild-type cells under basal conditions but no significant changes upon serum removal (Figure 7A). These results support that the higher content of LAMP-2A in lysosomes from Atg5−/− during serum deprivation did not result from de novo synthesis of this protein. Instead, we found that degradation rates of LAMP-2A in these lysosomes were slower than in lysosomes from wild-type cells both in the presence and in the absence of serum (Figure 7C), supporting that the observed increased content of LAMP-2A in lysosomes from Atg5−/− cells during starvation results from exacerbation of the normal regulatory mechanism.
The moderate increase in the amount of LAMP-2A synthesized de novo in Atg5−/− cells under basal conditions compared with wild-type cells (Figure 7A), correlates well with the very modest increase in the levels of this protein in their lysosomes (Figure 5A). However, these findings contrast with the twofold increase in CMA activity observed in these cells (Figure 2A). Our previous studies have revealed an almost linear correlation between levels of LAMP-2A at the lysosomal membrane and CMA activity (Cuervo and Dice, 2000a). We have found, however, that the distribution of LAMP-2A at the lysosomal membrane is not homogeneous but that instead it dynamically associates to membrane microdomains of discrete protein and lipid composition (Kaushik et al., 2006). Inclusion of LAMP-2A in these membrane microdomains reduces levels of CMA activity whereas mobilization outside these regions increases uptake of CMA substrates by lysosomes. To determine whether changes in the subcompartmentalization of LAMP-2A at the lysosomal membrane, rather than total levels of LAMP-2A, were behind the observed increase in CMA activity in lysosomes from Atg5−/− cells under basal conditions, we compared the distribution of LAMP-2A between DR and DS regions in these membranes. As shown in Figure 7D, the percentage of LAMP-2A localized in the detergent-resistant microdomains in Atg5−/− cells was half to the one observed in wild-type cells, suggesting that changes in the organization of LAMP-2A at the membrane could explain in part the higher CMA activity in Atg5−/− cells. Whether the lower cholesterol content observed in Atg5−/− cells is behind the reduced amount of LAMP-2A in the cholesterol-enriched membrane microdomains of the lysosomal membrane in these cells remains to be elucidated.
In summary, changes in the lysosomal compartment, namely, increased levels of LAMP-2A, are behind the up-regulated CMA, but the mechanisms responsible for these changes are different for the constitutive (basal conditions) and the inducible (serum removal) activation of CMA.
Mediators of the Cross-Talk between Autophagic Pathways
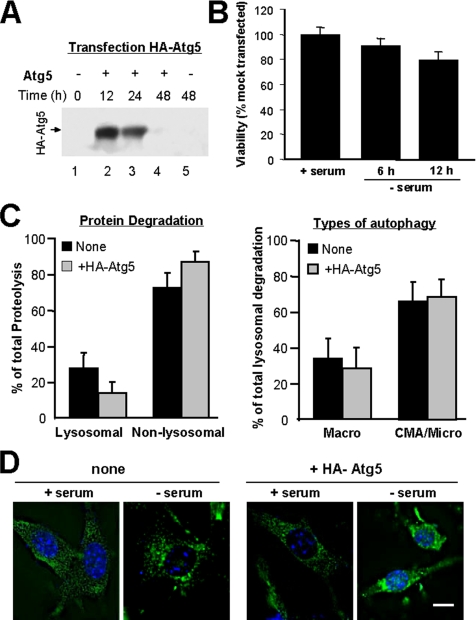
Our previous findings in cells with impaired CMA (Massey et al., 2006a) and the current observations in cells defective in macroautophagy support a direct cross-talk between these two types of autophagy. To gain insight into the mechanism underlying this cross-talk, and because knock-down of Atg5 was sufficient to up-regulate CMA, we first considered the possibility of Atg5 being a physiological inhibitor of CMA. To determine the possible effect of an increase in intracellular levels of Atg5 on CMA, we transiently transfected mouse fibroblasts with cDNA for an HA-tagged form of Atg5 (Figure 8A). Despite recent descriptions of the proapoptotic properties of an Atg5 derived fragment (Yousefi et al., 2006), under our experimental conditions we did not detect any cleavage product or changes in viability of the transfected cells during short or prolonged serum deprivation when compared with mock-transfected cells (Figure 8B). Because maximal levels of expression were observed at 12 h after transfection (Figure 8A), we adjusted our metabolic labeling experiments to be able to analyze rates of long-lived protein degradation at 12 and 24 h after transfection (see Materials and Methods). As shown in Figure 8C, the total rates of protein degradation, the percentage of lysosomal versus nonlysosomal degradation and the contribution of 3-MA–sensitive and -insensitive forms of autophagy to degradation of long-lived proteins was comparable in transfected and mock-transfected cells. Moreover, we did not find differences in the intracellular pattern of distribution of the LAMP-2A–enriched lysosomes under basal conditions or in their mobilization toward the perinuclear region during serum deprivation between mock and HA-Atg5–transfected cells (Figure 8D). These results do not support that a mere increase in intracellular levels of Atg5 is enough to inhibit CMA, and they made us rule out that the up-regulation of CMA in Atg5−/− cells resulted directly from the lack of Atg5 in these cells.
Figure 8.
Effect of Atg5 overexpression on CMA. (A) Cell lysates from NIH3T3 mouse fibroblasts (150 μg of protein) were subjected to SDS-PAGE and immunoblot for HA at the indicated times after transient transfection with the cDNA for HA-Atg5. (B) Viability of the HA-Atg5–transfected cells at the indicated times after transfection was determined by the MTT assay. Values are expressed as percentage of activity in cells subjected to mock transfection (no cDNA), and they are mean + SE of three different experiments. (C) Degradation of long-lived proteins in mouse fibroblasts transiently transfected with the cDNA for HA-Atg5 was determined as described in Figure 2A. The percentage of lysosomal and nonlysosomal degradation (left) was calculated from its sensitivity or insensitivity, respectively, to 20 mM ammonium chloride. The percentage of lysosomal degradation sensitive (Macro) or insensitive (CMA/Micro) to 3-methyladenine is shown on the right. Values are the mean + SE of three different experiments. (D) Distribution of CMA-active lysosomes in mouse fibroblasts transiently transfected with the cDNA for HA-Atg5 maintained in the presence and absence serum was determined by immunofluorescence against LAMP-2A. Mounting media contains DAPI to highlight the cell nuclei. Representative cells for each condition are shown. Bar, 5 μm.
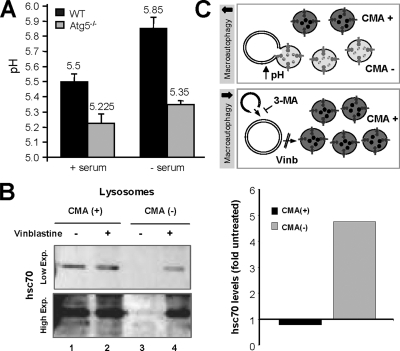
Because our comparative analysis of CMA components in lysosomes from wild-type and Atg5−/− cells and from cells treated or not with 3-MA revealed the presence of significantly larger amount of lysosomal hsc70 in macroautophagy-impaired cells both under basal and nutrient deprivation conditions (Figures 4–6), we then analyzed the possible contribution of macroautophagy to modulate lysosomal levels of hsc70. Although substrate binding to the receptor is the limiting step in CMA, the presence of hsc70 in the lysosomal lumen is absolutely necessary for substrate uptake and higher content of luminal hsc70 has been described under conditions of maximal activation, supporting a possible role in the accelerated transport of substrates (Cuervo et al., 1995; Agarraberes et al., 1997). Hsc70 is the constitutive member of the hsp70 family of chaperones, and consequently levels of this protein are not transcriptionally regulated. Instead, we have previously described that changes in levels of luminal hsc70 in lysosomes result from variations in the lysosomal pH (Cuervo et al., 1997). Thus, hsc70 is more stable in the lysosomal lumen when pH values are around or <5.2–5.5, but an increase in pH values (of just 0.4 U) accelerates its degradation by luminal proteases. To determine whether changes in the acidification of lysosomes in Atg5−/− cells could be behind their higher hsc70 content, we compared the luminal pH of lysosomes from wild-type and Atg5−/− cells. Although fluorescence microscopy of cells incubated with the pH-sensitive probe LysoSensor was not sensitive enough to reveal significant differences between both groups of cells (Supplemental Figure 6), direct measurement of the pH of isolated lysosomes by the FITC-dextran method (as described under Materials and Methods) revealed a difference of 0.3 and 0.5 pH units between lysosomes from wild-type and Atg5−/− cells maintained in the presence or absence of serum, respectively (Figure 9A).
Figure 9.
Macroautophagy-induced changes in lysosomal compartments related to CMA. (A) Intralysosomal pH. The pH of lysosomes isolated from wild-type and Atg5−/− cells maintained in the presence or absence of serum for 16 h was measured by the FITC-dextran method. Values are mean of the pH values obtained with lysosomes from three different experiments. (B) Levels of hsc70 in two populations of lysosomes isolated from liver of 6-h starved rats treated or not with vinblastine were determine by immunoblot with a specific antibody against hsc70. Left, two different exposures of a representative immunoblot are shown to better appreciate the absence of hsc70 in the population of lysosomes usually inactive for CMA [CMA (−)]. Right, changes in the lysosomal levels of hsc70 in both groups of lysosomes in response to vinblastine were calculated after densitometric quantification of immunoblots from two different experiments. Values are expressed as folds the hsc70 in lysosomes from untreated animals, which were given an arbitrary value of 1. (C) Hypothetical model of the mechanism that lead to increased number of CMA-active (CMA+) lysosomes when macroautophagy is blocked.
An appealing explanation for the higher lysosomal acidification of lysosomes from Atg5−/− cells is that the absence of fusion of this compartment with autophagosomes prevents the typical dissipation of part of the acidic pH observed during autophagosome/lysosome fusion. To test this possibility, we directly analyzed changes in the lysosomal levels of hsc70 under conditions in which autophagosomes still form, but their fusion to lysosomes is prevented. For these studies, we isolated lysosomes from livers of 6-h starved rats (to activate macroautophagy) treated or not with vinblastine (to prevent fusion of autophagosomes to lysosomes). We used this animal model, because it allows us to physically separate lysosomes that contain hsc70 in their lumen and hence are active for CMA, from those with undetectable levels of the chaperone and consequently unable to perform CMA. We have previously shown that the group of lysosomes with low hsc70 content has a slightly higher luminal pH, enough to render hsc70 unstable in this compartment (Cuervo et al., 1997). Treatment with vinblastine did not change the amount of hsc70 associated to CMA-competent lysosomes, but it significantly increased the amount of hsc70 in the group of lysosomes that usually lacks this chaperone (Figure 9B). Our studies thus support that blockage of macroautophagy leads to CMA activation, at least in part, by increasing the intracellular pool of lysosomes competent for CMA, as demonstrated previously by our immunofluorescence studies (Figure 6) and that this lysosomal switch from CMA-incompetent to CMA-competent is attained through the stabilization of hsc70 in their lumen (Figure 9C, see model).
In summary, this work presents evidence supporting that the cross-talk between macroautophagy and CMA is bidirectional and that changes in the lysosomal properties as a result of the interactions between macroautophagy-related compartments and lysosomes, contribute to modulate their CMA activity.
DISCUSSION
In this work, we present evidence of a direct cross-talk between two different forms of autophagy, macroautophagy and CMA, both maximally activated in response to stress. Blockage of macroautophagy both pharmacologically or through knockdown of an essential macroautophagy gene results in activation of CMA even under basal conditions. CMA up-regulation is attained through an increase in the lysosomal levels of the critical components of the translocation complex, LAMP-2A and hsc70. We have previously shown that blockage of CMA in cultured cells activates macroautophagy (Massey et al., 2006a). The results compiled in the present work establish that the cross-talk between macroautophagy and CMA acts in both directions and that changes in the activity of one of these pathways will affect the contribution of the other pathway to protein breakdown.
An interesting aspect in both studies is that blockage of either of the stress-related pathways results in compensatory activation of the other not only during stress but also under basal conditions. Previous reports on macroautophagy have proposed a different mechanism of regulation for the constitutive versus the inducible form of this autophagic pathway (Sarkar et al., 2005; Yamamoto et al., 2006). Although the molecular intricacies of these regulatory differences are still unknown, it is interesting that the compensatory activation of CMA in response to macroautophagy blockage also seems to obey to different mechanisms depending on whether cells are under basal or stress conditions. Thus, although in both cases up-regulation of CMA is attained through an increase in the number of the essential CMA translocation components (LAMP-2A and hsc70) and the total amount of lysosomes competent for CMA, the increase in LAMP-2A under basal conditions results mainly from de novo synthesis of the receptor protein, whereas decreased breakdown of the LAMP-2A molecules already at the lysosomal membrane seems the main mechanism for elevated LAMP-2A levels during nutrient deprivation. These results are in agreement with our previous observation that different CMA stimuli modify levels of LAMP-2A through different mechanisms (Cuervo and Dice, 2000b; Kiffin et al., 2004). Changes in the LAMP-2A protein already in the lysosomal compartment seem a more conservative option than de novo synthesis of LAMP-2A when nutrients are scarce. As described previously during mild oxidative stress, the increase in synthesis of LAMP-2A in macroautophagy-defective cells associates to changes in its organization in the lipid microdomains at the lysosomal membrane (Kaushik et al., 2006). It is possible that the higher affluence of LAMP-2A molecules to lysosomes saturates the capability of the lipid microdomains, leaving most of the protein outside these regions, where we have shown that it becomes competent for CMA substrate binding and uptake (Kaushik et al., 2006).
It is important, however, to notice that there are some differences between the two mechanisms used in this work to block macroautophagy, 3-MA and Atg5 knockout, in their effect on CMA. Thus, although both interventions activated CMA under basal conditions, pharmacological blockage of macroautophagy during serum deprivation did not activate CMA beyond the increase in activity observed in the untreated cells (Figure 1A). In fact, 3-MA treatment during serum deprivation did not increase (or even reduced) levels of LAMP-2A and hsc70 in lysosomes. Although, one major difference between both treatments is the absence of Atg5 in the knockout cells, our results do not support a direct inhibitory function for this protein on CMA that could justify the higher CMA activation in absence of this protein (Figure 8). Instead, it is possible that the chronic blockage in the knockout cells, compared with the acute nature of the pharmacological inhibition, associates to permanent changes in the lysosomal compartment that favor CMA activation. For example, the persistent lack of fusion of lysosomes to autophagosomes seems to preserve hsc70 in the lumen of a larger number of lysosomes, increasing thus the pool of lysosomes available to be recruited for CMA under the stress condition. Furthermore, it is also possible that the persistent lack of fusion with autophagosomes leads to changes in the lipid composition of the lysosomal membrane that stabilize LAMP-2A in this compartment (i.e., we observed lower rates of degradation of this protein in the lysosomes from Atg5−/− cells). These and other possibilities require further investigation.
The observed changes in the amount of lysosomal hsc70 during macroautophagy blockage are of particular interest, because they offer the first clue about possible molecular mechanisms behind the cross-talk between the two stress-induced autophagic pathways. We have previously shown that the number of hsc70-containing lysosomes increases under conditions resulting in CMA activation and that a second pool of lysosomes lacking hsc70, could become competent for CMA once they acquire the missing chaperone in their lumen (Cuervo et al., 1997). However, it is not clear how the lumenal form of this chaperone reaches the lysosomal lumen. Based on the sequential activation of macroautophagy and CMA during starvation we initially proposed that hsc70 could be delivered to lysosomes via macroautophagy. In their nonselective engulfment of cytosolic regions, autophagosomes would also sequester cytosolic hsc70 and deliver it to lysosomes upon fusion. In fact, a recent proteomic analysis has revealed hsc70 as a protein highly enriched in autophagosomes (Øverbye et al., 2007). However, our current findings demonstrate the presence of hsc70 in lysosomes even in cells with impaired macroautophagy (altered autophagosome formation, Figures 4 and 5; or autophagosome/lysosome fusion; Figure 9), thus discarding macroautophagy as the main mechanism for delivery of hsc70 into lysosomes. Activation of macroautophagy seems instead to contribute to decrease the number of lysosomes active for CMA. On this respect, we present in this work evidence supporting that the transient dissipation of lysosomal pH associated to fusion of autophagosomes with lysosomes could be a modulator of levels of hsc70 in the lysosomal lumen (Figure 9C, see model). The mechanisms that modulate pH in the different lysosomal compartments still remain obscure. For example, although we have found similar levels of the different subunits of the proton pump ATPase in CMA+ and CMA− lysosomes, their lumenal pH is different in 0.4–0.6 U (Cuervo et al., 1997). Factors other than the proton pump may thus modulate lysosomal pH. We propose here that the more acidic pH in lysosomes from Atg5−/− could result from the lack of dissipation of this pH when lysosomes fuse with autophagosomes, and this higher acidification confers more stability to essential CMA components such as hsc70. In support of this hypothesis, we have found that blockage of lysosome/autophagosome fusion through vinblastine treatment also resulted in higher levels of hsc70 in the subset of lysosomes that normally fuse with autophagosomes.
For obvious reasons, compensation for an autophagic pathway by another is never complete. Thus, although macroautophagy could take care of the degradation of CMA substrates, it lacks the selectivity of this pathway, which could be important when particular proteins need to be spared from degradation. Alternatively, CMA would only be able to degrade cytosolic proteins, but could not compensate for macroautophagy-mediated degradation of organelles. However, despite these limitations, compensatory activation is beneficial for the cells. Thus, although CMA-impaired cells are more susceptible to different stressors, blockage of the compensatory activation of macroautophagy further increases this sensitivity, suggesting that up-regulation of macroautophagy in these cells contributes to maintain cell viability (Massey et al., 2006a). Likewise, we have found that cells with defective macroautophagy are more susceptible to different death stimuli that involve organelle compromise (i.e., Fas-ligand mediated apoptosis or endoplasmic reticulum stress), but they are more resistant to oxidative stress. Up-regulation of CMA in these cells seems responsible for this enhanced resistance, as blockage of this pathway renders these cells susceptible to oxidative stress (Wang et al., 2007).
Understanding the compensatory mechanisms between the autophagic pathways, their advantages and their limitations, is important in light of the recent interest in the development of interventions aimed to modulate one form of autophagy for therapeutic purposes in human diseases with altered autophagy such as neurodegenerative disorders or cancer.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to members of our laboratory for insight and critical revision of this manuscript. This work was supported by National Institutes of Health grants AG-021904, AG-25355, and DK-041918 (to A.M.C.) and by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to N.M.). A.C.M. is supported by National Institutes of Health training grant T32AG023475.
Abbreviations used:
- Atg
autophagy-related protein
- CMA
chaperone-mediated autophagy
- hsc70
heat-shock cognate protein of 70 kDa
- LAMP
lysosome-associated membrane protein
- MOPS
3-(N-morpholino)propanesulfonic acid.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-11-1155) on March 12, 2008.
REFERENCES
- Agarraberes F., Terlecky S., Dice J. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J. Cell Biol. 1997;137:825–834. doi: 10.1083/jcb.137.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auteri J., Okada A., Bochaki V., Dice J. Regulation of intracellular protein degradation in IMR-90 human diploid fibroblasts. J. Cell Physiol. 1983;115:159–166. doi: 10.1002/jcp.1041150210. [DOI] [PubMed] [Google Scholar]
- Blommaart E., Luiken J. J., Blommaart P. J., van Woerkom G.M., Meijer A. J. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 1995;270:2320–2326. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- Cuervo A. M. Autophagy: in sickness and in health. Trends Cell Biol. 2004;14:70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Cuervo A. M., Dice J. F. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- Cuervo A. M., Dice J. F. Regulation of lamp2a levels in the lysosomal membrane. Traffic. 2000a;1:570–583. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- Cuervo A. M., Dice J. F. Unique properties of lamp2a compared to other lamp2 isoforms. J. Cell Sci. 2000b;113:4441–4450. doi: 10.1242/jcs.113.24.4441. [DOI] [PubMed] [Google Scholar]
- Cuervo A. M., Knecht E., Terlecky S., Dice J. F. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am. J. Physiol. 1995;269:C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- Cuervo A. M., Dice J. F., Knecht E. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J. Biol. Chem. 1997;272:5606–5615. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- Cuervo A. M., Stefanis L., Fredenburg R., Lansbury P. T., Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- Dice J. Chaperone-mediated autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- Dice J. F., Chiang H.-L., Spencer E. P., Backer J. M. Regulation of catabolism of microinjected ribonuclease A: identification of residues 7–11 as the essential pentapeptide. J. Biol. Chem. 1986;262:6853–6859. [PubMed] [Google Scholar]
- Eskelinen E., et al. Disturbed cholesterol traffic but normal proteolytic function in LAMP-1/LAMP-2 double-deficient fibroblasts. Mol. Biol. Cell. 2004;15:3132–3145. doi: 10.1091/mbc.E04-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn P. F., Dice J. F. Ketone bodies stimulate chaperone-mediated autophagy. J. Biol. Chem. 2005;280:25864–25870. doi: 10.1074/jbc.M502456200. [DOI] [PubMed] [Google Scholar]
- Finn P. F., Mesires N., Vine M., Dice J. F. Effects of small molecules on chaperone-mediated autophagy. Autophagy. 2005;1:141–145. doi: 10.4161/auto.1.3.2000. [DOI] [PubMed] [Google Scholar]
- Fuertes G., Martin De Llano J., Villarroya A., Rivett A., Knecht E. Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem. J. 2003;375:75–86. doi: 10.1042/BJ20030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Iwata A., Christianson J. C., Bucci M., Ellerby L. M., Nukina N., Forno L. S., Kopito R. R. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc. Natl. Acad. Sci. USA. 2005;102:13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadot M., Wattiaux R., Mainferme F., Dubois F., Claessens A., Wattiaux-De Coninck S. Soluble form of Lamp II in purified rat liver lysosomes. Biochem. Biophys. Res. Commun. 1996;223:353–359. doi: 10.1006/bbrc.1996.0898. [DOI] [PubMed] [Google Scholar]
- Jentoft N., Dearborn D. Protein labeling by reductive alkylation. Methods Enzymol. 1983;91:570–579. doi: 10.1016/s0076-6879(83)91052-2. [DOI] [PubMed] [Google Scholar]
- Kaushik S., Massey A. C., Cuervo A. M. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 2006;25:3921–3933. doi: 10.1038/sj.emboj.7601283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiffin R., Christian C., Knecht E., Cuervo A. Activation of chaperone-mediated autophagy during oxidative stress. Mol. Biol. Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D., Cuervo A., Seglen P. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., et al. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Komatsu M., et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M., et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Laemmli U. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liang X., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Lowry O., Rosebrough N., Farr A., Randall R. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Martinez-Vicente M., Cuervo A. M. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 2007;6:352–361. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- Marzella L., Ahlberg J., Glaumann H. Autophagy, heterophagy, microautophagy and crinophagy as the means for intracellular degradation. Virchows Archiv. B Cell Pathol. 1981;36:219–234. doi: 10.1007/BF02912068. [DOI] [PubMed] [Google Scholar]
- Massey A. C., Kaushik S., Sovak G., Kiffin R., Cuervo A. M. Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. USA. 2006a;103:5805–5810. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey A., Zhang C., Cuervo A. Chaperone-mediated autophagy in aging and disease. Curr. Top Dev. Biol. 2006b;73:205–235. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 2005;12:1535–1541. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Ohsumi Y., Yoshimori T. Autophagosome formation in mammalian cells. Cell. Struct. Funct. 2002;27:421–429. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimore G. E., Lardeux B. R., Adams C. E. Regulation of microautophagy and basal protein turnover in rat liver. Effects of short-term starvation. J. Biol. Chem. 1988;263:2506–2512. [PubMed] [Google Scholar]
- Nakai A., et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat. Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- Nixon R. Autophagy in neurodegenerative disease: friend, foe or turncoat? Trends Neurosci. 2006;29:528–535. doi: 10.1016/j.tins.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Ohkuma S., Moriyama Y., Takano T. Identification and characterization of a proton pump on lysosomes by fluorescein-isothiocyanate-dextran fluorescence. Proc. Natl. Acad. Sci. USA. 1982;79:2758–2762. doi: 10.1073/pnas.79.9.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y., Ishikawa T., Kato K. A rapid and simplified method for the preparation of lysosomal membranes from rat liver. J. Biochem. 1983;93:547–556. [PubMed] [Google Scholar]
- Ohsumi Y., Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin. Cell Dev. Biol. 2004;15:231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Øverbye A., Fengsrud M., Seglen P. Proteomic analysis of membrane-associated proteins from rat liver autophagosomes. Autophagy. 2007;3:300–322. doi: 10.4161/auto.3910. [DOI] [PubMed] [Google Scholar]
- Ravikumar B., Duden R., Rubinsztein D. C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Ravikumar B., et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Rubinsztein D. C., DiFiglia M., Heintz N., Nixon R. A., Qin Z.-H., Ravikumar B., Stefanis L., Tolkovsky A. M. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy. 2005;1:11–22. doi: 10.4161/auto.1.1.1513. [DOI] [PubMed] [Google Scholar]
- Sarkar S., Flot R. A., Berger Z., Imarisio S., Cordenier A., Pasco M., Cook L., Rubinsztein D. Lithium induces autophagy by inhibiting inositol monophosphatase. J. Cell Biol. 2005;170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen P., Gordon P. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated hepatocytes. Proc. Nat. Acad. Sci. USA. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibatani T., Nazir M., Ward W. Alteration of rat liver 20S proteasome activities by age and food restriction. J. Gerontol. A. Biol. Sci. Med. Sci. 1996;51:B316–322. doi: 10.1093/gerona/51a.5.b316. [DOI] [PubMed] [Google Scholar]
- Shintani T., Klionsky D. J. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storrie B., Madden E. Isolation of subcellular organelles. Methods Enzymol. 1990;182:203–225. doi: 10.1016/0076-6879(90)82018-w. [DOI] [PubMed] [Google Scholar]
- Terlecky S., Dice J. Polypeptide import and degradation by isolated lysosomes. J. Biol. Chem. 1993;268:23490–23495. [PubMed] [Google Scholar]
- Terlecky S. R., Chiang H.-L., Olson T. S., Dice J. F. Protein and peptide binding and stimulation of in vitro lysosomal proteolysis by the 73-kDa heat shock cognate protein. J. Biol. Chem. 1992;267:9202–9209. [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Singh R., Massey A. C., Kane S., Kaushik S., Grant T., Xiang Y., Cuervo A. M., Czaja M. J. Loss of macroautophagy promotes or prevents fibroblast apoptosis depending on the death stimulus. J. Biol. Chem. 2007;283:4766–4777. doi: 10.1074/jbc.M706666200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing S., Chiang H. L., Goldberg A. L., Dice J. F. Proteins containing peptide sequences related to KFERQ are selectively depleted in liver and heart, but not skeletal muscle, of fasted rats. Biochem. J. 1991;275:165–169. doi: 10.1042/bj2750165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Cremona M., Rothman J. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J. Cell Biol. 2006;172:719–731. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D. J. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi S., Perozzo R., Schmid I., Ziemiecki A., Schaffner T., Scapozza L., Brunner T., Simon H. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.